Abstract
Biodiesel production is a field of outstanding prospects due to the renewable nature of its feedstock and little to no overall CO2 emissions to the environment. Data-based soft sensors are used in realizing stable and efficient operation of biodiesel production. However, the conventional data-based soft sensors cannot grasp the effect of process uncertainty on the process outcomes. In this study, a framework of data-based soft sensors was developed using ensemble learning method, i.e., boosting, for prediction of composition, quantity, and quality of product, i.e., fatty acid methyl esters (FAME), in biodiesel production process from vegetable oil. The ensemble learning method was integrated with the polynomial chaos expansion (PCE) method to quantify the effect of uncertainties in process variables on the target outcomes. The proposed modeling framework is highly accurate in prediction of the target outcomes and quantification of the effect of process uncertainty.
1. Introduction
Extensive use of fossil fuels is causing environmental issues, i.e., global warming and pollution, and depletion of energy resources [1]. These challenges drive the quest for exploring alternative energy resources that can help in the reduction of fossil fuels consumption and environmental impact. Bioenergy is one of the viable alternatives that is shifting the paradigm from conventional fuels to more sustainable resources. Biodiesel is one of the major bio-based fuels, and a drastic increase in its production has been reported in the last two decades, as shown in Figure 1 [2,3]. Efficient design and operation of biodiesel production process are investigated to minimize the consumption of raw materials and utilities and also produce a high quality product. Data-based soft sensors are used to realize stable operation of the biodiesel production. The data-based soft sensors are more efficient than the model-based soft sensors in capturing the non-linearity of complex processes and prediction of desired outcomes; an extensive review on data-based soft sensors can be found in [4]. Their applications in the biodiesel production process include online prediction, optimization, and control.
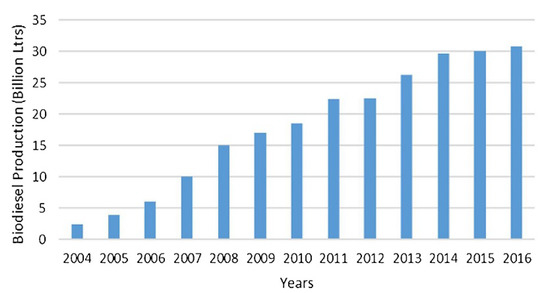
Figure 1.
Worldwide production of biodiesel [3].
Studies on online prediction mostly take kinematic viscosity, density, and cetane number of biodiesel as their target outcomes. For instance, Meng et al. (2013) developed an artificial neural networks (ANN) model to predict the biodiesel kinematic viscosity [5]. Rocabruno-Valdés et al. (2015) used an ANN model to predict density, dynamic viscosity, and cetane number of biodiesel [6]. Mostafaei et al. (2016) evaluated the efficiency of the response surface methodology (RSM) and neuro-fuzzy inference system (ANFIS) in modeling the yield achieved in an ultrasonic reactor [7]. Miraboutalebi et al. (2016) used random forest and ANN to predict cetane number of biodiesel [8]. Raman et al. (2018) developed an ANN model for estimation of densities of vegetable oil based ethyl esters biodiesel [9].
The concept of data-based soft sensing is also extended to parametric analysis, optimization, and control of the biodiesel production process. Wali et al. (2013) investigated online intelligent controllers for real-time temperature control in an advanced biodiesel microwave reacting system [10]. Fayyazi et al. (2014) determined optimum temperature, catalytic concentration, and reaction time in biodiesel production processes using genetic algorithm [11]. Sikorski et al. (2016) applied polynomial and high-dimensional model representation (HDMR) fitting to analyze the effect of process variables on the heat duties of equipment in a biodiesel production process [12]. Cheng et al. (2016) proposed a genetic algorithm-based evolutionary support vector machine (GA-ESVM) to get optimum mixture properties for higher biodiesel yield [13].
