Abstract
The identification of corn leaf diseases in a real field environment faces several difficulties, such as complex background disturbances, variations and irregularities in the lesion areas, and large intra-class and small inter-class disparities. Traditional Convolutional Neural Network (CNN) models have a low recognition accuracy and a large number of parameters. In this study, a lightweight corn disease identification model called DFCANet (Double Fusion block with Coordinate Attention Network) is proposed. The DFCANet consists mainly of two components: The dual feature fusion with coordinate attention and the Down-Sampling (DS) modules. The DFCA block contains dual feature fusion and Coordinate Attention (CA) modules. In order to completely fuse the shallow and deep features, these features were fused twice. The CA module suppresses the background noise and focuses on the diseased area. In addition, the DS module is used for down-sampling. It reduces the loss of information by expanding the feature channel dimension and the Depthwise convolution. The results show that DFCANet has an average recognition accuracy of 98.47%. It is more efficient at identifying corn leaf diseases in real scene images, compared with VGG16 (96.63%), ResNet50 (93.27%), EffcientNet-B0 (97.24%), ConvNeXt-B (94.18%), DenseNet121 (95.71%), MobileNet-V2 (95.41%), MobileNetv3-Large (96.33%), and ShuffleNetV2-1.0× (94.80%) methods. Moreover, the model’s Params and Flops are 1.91M and 309.1M, respectively, which are lower than heavyweight network models and most lightweight network models. In general, this study provides a novel, lightweight, and efficient convolutional neural network model for corn disease identification.
1. Introduction
Corn is the world’s third largest food crop, playing an important role in the agricultural economy [1]. Plant diseases cause significant losses in corn yields [2]. Due to similar disease characteristics, it is hard to distinguish between diseases with the naked eye. Inexperienced growers often misjudge the disease, which leads to the incorrect use of pesticides, affecting the yield and quality of corn and endangering the environment [3]. Having experienced plant pathologists visit the planting site for guidance is costly and difficult to achieve. A lightweight CNN model can be expediently deployed through mobile phones or edge devices. Therefore, automatically identifying leaf diseases through image processing techniques would be of great help to farmers.
With the advancement of digital image processing technology and deep learning methods, humans can automatically identify crop leaf diseases through machine learning algorithms and CNN methods. Increasing studies have been conducted regarding this issue. For instance, Aravind et al. [4] proposed a multi-classification Support Vector Machine (SVM) based on feature bags, which classified leaf spot disease, rust, leaf blight, and healthy leaves in the Plantvillage dataset with an accuracy rate of 83.7%. In addition, Budiarianto et al. [5] found that RGB features are the most accurate of most classifiers in the commonly used machine learning algorithms.
Using machine learning algorithms to identify crop diseases requires the manual design of features, which is laborious and inconvenient. It is difficult to cope with the identification of multiple crop diseases with different characteristics. Since the CNN method can seamlessly integrate pattern classification while extracting features and improving the efficiency of automatic disease identification, it has been adopted extensively in recent years. By optimizing the LeNet model, Ramar et al. [6] achieved the best accuracy (equal to 97.89%) in classifying three types of diseases in corn in the Plantvillage dataset. Panigrahi et al. [7] proposed a CNN model with both convergence speed and accuracy. The improved CNN model achieved 98.78% of average accuracy. Mohanty et al. [8] obtained 99.35% accuracy in Plantvillage by fine-tuning migration learning on Googlenet. Mishra et al. [9] proposed a CNN model for hardware devices with 88.46% accuracy for the real-time detection of maize diseases in raspberry pi 3.
In the above studies, the crop disease dataset images usually have a simple background. It is significantly different from the real environment. Saleem et al. [10] pointed to the importance of datasets with realistic conditions in plant disease detection and classification. Similarly, in Ferentinos’s study, it was demonstrated that the model trained by a simple background image dataset does not work under real conditions [11]. Noise and interference in images collected in natural light make it difficult to distinguish disease features. Therefore, it is difficult to achieve the accuracy of exact disease identification using previous models. Thus, to enhance the details of corn disease characteristics and reduce the complex background noise, Lv et al. [12] proposed an image preprocessing algorithm, called WT-DIR, for a dataset under real conditions. It has an accuracy rate of 98.62% in the DMS-Robust AlexNet network model. Moreover, Zeng et al. [13] improved the ResNet50 method by replacing convolutional kernels, activation functions, and loss functions. The proposed CNN algorithm, called SKPSNet-50, achieved an accuracy of 92.6% in the corn disease dataset taken in real environments.
Furthermore, for complex background images, the attention mechanism can increase the pertinence of the model to focus on the disease area, improving the model’s ability to learn the characteristics of diseases. Hence, Akshay et al. [14] proposed an Attention Dense Learning (ADL) mechanism. By stacking five ADLs into a CNN, called DADCNN-5, the simulation achieved a 97.33% accuracy rate in the dataset of complex background images captured by mobile phones. In addition, Zhu et al. [15] achieved an accuracy of 96.58% in a dataset of complex backgrounds by using a transformer-embedded convolutional neural network.
