Saturation Modeling of Gas Hydrate Using Machine Learning with X-Ray CT Images
Abstract
1. Introduction
2. Methodology
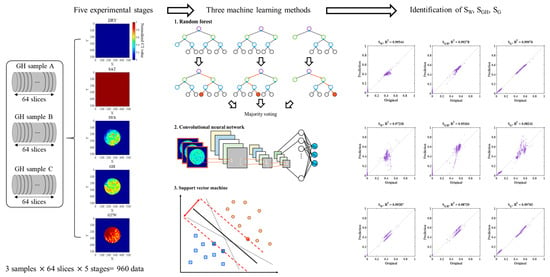
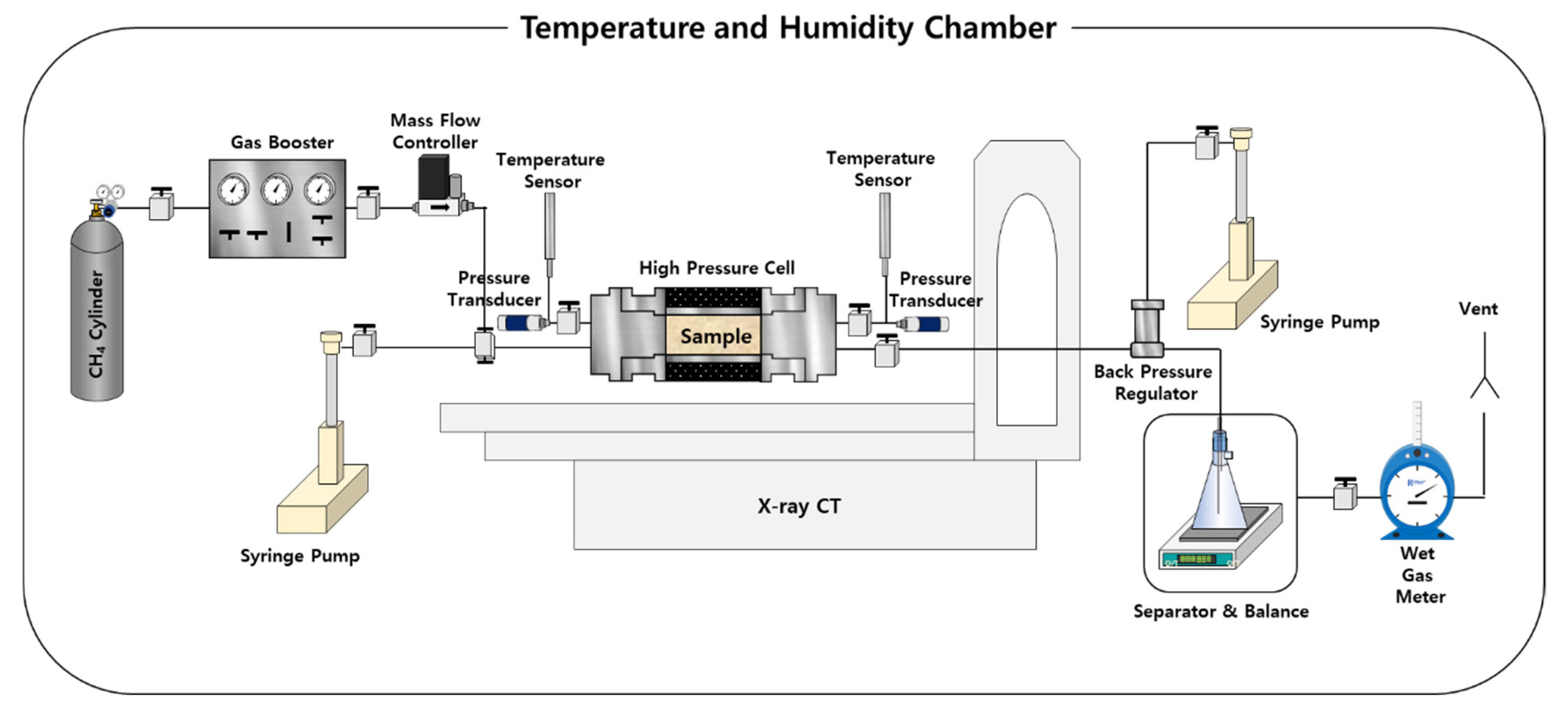
2.1. GH Experiment with CT Imaging
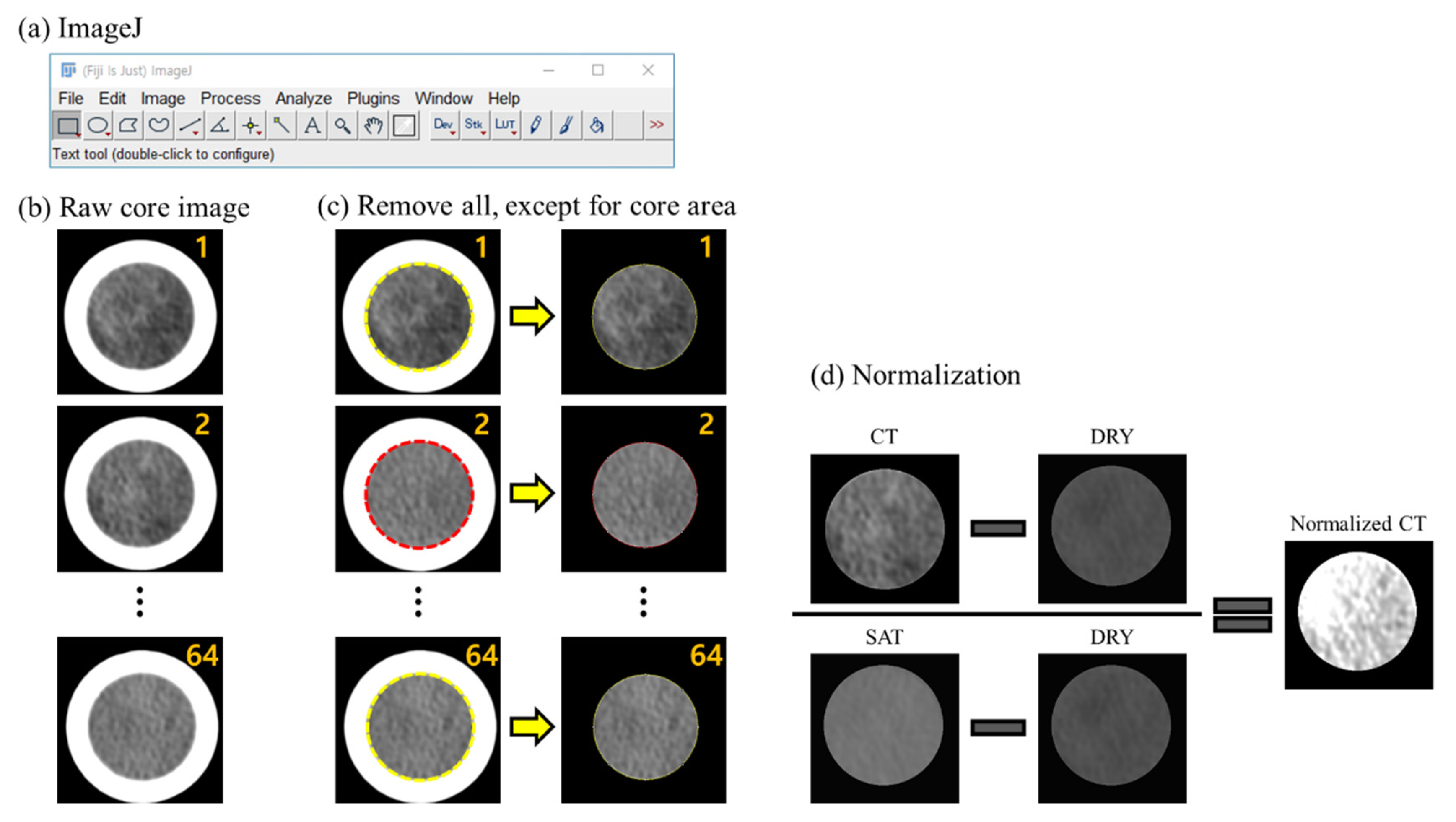
2.2. Data Acquisition and Pre-Process
2.3. Machine Learning Methodologies
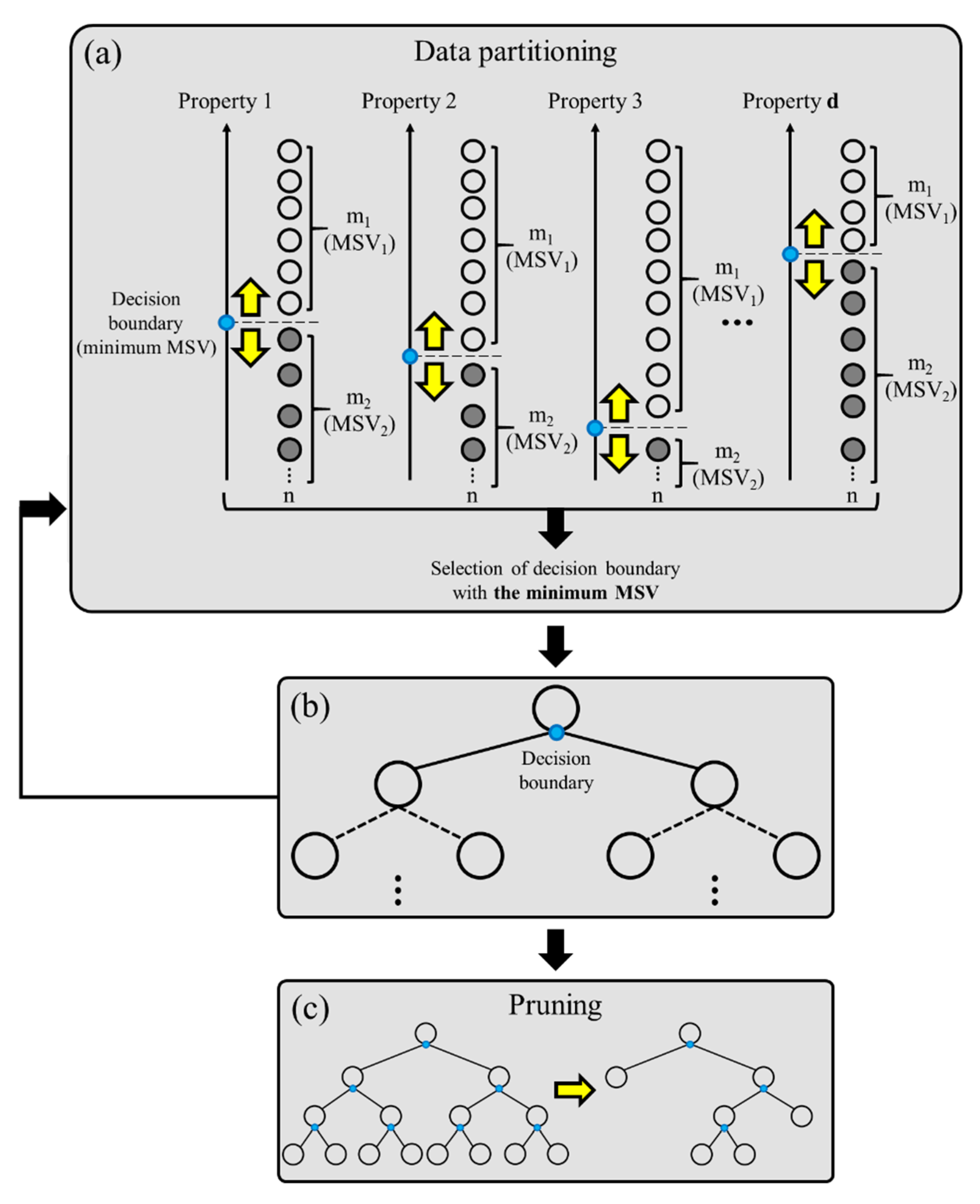
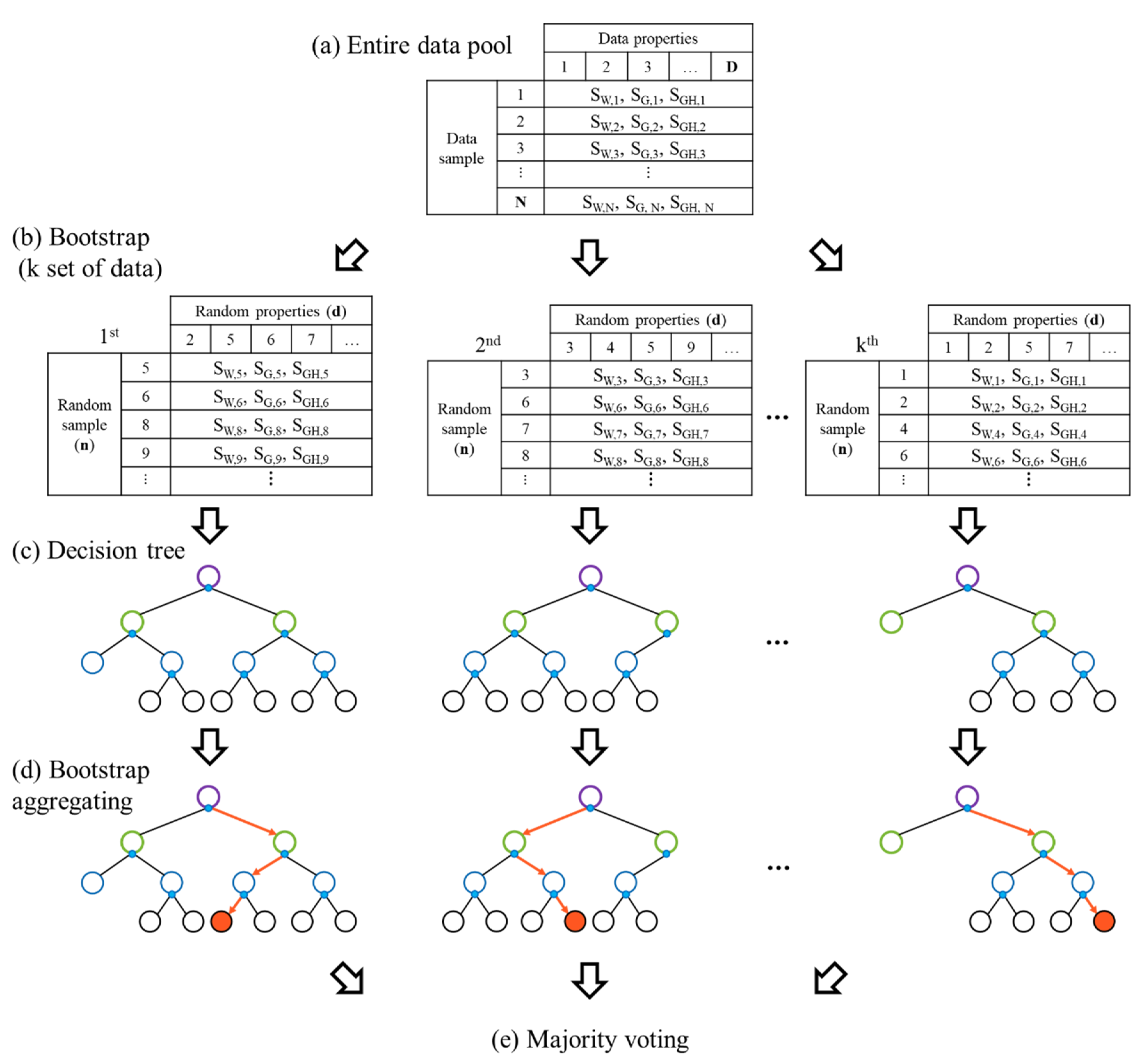
2.3.1. Random Forest
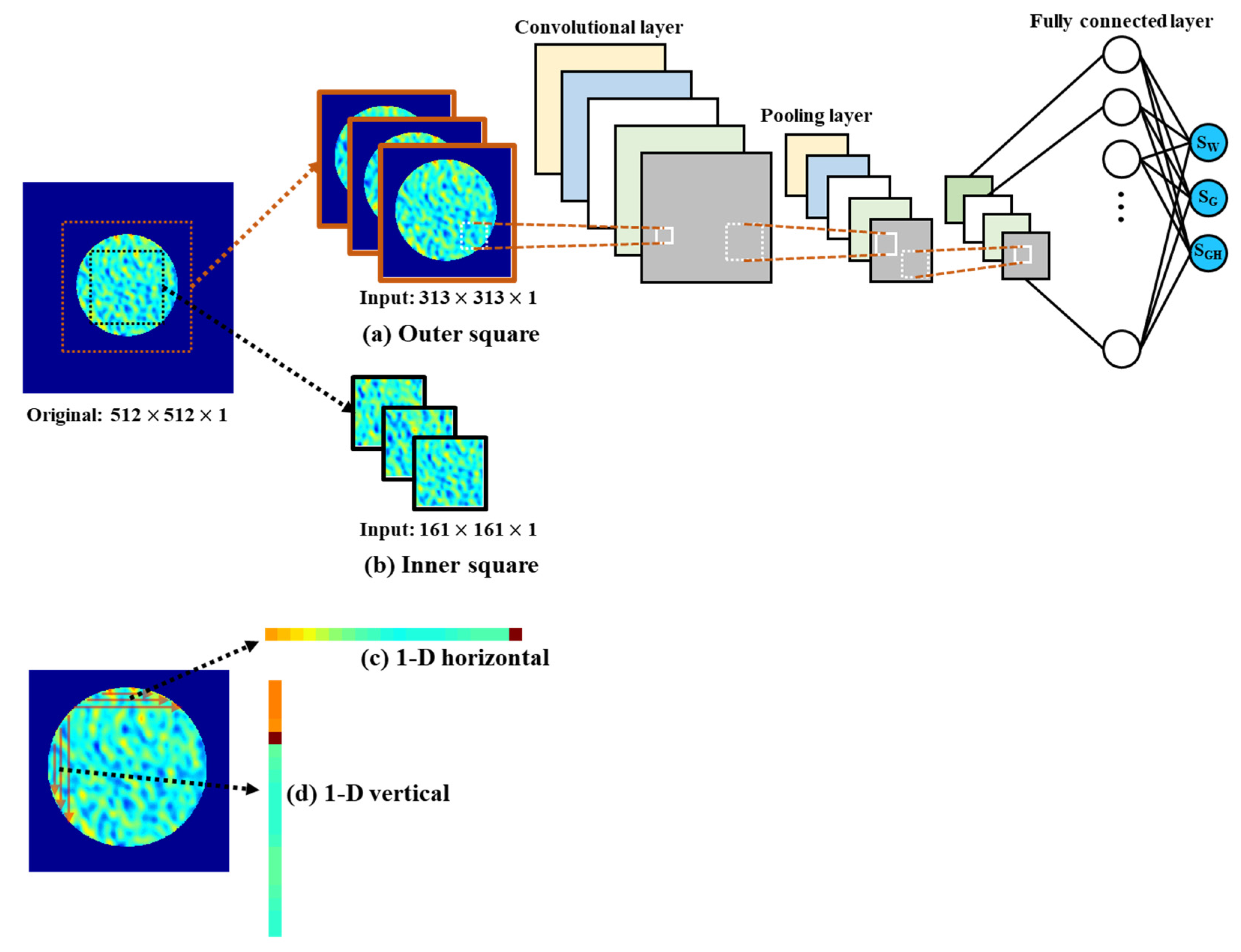
2.3.2. Convolutional Neural Network
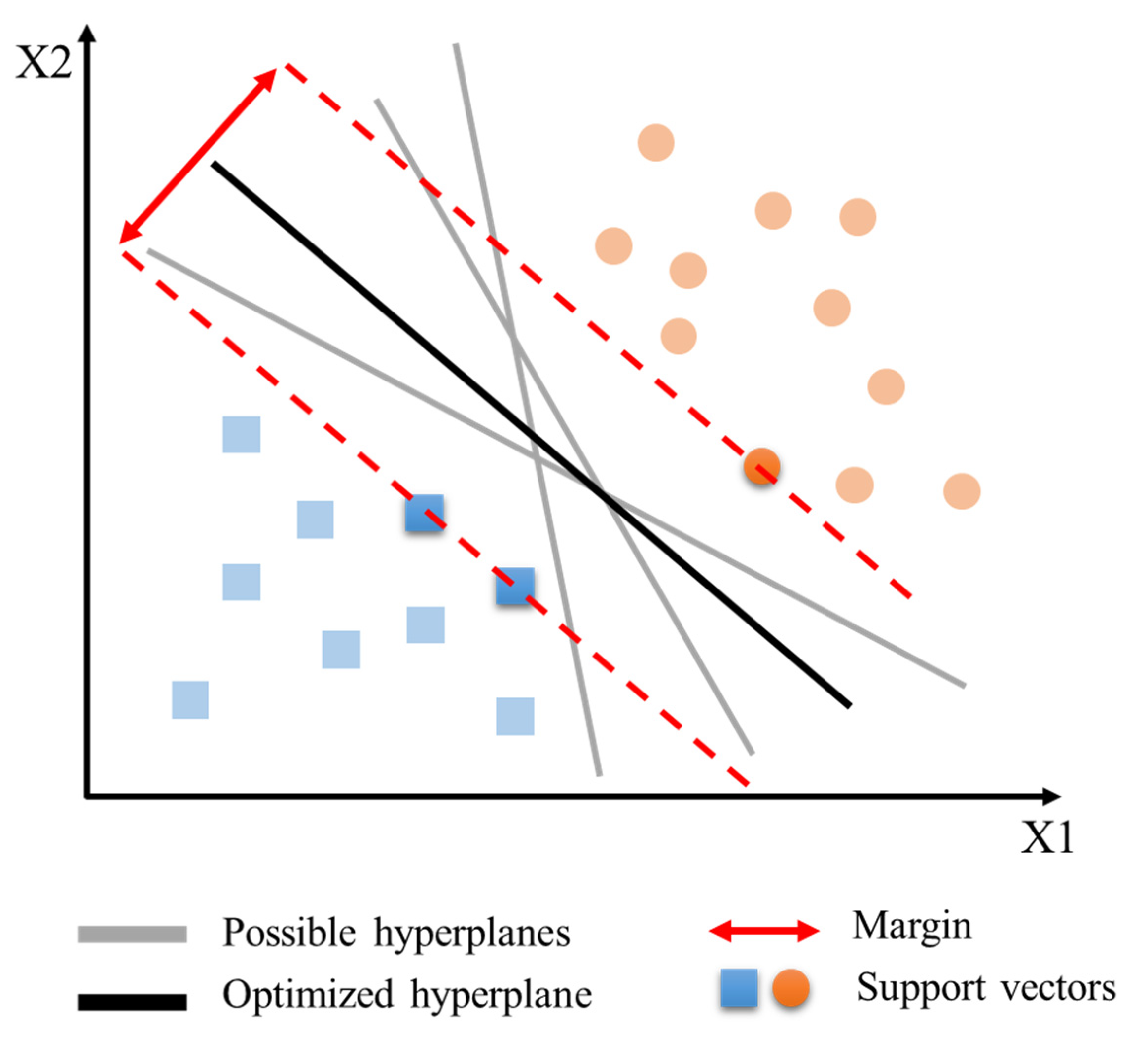
2.3.3. Support Vector Machine
3. Results
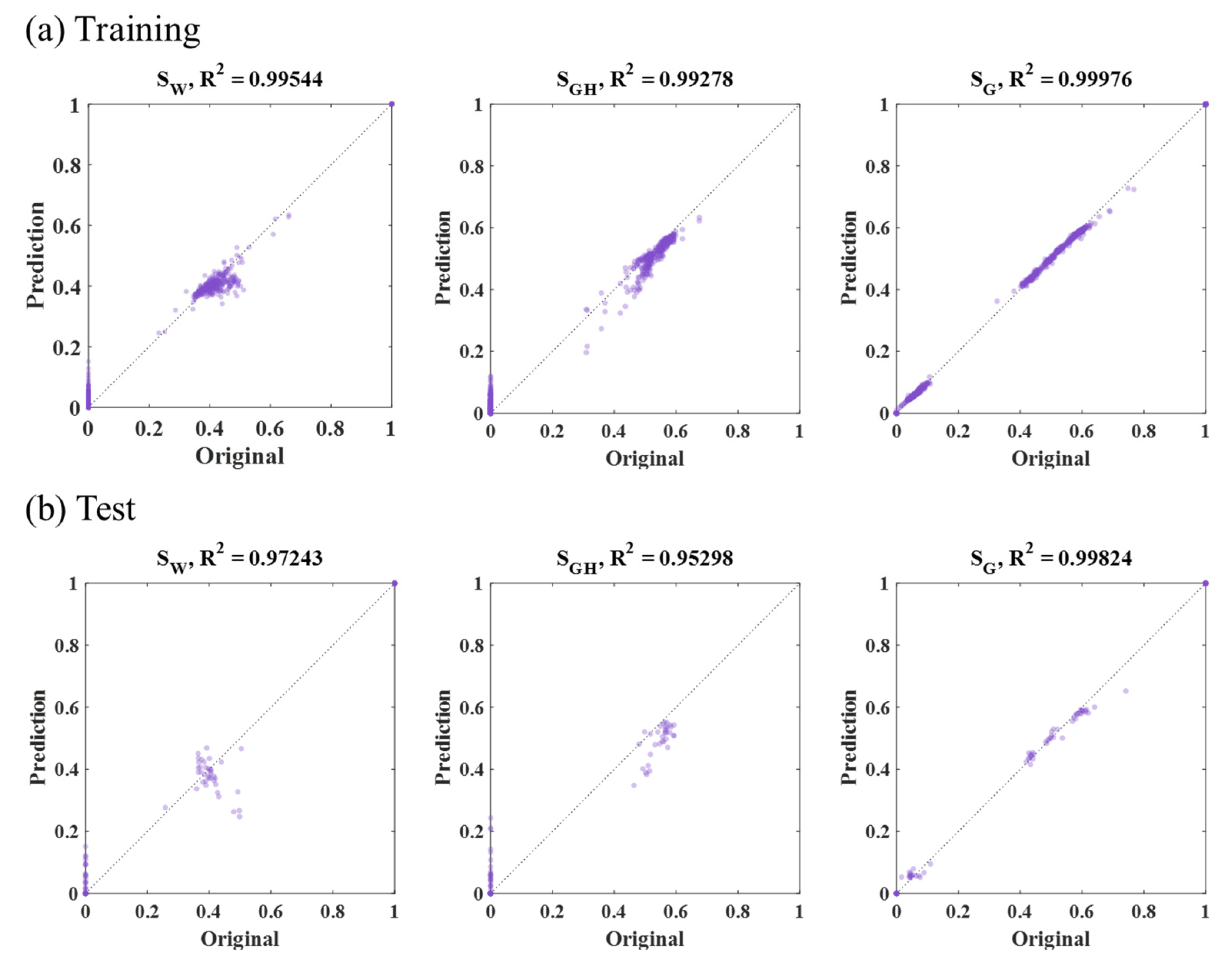
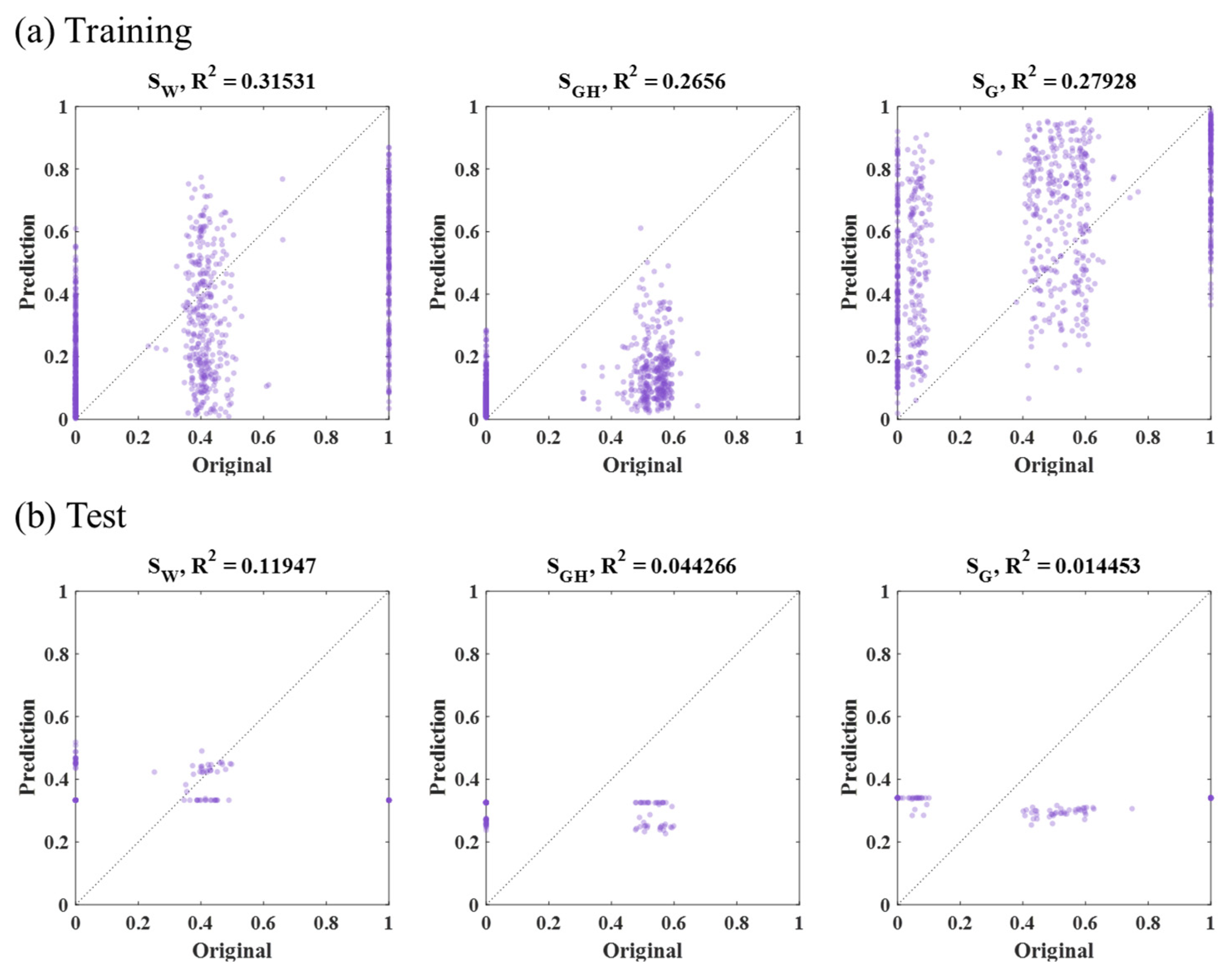
3.1. RF Results
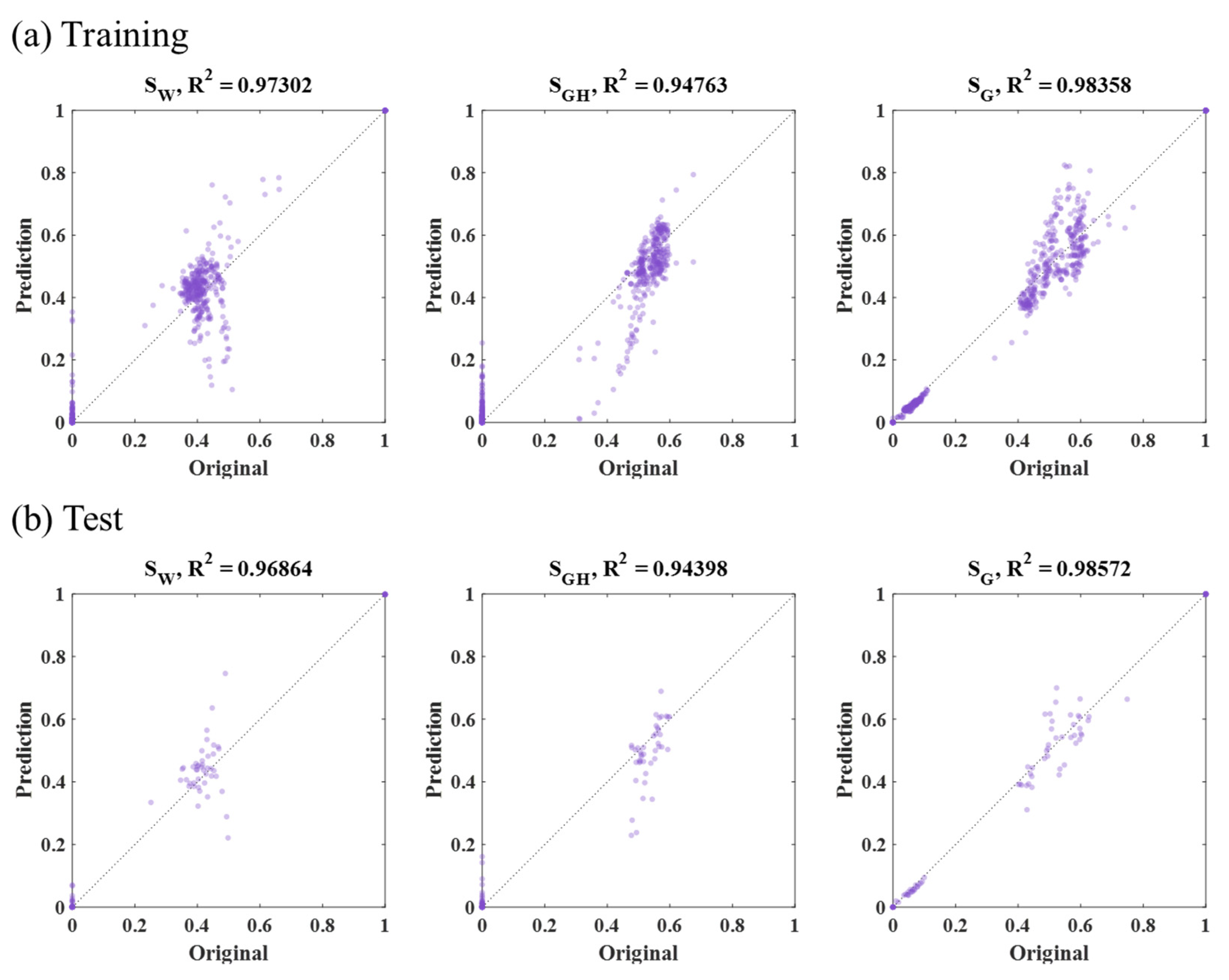
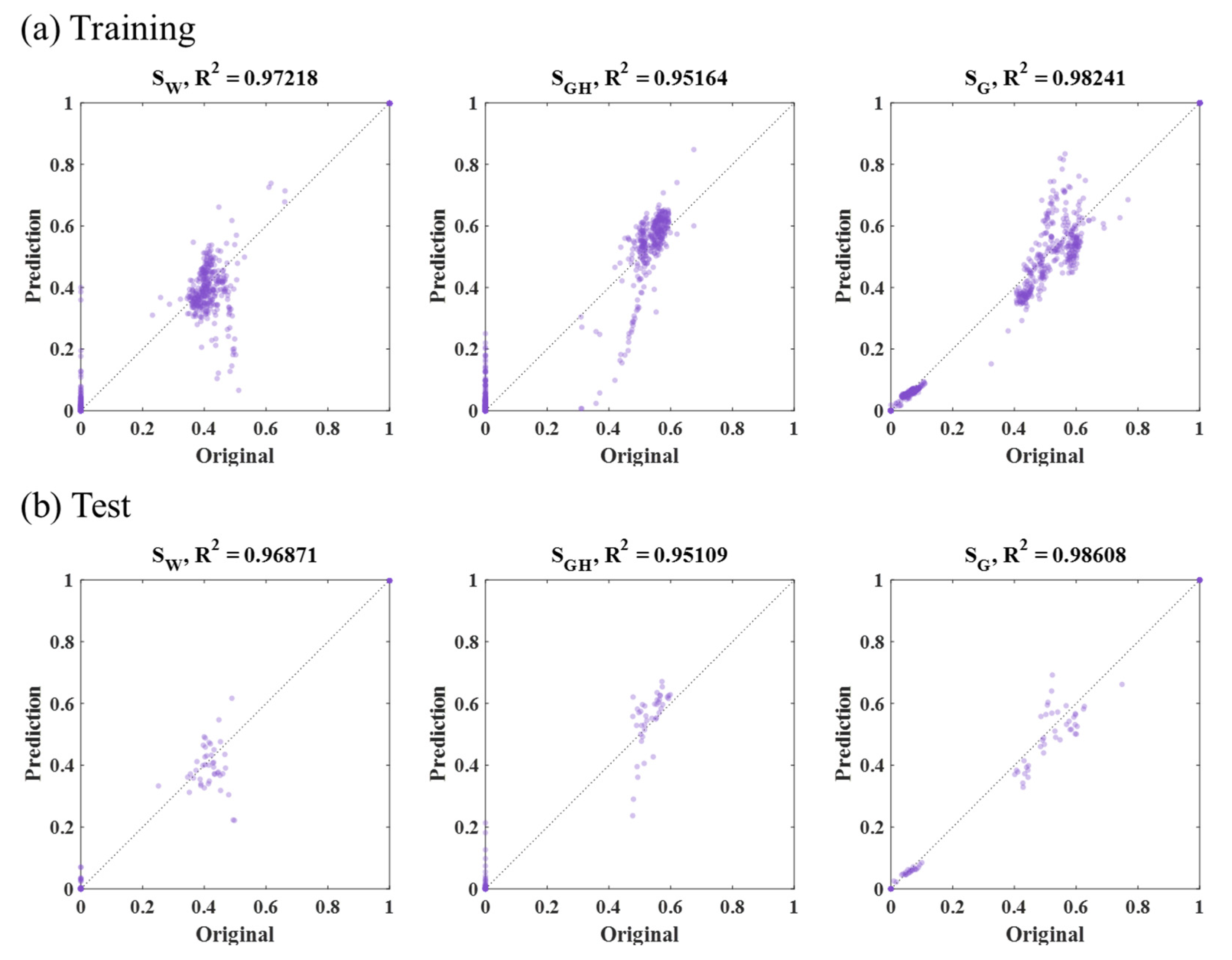
3.2. CNN Results
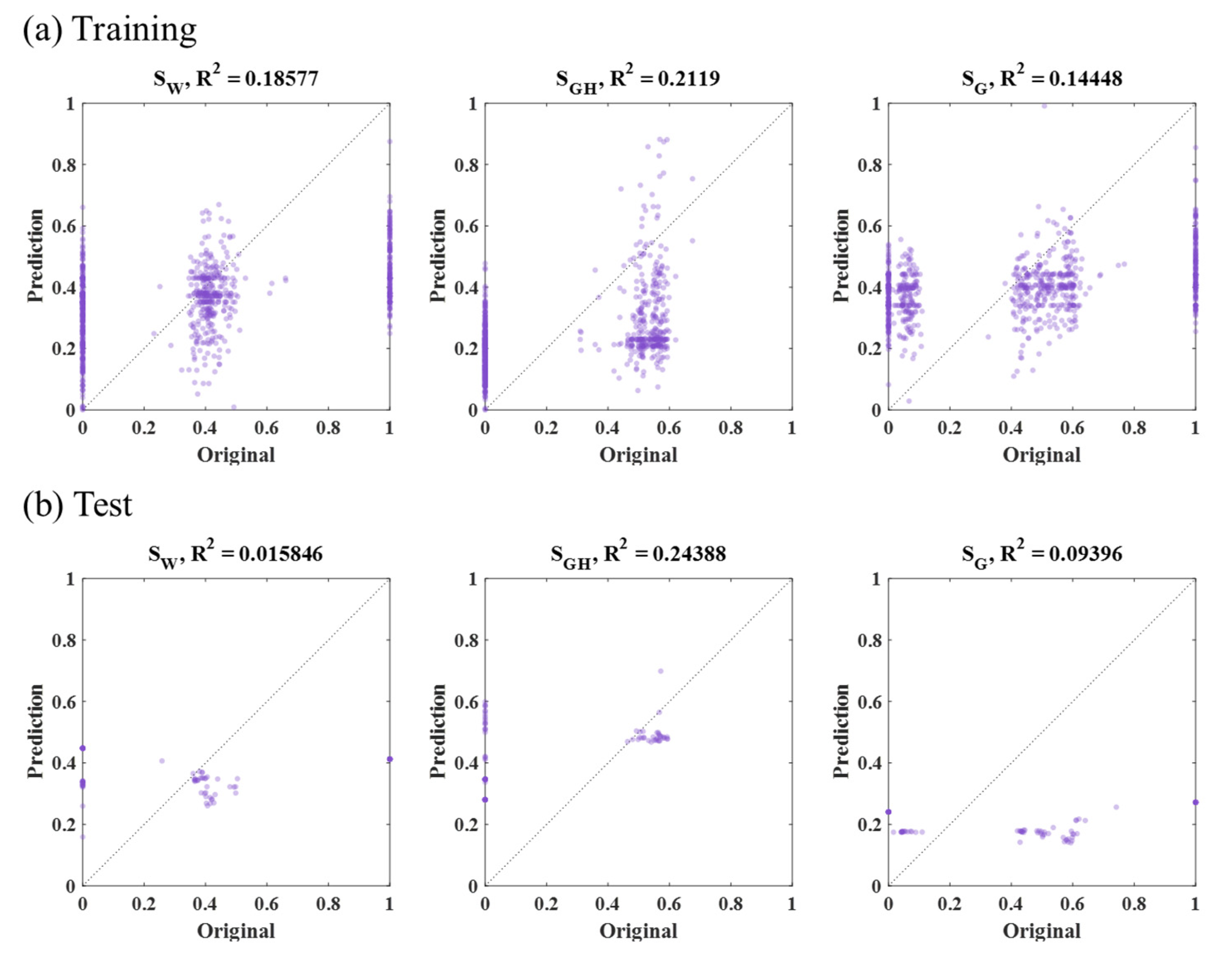
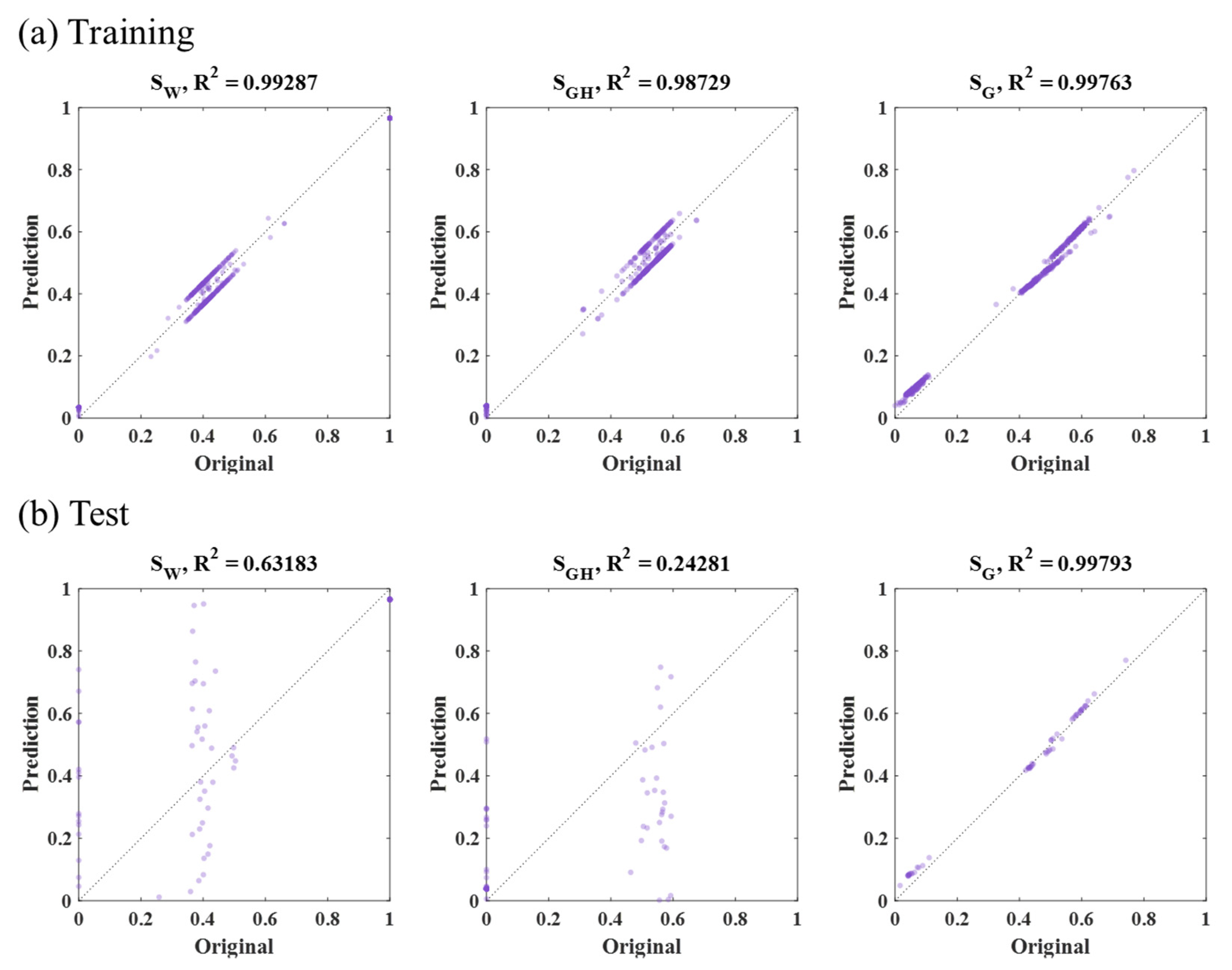
3.3. SVM Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burdine, N.T. Relative permeability calculations from pore size distribution data. Soc. Pet. Eng. 1953. [Google Scholar] [CrossRef]
- Beard, D.C.; Weyl, P.K. Influence of texture on porosity and permeability of unconsolidated sand. AAPG Bull. 1973, 57, 349–369. [Google Scholar] [CrossRef]
- Katz, A.J.; Thompson, A.H. Quantitative prediction of permeability in porous rock. Phys. Rev. B 1986, 34. [Google Scholar] [CrossRef] [PubMed]