The efficiency of the data-based soft sensing methods is deteriorated due to uncertainty in process conditions. To realize a more robust sensing system, some uncertainty quantification mechanism should be incorporated in the soft sensor framework [14]. Uncertainty analysis is used to quantify the impact of uncertainty in model input on the model output [15]. It has been the focus of research and several methods are reported in the literature [16]. Scenario analysis, multiple model simulation methods, inverse modeling method, sensitivity analysis, and sampling-based method are commonly used for uncertainty analysis. The scenario analysis quantifies the impact of uncertainty associated with future developments on the model performance or relevance [17]. The multiple model simulation evaluates several modeling structures to determine the overall impact of structural uncertainty on the model performance [18,19]. The inverse modeling method assesses the effect of uncertainty in process parameters on the model performance and helps in optimizing the parameters [20,21]. Sensitivity analysis (SA) derives a hierarchy of model input in terms of their contribution to the model output uncertainty [22]. The sample-based uncertainty analysis methods, i.e., Monte Carlo and polynomial chaos expansion (PCE), quantify the collective impact of uncertainty in model input on its output [15].
In this study, an integrated framework of data-based soft sensing and uncertainty analysis was proposed. Data-based soft sensors were developed using ensemble learning method, i.e., boosting, to predict composition, quantity, and quality of fatty acid methyl esters (FAME) in the outlet streams of biodiesel production process; cetane number of the FAME was used as a quality parameter. The cetane number relates to the ignition delay time of a fuel and is applied to alternative diesel fuels such as biodiesel and its components [23]. An increase in cetane number reduces the ignition delay period and stabilizes running of the engine. However, an excessive rise in cetane number causes too much reduction in the ignition delay. As a result, the fuel does not have proper time to spread into the combustion chamber and performance of the engine decreases. Prediction of composition helps in maintaining desired level of mole fraction of components while prediction of cetane number assists in realizing high quality of FAME. Prediction of quantity, i.e., flow rate, of FAME is important for evaluating conversion efficiency of the process. Polynomial chaos expansion (PCE) method was incorporated into the development of the soft sensor to quantify the effect of process uncertainties on the target outcomes, i.e., composition, quantity, and quality of FAME. PCE is computationally less expensive than other sampling method such as Monte Carlo. The incorporation of PCE transformed the prediction of the ensemble model from deterministic to stochastic format where the effect of process uncertainties is visualized in the form of predictive distributions.
2. Process and Data Description
A process flow diagram of biodiesel production from vegetable oil is shown in Figure 2.
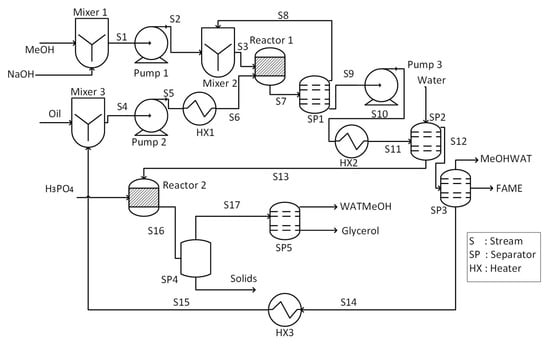
Figure 2.
Process flowsheet of biodiesel production from vegetable oil.
It is a base catalyzed process that utilizes NaOH as a catalyst. The main steps in the process involve transesterification reaction, biodiesel separation, glycerol separation, and recovery of methanol. Vegetable oil feed along with recycled oil is heated to reaction temperature using heater (HX1) and then fed to the reactor (Reactor 1). In addition, a mixture of NaOH and methanol (MeOH) is also charged into the reactor where transesterification reaction occurs. Effluent of the reactor is separated into two component by the first separator (SP1); excess methanol is recovered as the top stream and recycled, while bottom stream, i.e., fatty acid methyl esters (FAME), is sent to the second separator (SP2) for further purification.
Water washing is performed in SP2 to remove glycerol, catalyst, and unconverted methanol from FAME. The top stream (S12) is sent to the third separator (SP3) where unconverted oil is separated to get further purified FAME as a product. The bottom stream of SP2 is fed to the other reactor (Reactor 2) where phosphoric acid is used to neutralize the stream. Solids are removed from the effluent of Reactor 2 by filtration in SP4. Then, another separator (SP5) is used to remove water from the top stream of SP4 and produce pure glycerol as a by-product.
3. Fundamentals of Modeling and Analysis Methods
3.1. Soft-Sensor Development
Boosting, which is an ensemble learning technique, is adopted in this study for the soft sensor development. Boosting is based on the idea of developing a robust model by combining several weak models [24]. The concept of boosting is demonstrated in Figure 3 [25]. The models are developed in a series of rounds where the focus on incorrectly predicted target samples is increased with the help of increasing their respective weight. Several boosting mechanism are developed on the basis of variation in their methods. Least Squares Boosting (LSBoost) was adopted in this study [26].