Although the above deep learning models show satisfying accuracies in plant dis-ease identification, they are heavyweight and require abundant computational resources. Therefore, it is necessary to design lightweight neural networks. For instance, Chen et al. [16] simplified the DenseNet algorithm by replacing the standard convolutions with depthwise separable convolutions and proposed a neural network called MS-DNet. The number of parameters in MS-DNet is approximately 0.36M, which is less than the parameter number of existing DenseNet. The proposed model obtained an accuracy rate of 98.32%. Based on MobileNet v2 as the backbone, Chen et al. [17] embedded the attention modules and optimized the loss function. The improved model achieved 98.48% accuracy in rice disease identification under complex background conditions. Meanwhile, the lightweight model named DISE-NET was proposed for the classification of maize small leaf spots by Yin et al. [18]. The dilated inception module and the attention module were designed to enhance the multi-scale feature extraction capability of DISE-NET. Recently, Zeng et al. [19] proposed a lightweight model for mobile deployment called LDSNet, which obtained an accuracy of 95.4% in classifying corn leaf diseases in the field. In addition, Lin et al. [20] presented a lightweight CNN model named GrapeNet based on residual blocks, Residual Feature Fusion Blocks (RFFBs), and Convolution Block Attention Modules (CBAMs) for grape leaf disease identification. The experiment result showed the GrapeNet model achieved the best classification performance with an accuracy of 86.29%.
In general, large-scale data are necessary to ensure the performance of deep learning models [21]. In routine computer vision tasks, researchers have built large-scale datasets, such as ImageNet [22] and COCO [23]. However, due to the time-consuming nature of collecting and annotating datasets, there are few large-scale and open-access datasets for crop disease. Thus, data augmentation is an efficient way to mitigate data shortfalls. For example, Pan et al. [24] expanded 985 images of corn northern blight and healthy corn leaf to 30,655 through traditional offline data enhancement methods, such as image segmentation, sizing, cropping, and transformation. Richey et al. [25] used various photometric and geometric enhancements to expand the number of images. Nevertheless, offline data augmentation could result in low diversity in crop disease images and lead to overfitting of the model. Alternatively, augmenting data using the Generative Adversarial Network (GAN) method is very efficient. It could enrich the disease characteristics of the dataset [26]. Chen et al. [27] used boost DCGAN and traditional data augmentation methods combined to obtain a better featured-image dataset of corn diseases. However, GAN requires high computational power from the computer and the process is complex. Online data augmentation is a random augmentation of the original data before each training epoch; this method is flexible and simple, and the data are different for each epoch. Traditional online augmentation helps to create many virtual images by randomly rotating, moving, cropping, and flipping the original image. For example, Albarrak, Gulzar, and Hamid et al. [28,29,30] achieved good results in augmenting datasets, such as seeds and fruits, by traditional online data augmentation. However, the traditional data augmentation method based on geometric transformation loses some feature information about the lesion area. Hence, it is necessary to explore novel approaches for crop disease image online data augmentation.