- Akin, S.; Kovscek, A.R. Computed tomography in petroleum engineering research. Geol. Soc. Lond. 2003, 215, 23–38. [Google Scholar] [CrossRef]
- Sondergeld, C.H.; Newsham, K.E.; Comisky, J.T.; Rice, M.C.; Rai, C.S. Petrophysical considerations in evaluating and producing shale gas resources. Soc. Pet. Eng. 2010. [Google Scholar] [CrossRef]
- KIGAM. Studies on Gas Hydrate Development and Production Technology; Report GP2012-02502014(3); Pacific Northwest National Laboratory: Daejeon, Korea, 2014; p. 330.
- Lee, M.; Suk, H.; Lee, J.; Lee, J. Quantitative Analysis for Gas Hydrate Production by Depressurization Using X-ray CT. In Proceedings of the 2018 Joint International Conference of the Geological Science & Technology of Korea, KSEEG, Busan, Korea, 17–20 April 2018; p. 363. [Google Scholar]
- Suk, H.; Ahn, T.; Lee, J.; Lee, M.; Lee, J. Development of gas hydrate experimental production system combined with X-ray CT. J. Korean Soc. Miner. Energy Resour. Eng. 2018, 55, 226–237. [Google Scholar] [CrossRef]
- KIGAM. Field Applicability Study of Gas Hydrate Production Technique in the Ulleung Basin; Report GP2016-027-2016(1); Pacific Northwest National Laboratory: Daejeon, Korea, 2016; pp. 37–73.
- KIGAM. Gas Hydrate Exploration and Production Study; Report GP2016-027-2017(2); Pacific Northwest National Laboratory: Daejeon, Korea, 2017; pp. 164–199.