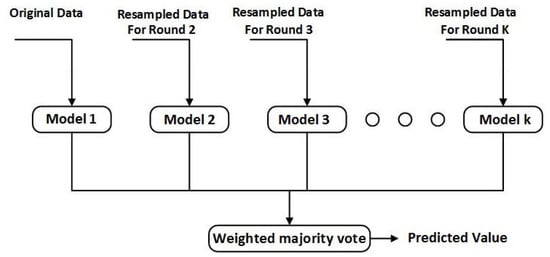
Figure 3.
The framework of boosting.
3.2. Uncertainty Analysis
Uncertainty analysis quantifies the accumulated impact of uncertainties of all input variables of a model [27]. Uncertainty has several dimensions, i.e., location, level, and nature; an extensive review of dimensions of uncertainty in process modeling was done by Ahmad et al. [14]. There are several methods available for uncertainty analysis of a process model. This study utilized sampling-based methods, i.e., polynomial Chaos Expansion (PCE), for uncertainty analysis of the process. The sampling-based uncertainty analysis helps in determining collective impact of uncertainties in all process variables on the process output.
In PCE, a random variable x is represented as a function (f()) of another random variable (ξ) [28,29]:
The PCE seeks an appropriate function f(), by describing x through deterministic and stochastic components:
where and are the deterministic and the stochastic components, respectively. is a polynomial that satisfy the following condition:
where is the inner product of and , and is the probability distribution function (PDF) of ξ.
To implement the PCEs, the mode strengths should be estimated by intrusive or non-intrusive (black box) methods [30,31]. In the current study, we implemented a non-intrusive approach where an ensemble model was used as a black-box system.
4. Proposed Modeling and Analysis Framework
The proposed modeling strategy is shown in Figure 4 and summarized as follows:
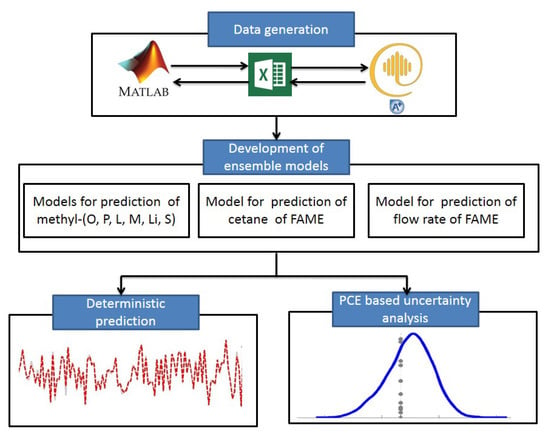
Figure 4.
Proposed modeling framework.
- Data generation: Data are generated by inserting variations in the steady state values of the process model through interfacing of MATLAB®, Excel® and Aspen® environments. Lists of process input and output variables used for the model development are shown in Table 1 and Table 2, respectively.
 Table 1. Process input variables.
Table 1. Process input variables. Table 2. Process output variables used for ensemble learning.
Table 2. Process output variables used for ensemble learning. - Soft-sensor Design: The generated data are used to develop the soft-sensors through the ensemble learning method. The number of decision trees, i.e. weak learners, in the ensemble models is optimized.
- PCE based uncertainty analysis: The ensemble model developed in Step 2 is used within the PCE framework. PCE level and the number of terms are optimized. A uniform uncertainty in all input variables is assumed and PCE based random variables are generated for each of the input variables. The PCE based generated random variables are fed to the ensemble model and predictive distributions of respective outputs are obtained.
MATLAB®, Excel®, and Aspen® were interfaced to generate 525 data samples. In the ensemble modeling, the weak learners and their optimized number for the respective models are shown in Table 3. In the PCE based method, Hermite function was used for a level of 6 and initial 20 terms.

Table 3.
Summary of soft sensors predictions.
5. Results and Discussion
This section covers the application of the proposed integrated framework of data-based soft sensing and uncertainty analysis to the biodiesel production process from vegetable oil. Seven ensemble models (soft sensors) were developed, one for flow rate of FAME, one for cetane number, and one each for prediction of the mole fractions of the components, i.e., Methyl-Li, Methyl-M, Methyl-O, Methyl-S and Methyl-P.