Table 1 summarizes the main work of the above literature.

Table 1.
Comparative analysis of the related work on plant disease identification.
The identification of corn leaf diseases in a real field environment faces several difficulties, such as complex background disturbances, variations and irregularities in the lesion areas, and large intra-class and small inter-class disparities. In addition, in crop recognition tasks, traditional CNN models with a large number of parameters require more computational resources and are difficult to widely scale up.
To address the above issues, we designed a lightweight CNN model called DFCANet. Inside, the DFCA block improves the feature extraction ability by fusing low-level characteristic information and high-level feature information together. Meanwhile, in order to focus on the lesion area in a complex background, the attention mechanism was applied. Moreover, the DS block can retain useful information better while effectively suppressing noise information by extending the channel dimension and using different down-sampling methods. The main aims of the study are as follows:
- (1)
- Proposing a lightweight convolutional neural network model, called DFCANet, based on DFCA blocks and DS blocks, which are used to identify corn diseases in real environments.
- (2)
- Exploring an online data augmentation method for images of corn leaf diseases.
- (3)
- Comparing the DFCANet with other classical network models to prove the performance advantages of DFCANet and conduct ablation experiments to verify the validity of the different module designs.
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
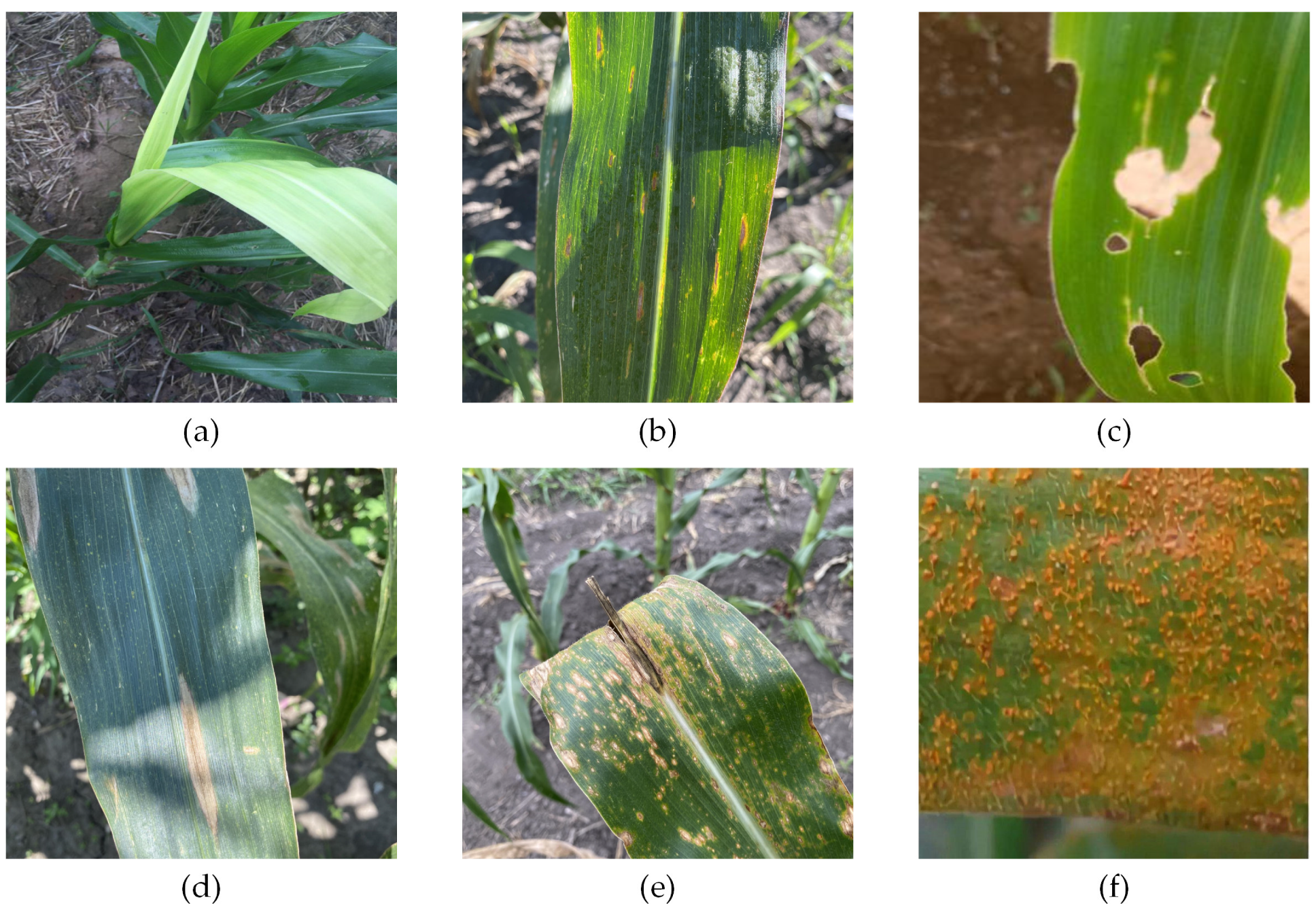
We acquired corn disease data from four different pathways, including three public datasets and web collections. The three public datasets were CD&S [31], PlantDoc [32], and Corn-Disease (https://github.com/FXD96/Corn-Diseases, accessed on 10 August 2022.). In addition, we collected some images from a search engine (https://image.baidu.com/). We obtained images of three types of corn diseases images from the CD&S [31] dataset, namely Northern Leaf Blight (NLB), Gray Leaf Spot (GLS), and Northern Leaf Spot (NLS). The CD&S dataset was acquired under field conditions at Purdue University’s Agronomy Center for Research and Education (ACRE) in West Lafayette, Indiana. We obtained images of corn rust leaves under real conditions from the PlantDoc [32] dataset. Additionally, we obtained images of corn leaves infected by the fall armyworm on Corn-Disease. Finally, we crawled the web of healthy corn leaves and a small number of other disease images to balance the data distribution. To summarize, we collected 3271 images, including 537 images of healthy leaves, 688 images of NLB disease, 551 images of NLS disease, 618 images of GLS disease, 445 images of corn rust, and 432 images of corn leaf infected by the fall armyworm. Figure 1 shows a sample corn disease dataset in a real environment.
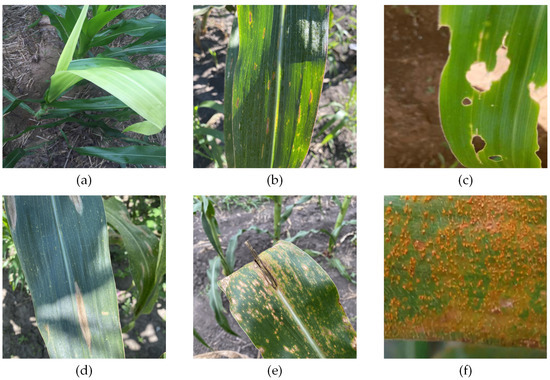
Figure 1.
Example of corn leaves. (a) Healthy leaf. (b) Gray leaf spot leaf. (c) Corn leaf infected by fall armyworm. (d) Northern leaf blight leaf. (e) Northern leaf spot leaf. (f) Corn rust leaf.