- Wang, J.; Zhao, J.; Yang, M.; Li, Y.; Liu, W.; Song, Y. Permeability of laboratory-formed porous media containing methane hydrate: Observations using X-ray computed tomography and simulations with pore network models. Fuel 2015, 170–179. [Google Scholar] [CrossRef]
- Mikami, J.; Masuda, Y.; Uchida, T.; Satoh, T.; Takeda, H. Dissociation of natural gas hydrate observed by X-ray CT scanner. Ann. N. Y. Acad. Sci. 2006, 912. [Google Scholar] [CrossRef]
- Kneafsey, T.J.; Tomutsa, L.; Moridis, G.; Seol, Y.; Freifeld, B.; Taylor, C.; Gupta, A. Methane hydrate formation and dissociation in a partially saturated core-scale sand sample. J. Pet. Sci. Eng. 2007, 56, 108–126. [Google Scholar] [CrossRef]
- Holland, M.; Schultheiss, P.; Roberts, J.; Druce, M. Observed Gas Hydrate Morphologies in Marine Sediments. In Proceedings of the 6th International Conference on Gas Hydrate (ICGH 2008), Vancouver, BC, Canada, 6–10 July 2008. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Y.; Chen, Y.; Yao, L.; Li, Q. The detection of tetrahydrofuran hydrate formation and saturation using magnetic resonance imaging technique. J. Nat. Gas Chem. 2010, 19, 224–228. [Google Scholar] [CrossRef]
- Seol, Y.; Kneafsey, T.J. Methane hydrate induced permeability modification for multiphase flow in unsaturated porous media. J. Geophys. Res. 2011, 116, B08102. [Google Scholar] [CrossRef]
- Wu, N.; Liu, C.; Hao, X. Experimental simulations and methods for natural gas hydrate analysis in China. China Geol. 2018, 1, 61–71. [Google Scholar] [CrossRef]