Cetane number for biodiesel was calculated using the following equation:
where represents cetane number of biodiesel; t refer to the type of methyl ester component, i.e., Methyl-Li, Methyl-M, Methyl-O, Methyl-S and Methyl-P; and X represents mass fraction.
A total of 525 datasets comprising the input and output data were generated for each model; data were divided into training (80%) and validation (20%) sets. Training sets were used for model development, while validation sets were used to evaluate models’ accuracy.
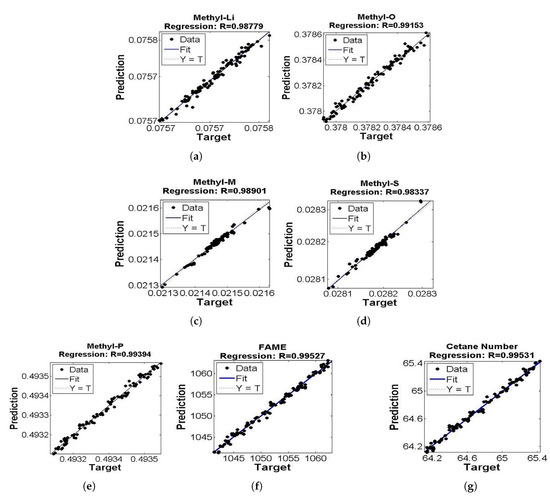
Figure 5.
(a–g) Regression performance of ensemble model of Methyl-Li, Methyl-O, Methyl-M, Methyl-S, Methyl-P, Fame and Cetane number.
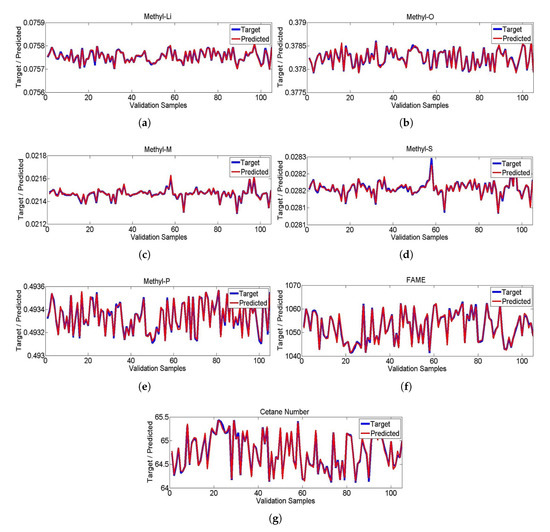
Figure 6.
(a–g) Target and predicted values of Methyl-Li, Methyl-O, Methyl-M, Methyl-S, Methyl-P, Fame and Cetane number.
A correlation coefficient between the target and predicted values of soft sensors was used to evaluate prediction accuracy. The correlation coefficients of soft sensor for Methyl-Li, Methyl-O, Methyl-M, Methyl-P, and Methyl-S components were 0.9877, 0.9915, 0.9890, 0.9939 and 0.9833, respectively. Root-mean-square errors (RMSE) for Methyl-Li, Methyl-O, Methyl-M, Methyl-P and Methyl-S components were 3.954 × 10−6, 2.391 × 10−5, 6.906 × 10−6, 1.488 × 10−5, and 4.188 × 10−6, respectively. Squared errors of prediction (SSE) for Methyl-Li, Methyl-O, Methyl-M, Methyl-P and Methyl-S components were 1.61 × 10−9, 5.89 × 10−8, 4.912 × 10−6, 2.279 × 10−8, and 1.807 × 10−9, respectively.
The correlation coefficient of flow rate of FAME was 0.9952, as shown in Figure 5 and Figure 6. RMSE and SSE values for FAME were 0.6025 and 37.99, respectively. Similarly, the correlation coefficient for cetane number was 0.9953 as shown in Figure 5 and Figure 6. RMSE and SSE were 0.0396 and 0.1407, respectively.
Table 3 shows a summary of prediction accuracy (correlation coefficient), RMSE and SSE values for all the seven soft sensors. Actual values of target variables in validation datasets and percent errors in their corresponding predicted values are shown in Table 4.