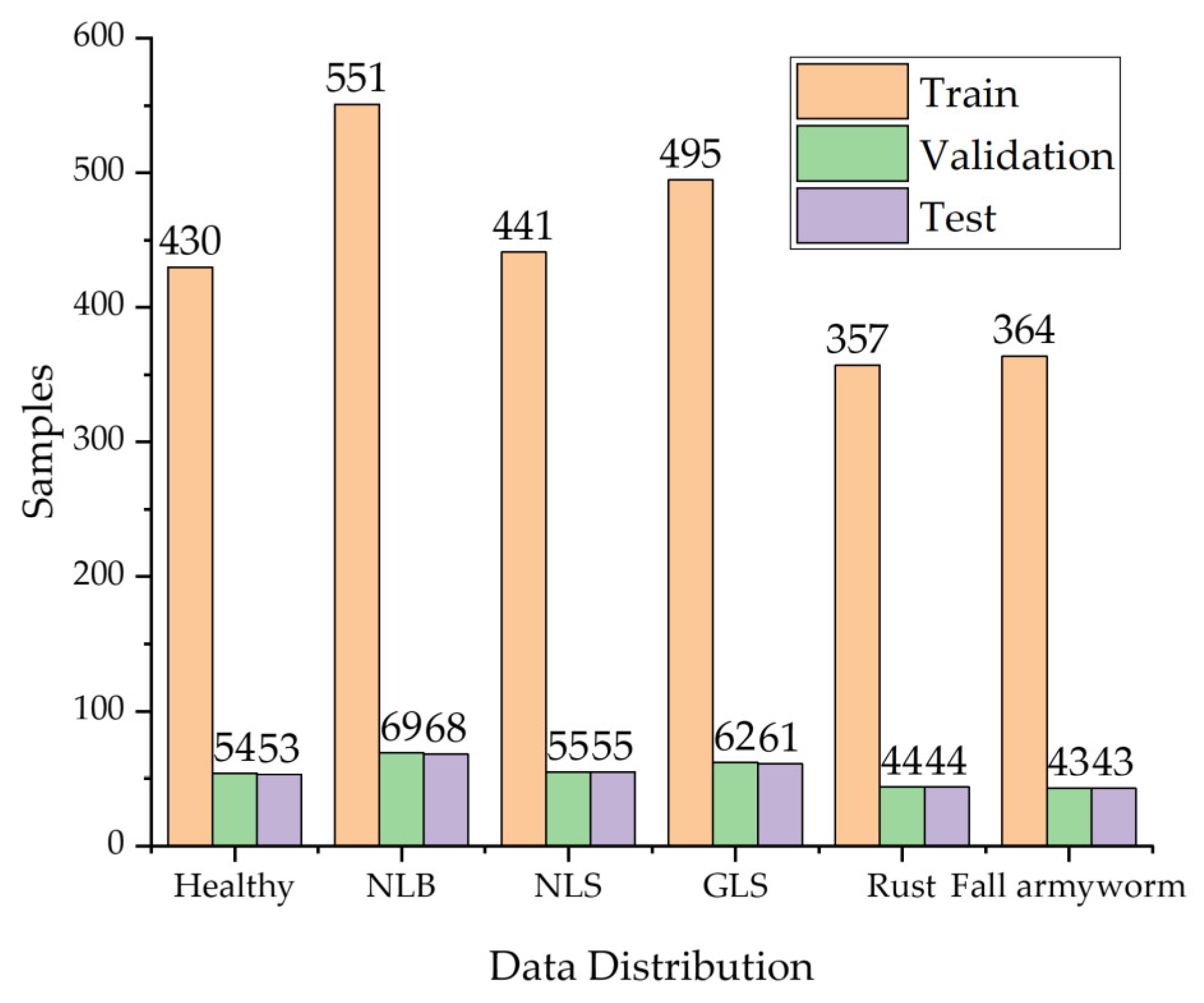
The data distribution of the dataset in this paper is shown in Figure 2. The training set, the validation set, and the test set were divided in the ratio of 8:1:1. In more detail, the validation set was used to save the model files with the highest accuracy in training, and the test set was used to test the performance of the model.
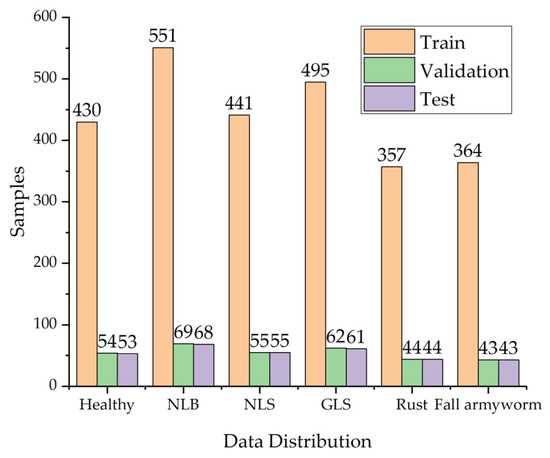
Figure 2.
Data distribution of corn leaf disease images.
Careful screening and supplementary data make its distribution more balanced, thus avoiding the overfitting of a certain type of disease in model training. The learning of deep learning models requires a large amount of data, but under real-world environmental conditions, the cost of collecting data is high, and the incidence of some crop diseases is low, resulting in a small number of crop disease images collected. Therefore, the data augmentation of the images is necessary. In deep learning, this process can be split into offline data augmentation and online data augmentation. On the one hand, offline data enhancement simply expands the amount of data by manipulating it (e.g., rotating, scaling, and contrasting). However, this method has poor flexibility, requires a vast storage capacity, and is prone to overfitting when the scaling up is too large. On the other hand, online data augmentation is carried out simultaneously in each batch of training, providing high flexibility and enhancing the generalization ability of the model. Figure 2 shows the data distribution of corn leaf disease images.
This article used a data augmentation method called KeepAugment [33], which avoids the disadvantages of traditional Cutout [34] and Random Erasing [35] that might erase diseased areas. Figure 3 illustrates some of the commonly used online data augmentation approaches, which can be seen to potentially mask out diseased areas of corn leaves.

Figure 3.
Classical online data augmentation with (a) original image, (b) Cutout, (c) GridMask [36] and (d) Random Erasing.