- Lei, L.; Seol, Y.; Choi, J.; Kneafsey, T. Pore habit of methane hydrate and its evolution in sediment matrix—Laboratory visualization with phase-contrast micro-CT. Mar. Pet. Geol. 2019, 104, 451–467. [Google Scholar] [CrossRef]
- Kim, S.; Kim, K.H.; Min, B.; Lim, J.; Lee, K. Generation of synthetic density log data using deep learning algorithm at the Golden field in Alberta, Canada. Geofluids 2020, 2020, 5387183. [Google Scholar] [CrossRef]
- Lee, K.; Lim, J.; Yoon, D.; Jung, H. Prediction of shale gas production at Duvernay Formation using deep-learning algorithm. SPE J. 2019, 24, 2423–2437. [Google Scholar] [CrossRef]
- Waldeland, A.; Jensen, A.C.; Gelius, L.-J.; Solberg, A.H.S. Convolutional neural networks for automated seismic interpretation. Lead. Edge 2018, 37, 529–537. [Google Scholar] [CrossRef]
- Huang, L.; Dong, X.; Clee, E.A. Scalable deep learning platform for identifying geologic features from seismic attributes. Lead. Edge 2017, 36, 249–256. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, X.; Yang, J.; Littile, E.; Zhou, Y. Deep learning-based method for SEM image segmentation in mineral characterization, an example from Duvernay Shale samples in Western Canada Sedimentary Basin. Comput. Geosci. 2020, 138, 104450. [Google Scholar] [CrossRef]