Table 4.
Actual values of target outcomes of soft sensors and errors exhibited during their prediction.
The non-intrusive PCE based predictive distributions of Methyl-Li, Methyl-O, Methyl-M, Methyl-P, and Methyl-S components, and FAME flow rate and cetane number are shown in Figure 7; the dotted gray lines show actual values of the respective components, while the blue lines show the PCE based predictive distributions. Actual values refer to the steady state values of the target variables of the process model. Table 5 shows mean absolute deviation percent (MADP) in the target variables of some selected validation datasets for 1% uncertainties in actual values of all input variables.
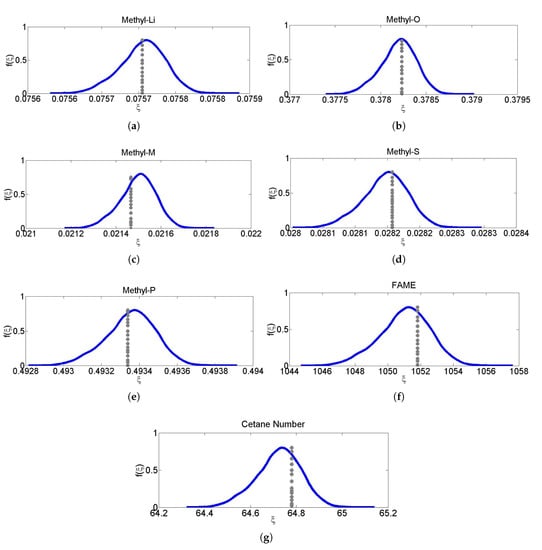
Figure 7.
(a–g) PCE based Predicted distribution of measured composition, cetane number and flow rate of FAME (blue lines), measured values of composition, cetane number and flow rate of fame (grey dotted line).

Table 5.
Mean absolute deviation percent (MADP) of output variables, i.e., compositions, flow rate and cetane number, from their respective measured values.
For Methyl-Li, Methyl-O, Methyl-M, Methyl-P, and Methyl-S components, MADP values were 0.27479, 0.32227, 2.41208, 0.1651, and 0.82135, respectively. For FAME flow rate and cetane number, MADP values were 0.96546 and 0.97013, respectively.
The boosting framework adopted in this study demonstrated high efficiency in predicting the desired outcomes. It is worth mentioning that boosting based soft sensors outperform other soft sensors based on single data-driven model such as ANN and other ensemble learning models such as RF [32,33]. The current soft sensing framework is more intuitive because the outputs cover all features of the product, i.e., quantity, quality and components affecting the quality. The multi-layer estimation of the process outputs promotes the efficiency of the process operation. In development of the soft sensors, many input variables were used, which enables them (soft sensors) to capture the actual dynamics of the process better than the soft sensors based on lesser number of input variables. Although the data used in the development of the soft sensors were extracted from an Aspen PLUS® model, the framework can be replicated for the process of a real biodiesel production plant where the raw material is vegetable oil.
The assumption of 1% uncertainty in all input variables was not based on reference information from plant operation but it helps in establishing a quantitative relation between the input uncertainty and their collective impact on the process outputs. MADP was used to quantify the deviation in the process output from their actual values. The deviation determined through the proposed framework can help in developing a control system for ensuring high yield and quality in a biodiesel production plant. In that context, parametric analysis of the process would be needed to identify the variables to be manipulated for maintaining desired values of the process outputs.
6. Conclusions
In this study, a data-based soft sensing mechanism was developed to predict composition, flow rate, and cetane number of fatty acid methyl esters (FAME). The non-intrusive polynomial chaos expansion (PCE) method was integrated in the soft sensors framework to quantify the effect of uncertainty on the soft sensors outcomes. A separate model (soft sensor) was developed for each of the components, flow rate and cetane. Prediction accuracies of Methyl-Li, Methyl-M, Methyl-O, Methyl-S, Methyl-P, FAME flow rate, and cetane number were 0.9877, 0.9890, 0.9915, 0.9833, 0.9939, 0.9952 and 0.9953, respectively. For 1% uncertainty in all input variables of the soft sensors, mean absolute deviation percent (MADP) values of 0.27479, 0.32227, 2.41208, 0.1651, 0.82135, 0.96546, and 0.97013 were noticed in the predicted values of Methyl-Li, Methyl-O, Methyl-M, Methyl-P, Methyl-S, FAME flow rate, and cetane number, respectively. The sensors are highly accurate in prediction and uncertainty quantification which make them suitable for real time applications.