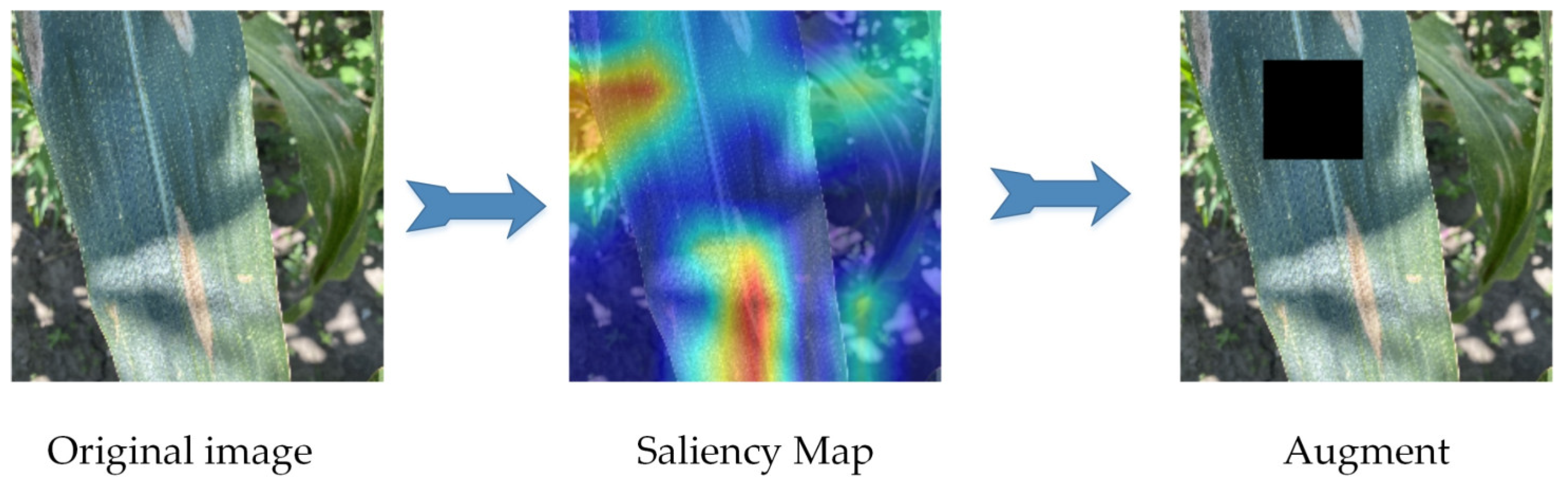
As shown in Figure 4, the KeepAugment data augmentation method detects important areas through a saliency map and preserves important areas in the image during the augmentation process, avoiding the erasure of disease features. The saliency map region was determined by calculating the backpropagation gradient to obtain the gradient of each pixel value and thus establishing the degree of influence of each pixel value on the category. Additionally, the division of the most important region and the least important region was determined by the sum of all the gradient values of this region being greater or lower than the corresponding threshold value.
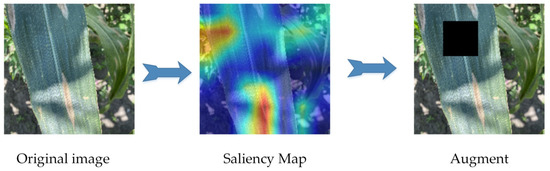
Figure 4.
KeepAugment data augmentation. The red area represents the location of interest to the model. KeepAugment data augmentation measures the vital areas of the leaf with the saliency map to avoid masking the area of the lesion.
2.2. DFCANet Model
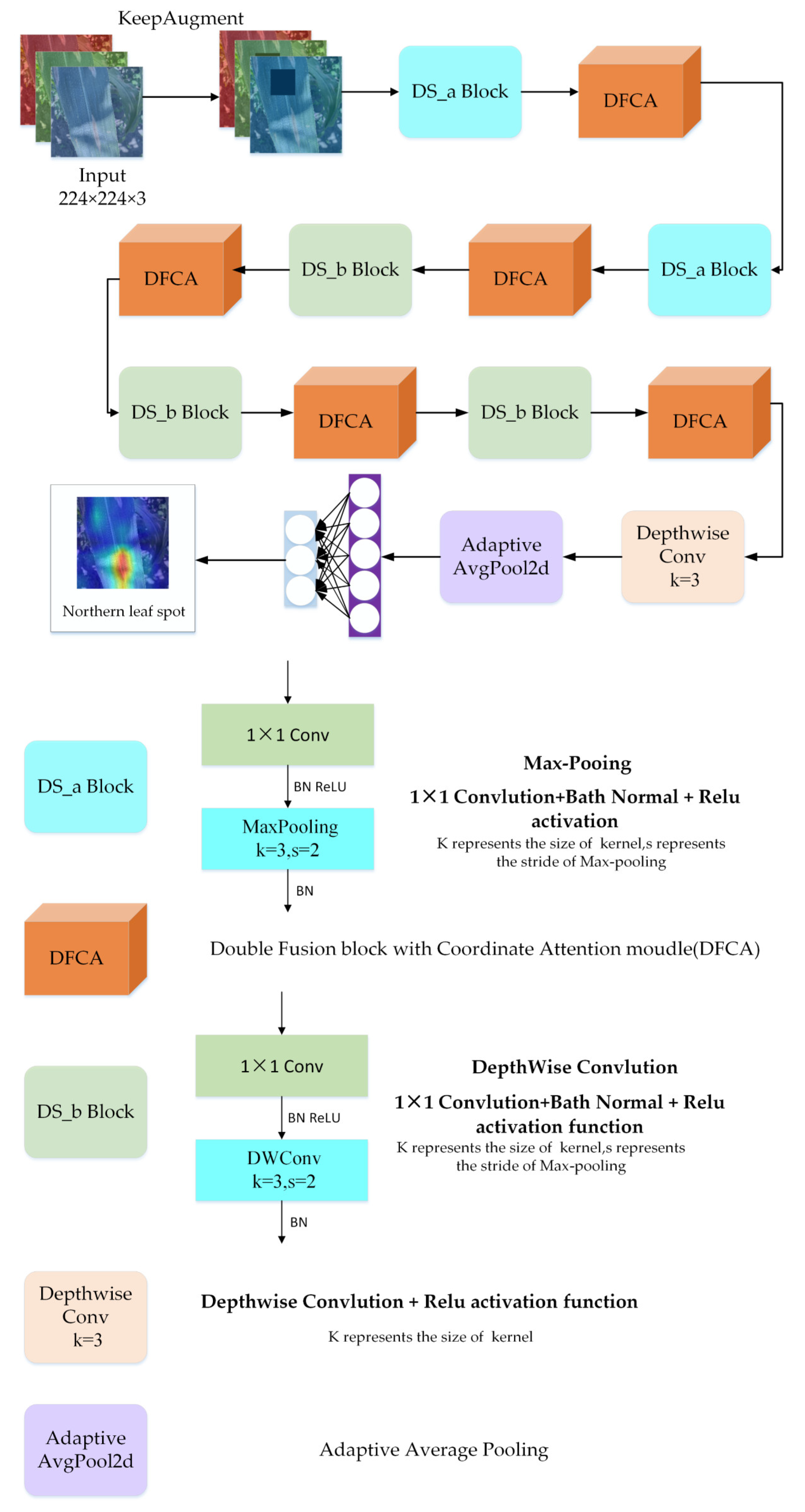
Figure 5 presents the structure of the DFCANet. It consists of a DFCA block, two different DS blocks, a depthwise convolution layer, an adaptive average pooling layer, and a classifier. All parts will be presented, in detail, in the remainder of this section.
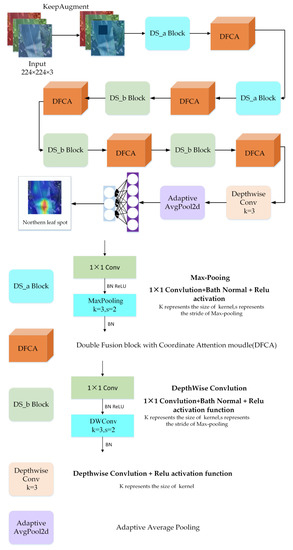
Figure 5.
Structure of DFCANet. It is composed of DS_a Blocks, DFCA modules, DS_b Blocks, a Depthwise Conv layer, an Adaptive Average Pooling layer, and a classifier.
2.2.1. DFCANet
As already shown, the DFCANet mainly consists of two blocks: A Double Fusion with Coordinate Attention [37] (DFCA) and a Down-Sampling block (DS). The complete architecture of DFCANet is shown in Table 2. The DFCA block is used to extract features, taking into consideration that the size of the feature map and the number of channels does not change. The DS block is used for down-sampling, expanding the number of channels and reducing the size of the feature map.