- Chauhan, S.; Rühaak, W.; Anbergen, H.; Kabdenov, A.; Freise, M.; Wille, T.; Sass, I. Phase segmentation of X-ray computer tomography rock images using machine learning techniques: An accuracy and performance study. Solid Earth 2016, 7, 1125–1139. [Google Scholar] [CrossRef]
- Karimpouli, S.; Tahmasebi, P. Segmentation of digital rock images using deep convolutional autoencoder networks. Comput. Geosci. 2019, 126, 142–150. [Google Scholar] [CrossRef]
- Krevor, S.; Pini, R.; Zuo, L.; Benson, S. Relative permeability and trapping of CO2 and water in sandstone rocks at reservoir conditions. Water Resour. Res. 2012, 48, W02532. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time-series. Handb. Brain Theory Neural Netw. 1995, 3361, 1995. [Google Scholar]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object Recognition with Gradient-Based Learning. In Shape, Contour and Grouping in Computer Vision; Springer: Berlin/Heidelberg, Germany, 1999; Volume 1681. [Google Scholar]
- Ji, S.; Xu, W.; Yang, M.; Yu, K. 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 221–231. [Google Scholar]
- Lin, B.; Wei, X.; Junjie, Z. Automatic recognition and classification of multi-channel microseismic waveform based on DCNN and SVM. Comput. Geosci. 2019, 123, 111–120. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60. [Google Scholar] [CrossRef]
- Jin, H.; Song, Q.; Hu, X. Auto-keras: Efficient neural architecture search with network morphism. arXiv 2018, arXiv:1806.10282v2. [Google Scholar]