Author Contributions
Conceptualization, I.A.; Software, I.A. and A.A.; Formal analysis, I.A. and A.A.; Methodology, I.A.; Supervision, I.A.; Writing—review & editing, A.A., U.I., M.K.K. and M.K.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atadashi, I.; Aroua, M.; Aziz, A.A.; Sulaiman, N. Refining technologies for the purification of crude biodiesel. Appl. Energy 2011, 88, 4239–4251. [Google Scholar] [CrossRef]
- Atadashi, I.; Aroua, M.; Aziz, A.A.; Sulaiman, N. Production of biodiesel using high free fatty acid feedstocks. Renew. Sustain. Energy Rev. 2012, 16, 3275–3285. [Google Scholar] [CrossRef]
- Jain, M.; Chandrakant, U.; Orsat, V.; Raghavan, V. A review on assessment of biodiesel production methodologies from Calophyllum inophyllum seed oil. Ind. Crop. Prod. 2018, 114, 28–44. [Google Scholar] [CrossRef]
- Kadlec, P.; Gabrys, B.; Strandt, S. Data-driven soft sensors in the process industry. Comput. Chem. Eng. 2009, 33, 795–814. [Google Scholar] [CrossRef]
- Meng, X.; Jia, M.; Wang, T. Neural network prediction of biodiesel kinematic viscosity at 313 K. Fuel 2014, 121, 133–140. [Google Scholar] [CrossRef]
- Rocabruno-Valdés, C.; Ramírez-Verduzco, L.; Hernández, J. Artificial neural network models to predict density, dynamic viscosity, and cetane number of biodiesel. Fuel 2015, 147, 9–17. [Google Scholar] [CrossRef]
- Mostafaei, M.; Javadikia, H.; Naderloo, L. Modeling the effects of ultrasound power and reactor dimension on the biodiesel production yield: Comparison of prediction abilities between response surface methodology (RSM) and adaptive neuro-fuzzy inference system (ANFIS). Energy 2016, 115, 626–636. [Google Scholar] [CrossRef]
- Miraboutalebi, S.; Kazemi, P.; Bahrami, P. Fatty Acid Methyl Ester (FAME) composition used for estimation of biodiesel cetane number employing random forest and artificial neural networks: A new approach. Fuel 2016, 166, 143–151. [Google Scholar] [CrossRef]
- Raman, A.A.A.; MK, A.; Sulaiman, N. Estimation of vegetable oil-based ethyl esters biodiesel densities using artificial neural networks. J. Appl. Sci. Data 2008, 8, 3005–3011. [Google Scholar] [CrossRef]
- Wali, W.; Hassan, K.; Cullen, J.; Shaw, A.; Al-Shamma’a, A. Real time monitoring and intelligent control for novel advanced microwave biodiesel reactor. Measurement 2013, 46, 823–839. [Google Scholar] [CrossRef]
- Fayyazi, E.; Ghobadian, B.; Najafi, G.; Hosseinzadeh, B. Genetic algorithm approach to optimize biodiesel production by ultrasonic system. Chem. Prod. Process Model. 2014, 9, 59–70. [Google Scholar] [CrossRef]
- Sikorski, J.J.; Brownbridge, G.; Garud, S.S.; Mosbach, S.; Karimi, I.A.; Kraft, M. Parameterisation of a biodiesel plant process flow sheet model. Comput. Chem. Eng. 2016, 95, 108–122. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Prayogo, D.; Ju, Y.H.; Wu, Y.W.; Sutanto, S. Optimizing mixture properties of biodiesel production using genetic algorithm-based evolutionary support vector machine. Int. J. Green Energy 2016, 13, 1599–1607. [Google Scholar] [CrossRef]
- Ahmad, I.; Kano, M.; Hasebe, S. Dimensions and Analysis of Uncertainty in Industrial Modeling Process. J. Chem. Eng. Jpn. 2018, 51, 1–11. [Google Scholar] [CrossRef]
- Helton, J.C.; Cooke, R.M.; McKay, M.D.; Saltelli, A. Sensitivity analysis of model output: SAMO 2004. Reliab. Eng. Syst. Saf. 2006, 10, 1105–1108. [Google Scholar] [CrossRef]