Table 2.
Architecture of DFCANet.
2.2.2. DFCA Block
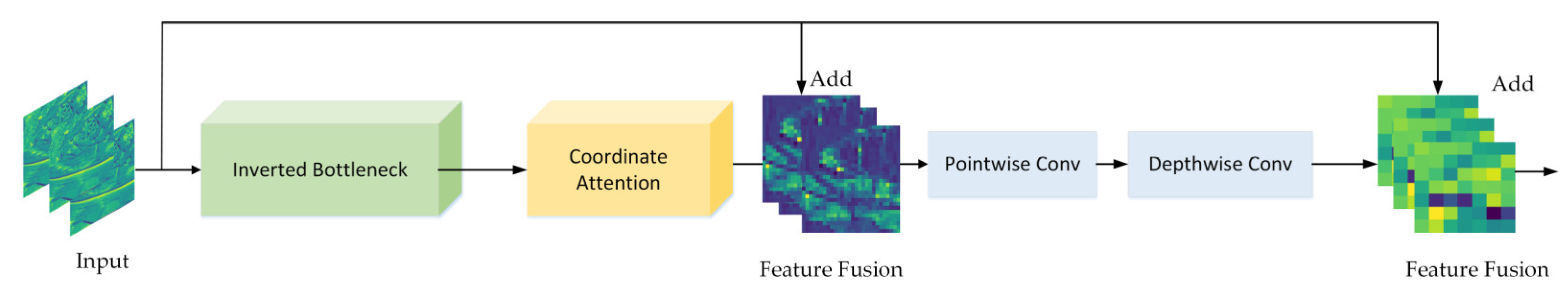
The DFCA block is shown in Figure 6. It mainly consists of three parts, namely, an inverted bottleneck, coordinate attention, and double fusion. Inspired by ConvNeXt [38], the inverted bottleneck was designed to better extract corn disease characteristics. The coordinate attention module looks for areas of disease characteristics and suppresses noise by recalibrating the channel weights of the input image. The feature information, extracted by CNN in several layers, is different and the feature fusion is to combine low-level extracted features with high-level extracted features to improve the recognition capability of the DFCA block. As shown in Figure 6, the low-level features are input features with a higher resolution and texture information. These features are transformed into mid-level features using feature extraction in the inverted bottleneck and recalibration in the attention module. The mid-level features have information on the location of diseased areas extracted by convolutional feature extraction in order to obtain high-level features with abstract semantic information. ResNet [39] completes the feature fusion by introducing quick identity connections and achieving widespread applications. Unlike ResNet, which only performs feature fusion in one stage, we performed two feature fusions to make it more thorough: The first phase consists of the fusion of low-level features with mid-level features, which can effectively locate the lesion area and ignore the background information, whereas the second phase involves low-level features with high-level features, which greatly enhances the model’s ability to extract subtle disease features.