- Cortes, C.; Vapnik, V.N. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory—COLT ‘92, Pittsburgh, PA, USA, 27–29 July 1992; p. 144. [Google Scholar] [CrossRef]
- Vapnik, V.; Golowich, S.E.; Smola, A.J. Support Vector Method for Function Approximation, Regression Estimation and Signal Processing. In Proceedings of the Advances in Neural Information Processing Systems 9, Denver, CO, USA, 2–5 December 1996; Moser, M.C., Jordan, M.I., Petsche, T., Eds.; MIT Press: Cambridge, MA, USA, 1997; pp. 281–287. [Google Scholar]
- Drucker, H.; Burges, C.C.; Kaufman, L.; Smola, A.J.; Vapnik, V.N. Support Vector Regression Machines. In Proceedings of the Advances in Neural Information Processing Systems 9, Denver, CO, USA, 2–5 December 1996; Moser, M.C., Jordan, M.I., Petsche, T., Eds.; MIT Press: Cambridge, MA, USA, 1997; pp. 155–161. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar]
- Chu, M.; Min, B.; Kwon, S.; Park, G.; Kim, S.; Huy, N.X. Determination of an infill well placement using a data-driven multi-modal convolutional neural network. J. Pet. Sci. Eng. 2019, 106805. [Google Scholar] [CrossRef]
- Ramos, G.A.R.; Akanji, L. Data analysis and neuro-fuzzy technique for EOR screening: Application in Angolan oilfields. Energies 2017, 10, 837. [Google Scholar] [CrossRef]
- Chen, X.; Espinoza, D.N.; Luo, J.S.; Tisato, N.; Flemings, P.B. Pore-scale evidence of ion exclusion during methane hydrate growth and evolution of hydrate pore-habit in sandy sediments. Mar. Pet. Geol. 2020, 117, 104340. [Google Scholar] [CrossRef]