- Refsgaard, J.C.; van der Sluijs, J.P.; Højberg, A.L.; Vanrolleghem, P.A. Uncertainty in the environmental modelling process—A framework and guidance. Environ. Model. Softw. 2007, 22, 1543–1556. [Google Scholar] [CrossRef]
- Van Der Heijden, K. Scenarios and forecasting: Two perspectives. Technol. Forecast. Soc. Chang. 2000, 65, 31–36. [Google Scholar] [CrossRef]
- Linkov, I.; Burmistrov, D. Model uncertainty and choices made by modelers: Lessons learned from the international atomic energy agency model intercomparisons. Risk Anal. Int. J. 2003, 23, 1297–1308. [Google Scholar] [CrossRef]
- Butts, M.B.; Payne, J.T.; Kristensen, M.; Madsen, H. An evaluation of the impact of model structure on hydrological modelling uncertainty for streamflow simulation. J. Hydrol. 2004, 298, 242–266. [Google Scholar] [CrossRef]
- Duan, Q.; Sorooshian, S.; Gupta, V.K. Optimal use of the SCE-UA global optimization method for calibrating watershed models. J. Hydrol. 1994, 158, 265–284. [Google Scholar] [CrossRef]
- Doherty, J.; Johnston, J.M. Methodologies for calibration and predictive analysis of a watershed model 1. JAWRA J. Am. Water Resour. Assoc. 2003, 39, 251–265. [Google Scholar] [CrossRef]
- Saltelli, A.; Chan, K.; Scott, E.M. Sensitivity Analysis; Wiley: New York, NY, USA, 2000; Volume 1. [Google Scholar]
- Knothe, G.; Matheaus, A.C.; Ryan, T.W., III. Cetane numbers of branched and straight-chain fatty esters determined in an ignition quality tester. Fuel 2003, 82, 971–975. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ahmad, I.; Mabuchi, H.; Kano, M.; Hasebe, S.; Inoue, Y.; Uegaki, H. Data-Based Ground Fault Diagnosis of Power Cable Systems. SICE J. Control Meas. Syst. Integr. 2013, 6, 290–297. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Helton, J.C.; Johnson, J.D.; Sallaberry, C.J.; Storlie, C.B. Survey of sampling-based methods for uncertainty and sensitivity analysis. Reliab. Eng. Syst. Saf. 2006, 91, 1175–1209. [Google Scholar] [CrossRef]
- Perez, R.A. Uncertainty Analysis of Computational Fluid Dynamics via Polynomial Chaos. Ph.D. Thesis, Virginia Tech, Blacksburg, VA, USA, 2008. [Google Scholar]
- Oladyshkin, S.; Nowak, W. Data-driven uncertainty quantification using the arbitrary polynomial chaos expansion. Reliab. Eng. Syst. Saf. 2012, 106, 179–190. [Google Scholar] [CrossRef]
- Rajabi, M.M.; Ataie-Ashtiani, B.; Simmons, C.T. Polynomial chaos expansions for uncertainty propagation and moment independent sensitivity analysis of seawater intrusion simulations. J. Hydrol. 2015, 520, 101–122. [Google Scholar] [CrossRef]
- Sudret, B.; Der Kiureghian, A. Stochastic Finite Element Methods and Reliability: A State-of-the-Art Report; Department of Civil and Environmental Engineering, University of California Berkeley: Berkeley, CA, USA, 2000. [Google Scholar]
- Ahmad, I.; Ali, G.; Bilal, M.; Hussain, A. Virtual sensing of catalytic naphtha reforming process under uncertain feed conditions. In Proceedings of the 2018 International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, 3–4 March 2018; pp. 1–6. [Google Scholar]
- Ahmad, I.; Mabuchi, H.; Kano, M.; Hasebe, S.; Inoue, Y.; Uegaki, H. Data-Based Fault Diagnosis of Power Cable System: Comparative Study of k-NN, ANN, Random Forest, and CART. In Proceedings of the 18th IFAC World Congress, Milano, Italy, 28 August–2 September 2011; pp. 12880–12885. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).