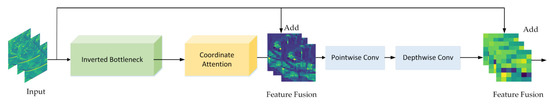
Figure 6.
Presentation of the DFCA block.
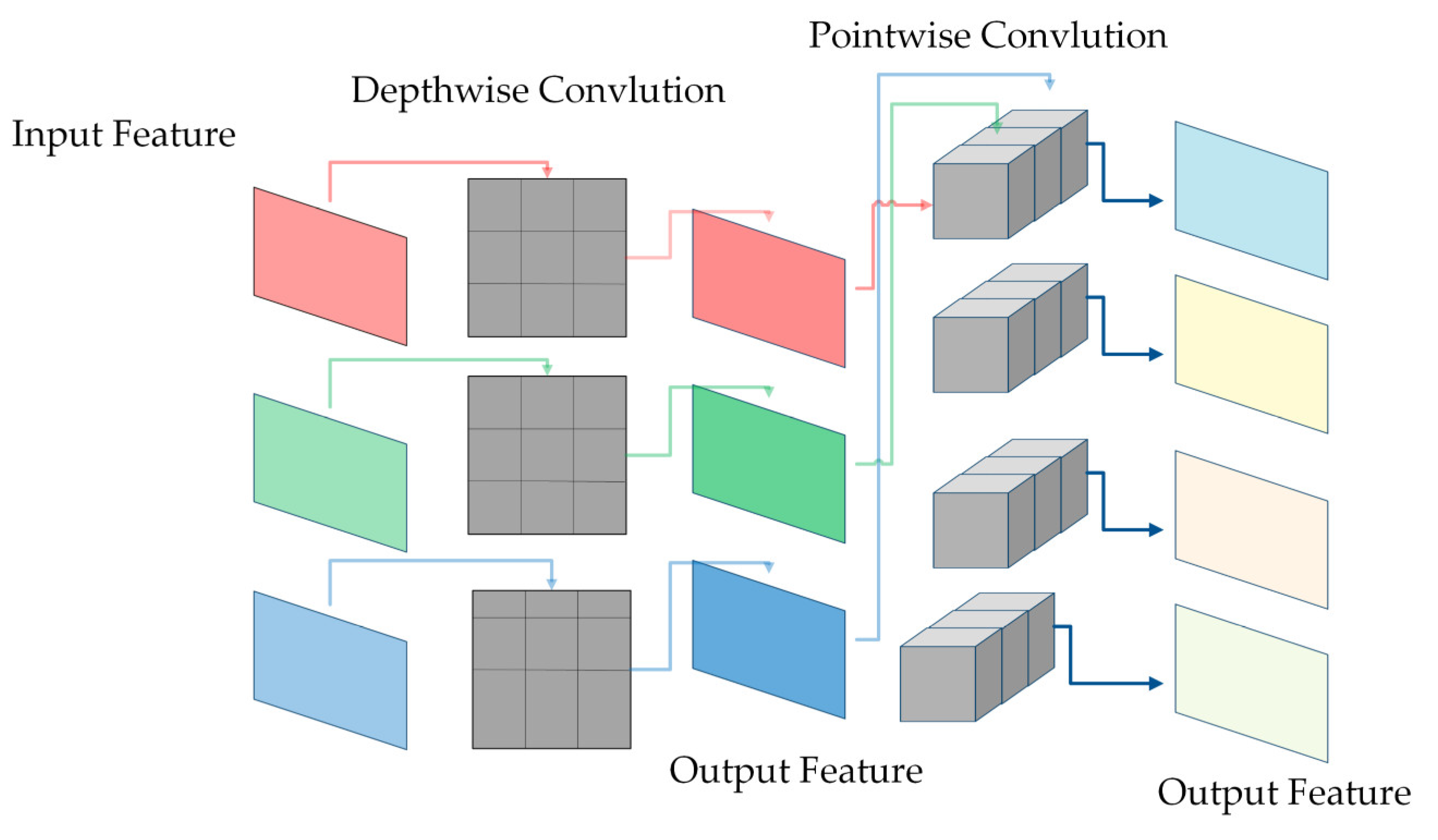
2.2.3. Depthwise and Pointwise Convolution
This study replaced ordinary convolution with depthwise convolution and pointwise convolution for lightweight. As shown in Figure 7, depthwise convolution convolutes each channel grouped into convolutions, allowing for a better collection of spatial features while significantly reducing the number of parameters. Additionally, point-by-point convolution sets the height and width of the convolutional kernel to one and the depth to the number of input channels. A lower parameter volume is maintained after being cascaded by deep convolution. The ratio of depthwise convolution and pointwise convolution calculation and ordinary convolutional calculation is as follows:
where represents the height and width of the convolutional kernel, represents the height and width of the input feature map, represents the number of the channel, and represents the number of channels of the output feature map. From the above equation, it is easy to see that depthwise convolution and pointwise convolution greatly reduce the computational effort of ordinary convolution.