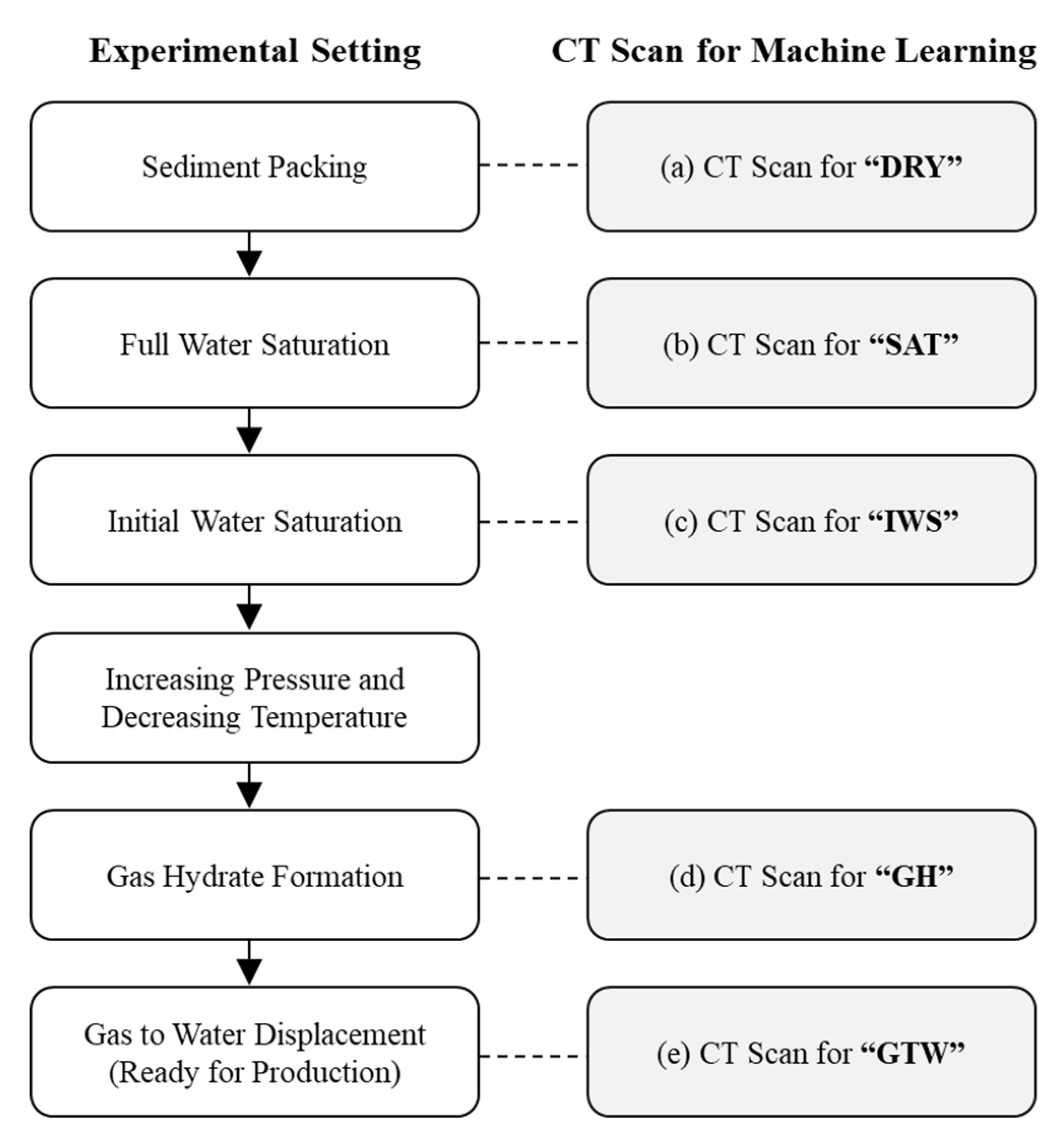
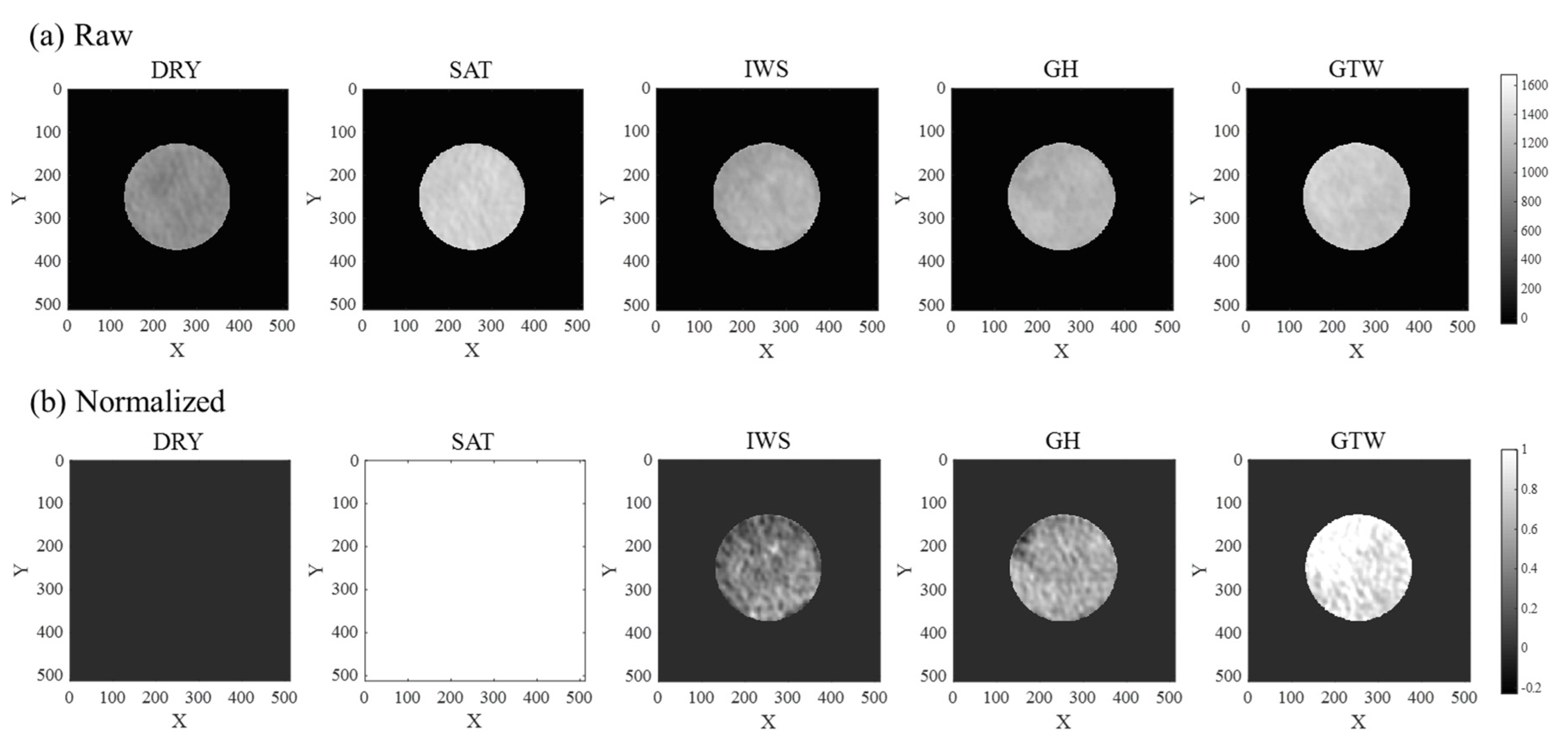
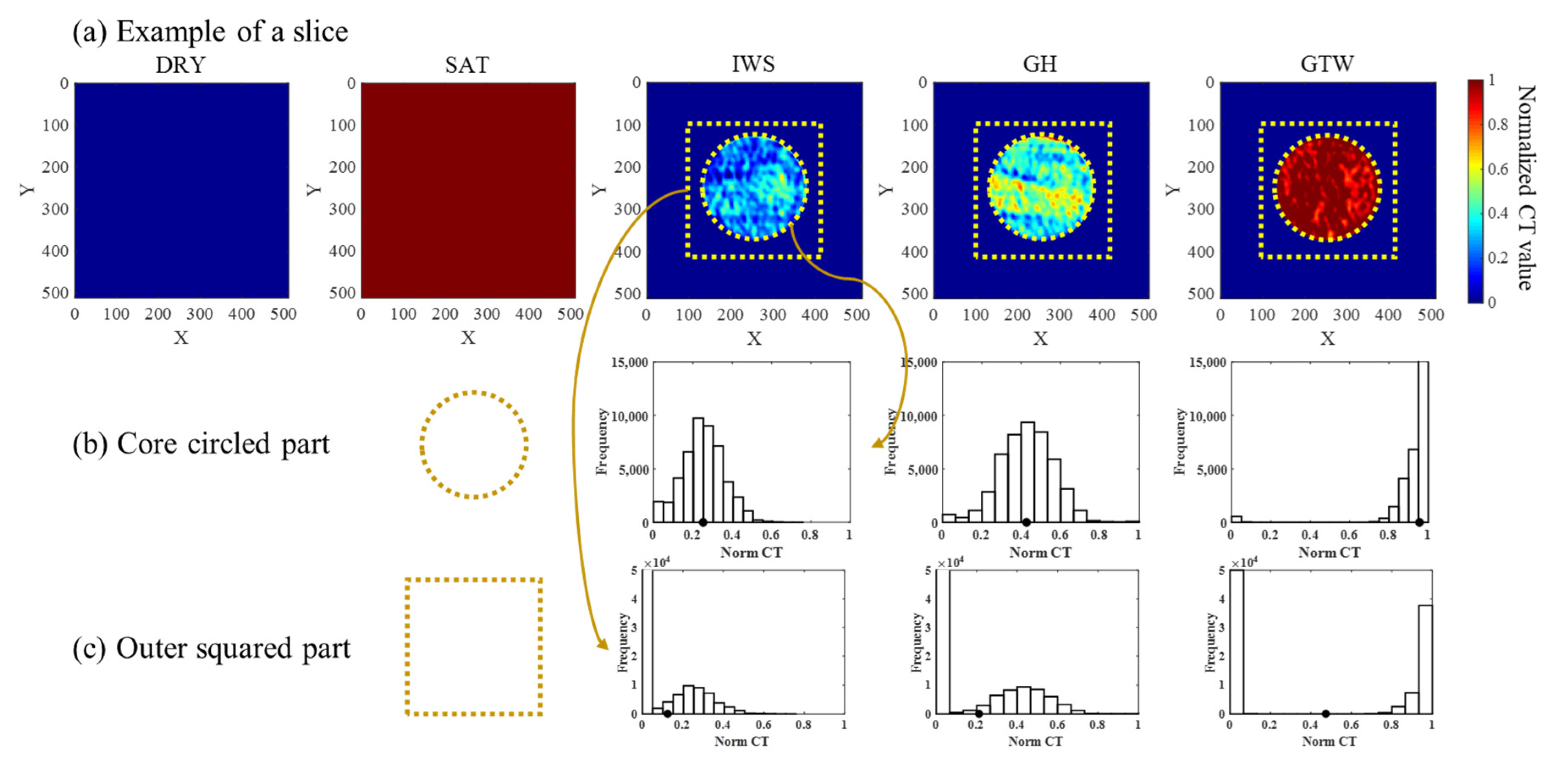
| Experimental Stage | SW (Density, g/cc) | SGH (Density, g/cc) | SG (Density, g/cc) |
|---|---|---|---|
| “IWS” (14.7 psi, 18 °C) | SW,IWS (1) | SGH,IWS = 0 | 1—SW,IWS (0.000678) |
| “GH” (2500 psi, 16 °C) | SW,GH = 0 | SGH,GH (0.91) | 1—SGH,GH (0.143) |
| “GTW” (2900 psi, 16 °C) | SW,GTW (1.008) | SGH,GTW = SGH,GH | 1—SW,GTW—SGH,GTW (0.167) |
| Method | List | Condition |
|---|---|---|
| Common condition | Training data | 864 |
| Test data | 96 (10% of the entire data) | |
| RF | Maximum depth | 10 |
| Number of trees | 200 | |
| Number of properties | 219 | |
| Data size of one sample | 47,996 | |
| CNN | Validation data | 96 (10% of the whole data) |
| Maximum trials | 50 | |
| Data size of one sample | 313 by 313 (Figure 8a) 161 by 161 (Figure 8b) 47,996 by 1 (Figure 8c) 47,996 by 1 (Figure 8d) | |
| SVM | Kernel function | Linear |
| Data pre-process | Standardize | |
| Data size of one sample | 47,996 |
| Method | Input Data | MSE (×10−4): SW, SGH, SG | Average (×10−4) | |||
|---|---|---|---|---|---|---|
| RF | Sample part (47,996 grid, Figure 10b) | Training | 7 | 7 | 3 | 6 |
| Test | 39 | 40 | 2 | 27 | ||
| CNN | 2-D outer square (313 by 313, Figure 8a) | Training | 1117 | 550 | 1176 | 948 |
| Test | 1388 | 1055 | 1752 | 1398 | ||
| 2-D inner square (161 by 161, Figure 8b) | Training | 857 | 513 | 850 | 740 | |
| Test | 2722 | 963 | 4990 | 2892 | ||
| 1-D horizontal (47,996, Figure 8c) | Training | 38 | 37 | 23 | 33 | |
| Test | 38 | 42 | 18 | 33 | ||
| 1-D vertical (47,996, Figure 8d) | Training | 39 | 36 | 25 | 33 | |
| Test | 36 | 39 | 19 | 31 | ||
| SVM | Sample part (47,996 grid, Figure 10b) | Training | 11 | 14 | 5 | 10 |
| Test | 652 | 645 | 4 | 434 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, K.; Lee, M.; Ahn, T.; Lee, J.; Suk, H.; Ning, F. Saturation Modeling of Gas Hydrate Using Machine Learning with X-Ray CT Images. Energies 2020, 13, 5032. https://doi.org/10.3390/en13195032
Kim S, Lee K, Lee M, Ahn T, Lee J, Suk H, Ning F. Saturation Modeling of Gas Hydrate Using Machine Learning with X-Ray CT Images. Energies. 2020; 13(19):5032. https://doi.org/10.3390/en13195032
Chicago/Turabian StyleKim, Sungil, Kyungbook Lee, Minhui Lee, Taewoong Ahn, Jaehyoung Lee, Hwasoo Suk, and Fulong Ning. 2020. "Saturation Modeling of Gas Hydrate Using Machine Learning with X-Ray CT Images" Energies 13, no. 19: 5032. https://doi.org/10.3390/en13195032
APA StyleKim, S., Lee, K., Lee, M., Ahn, T., Lee, J., Suk, H., & Ning, F. (2020). Saturation Modeling of Gas Hydrate Using Machine Learning with X-Ray CT Images. Energies, 13(19), 5032. https://doi.org/10.3390/en13195032