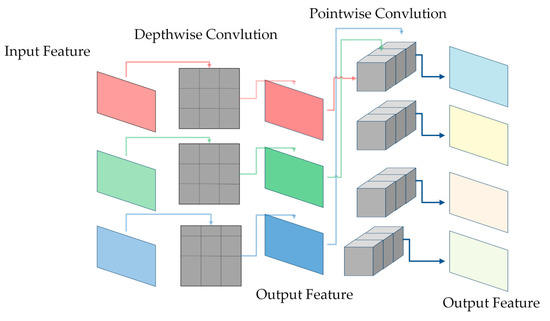
Figure 7.
Depthwise convolution and Pointwise convolution.
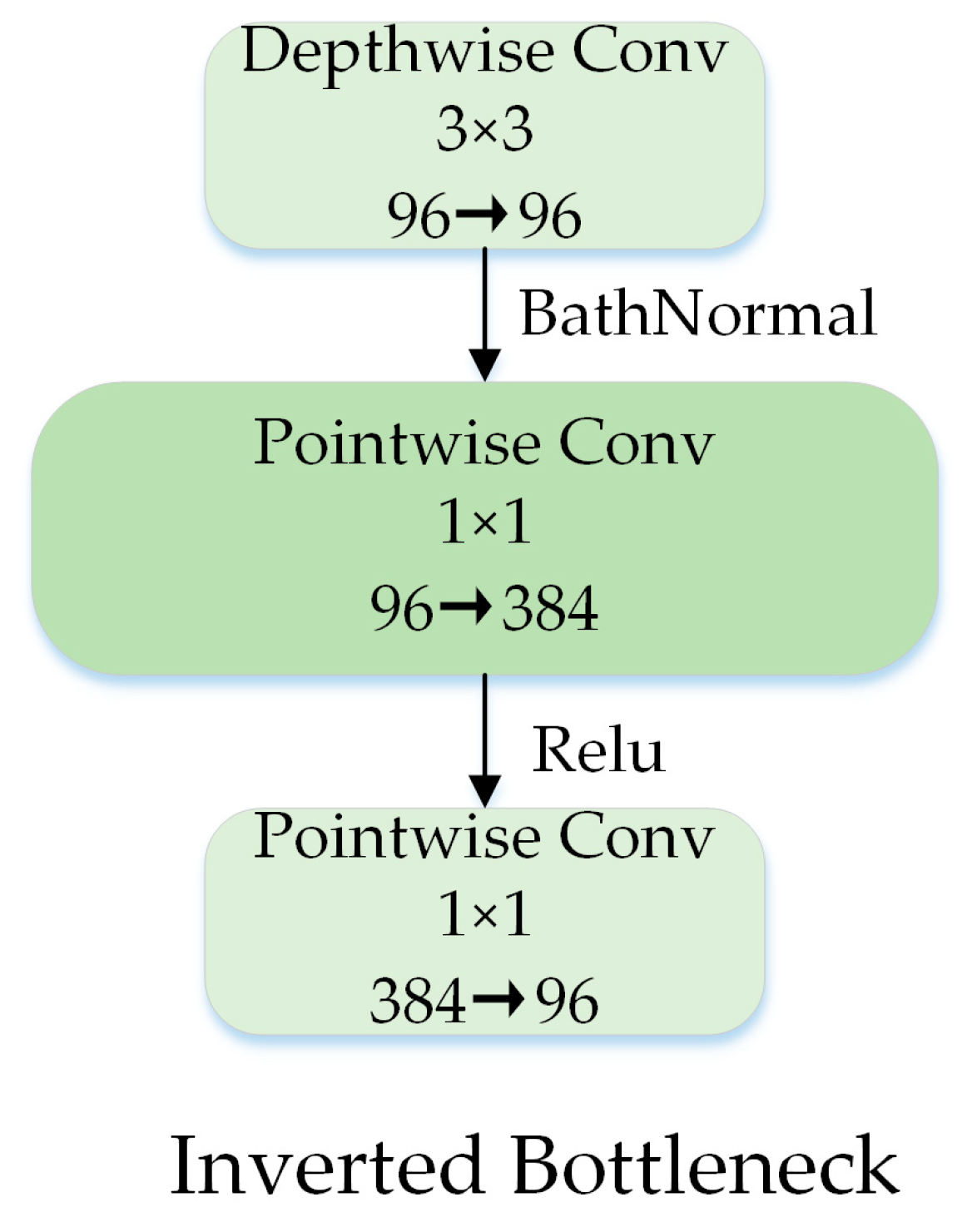
2.2.4. Inverted Bottleneck
The 1 × 1 pointwise convolution can extend the channel dimension. The inverted bottleneck structure can enrich the feature information, and it has been widely used since it was proposed in MobileNet V2 [40]. Drawing on ConvNeXt, we designed an inverted bottleneck structure as shown in Figure 8, placing the depthwise convolution in front of the pointwise convolution, which saved a considerable amount of computation time.
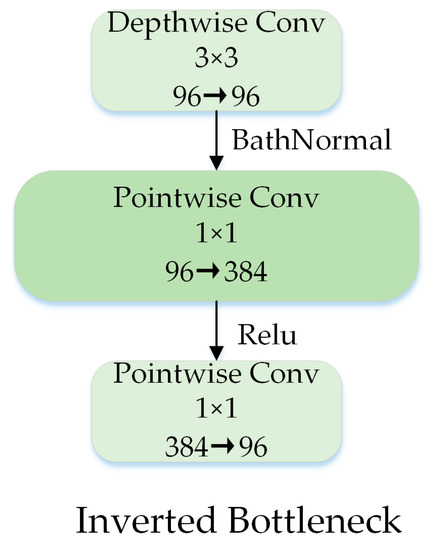
Figure 8.
Block designs for inverted bottleneck.
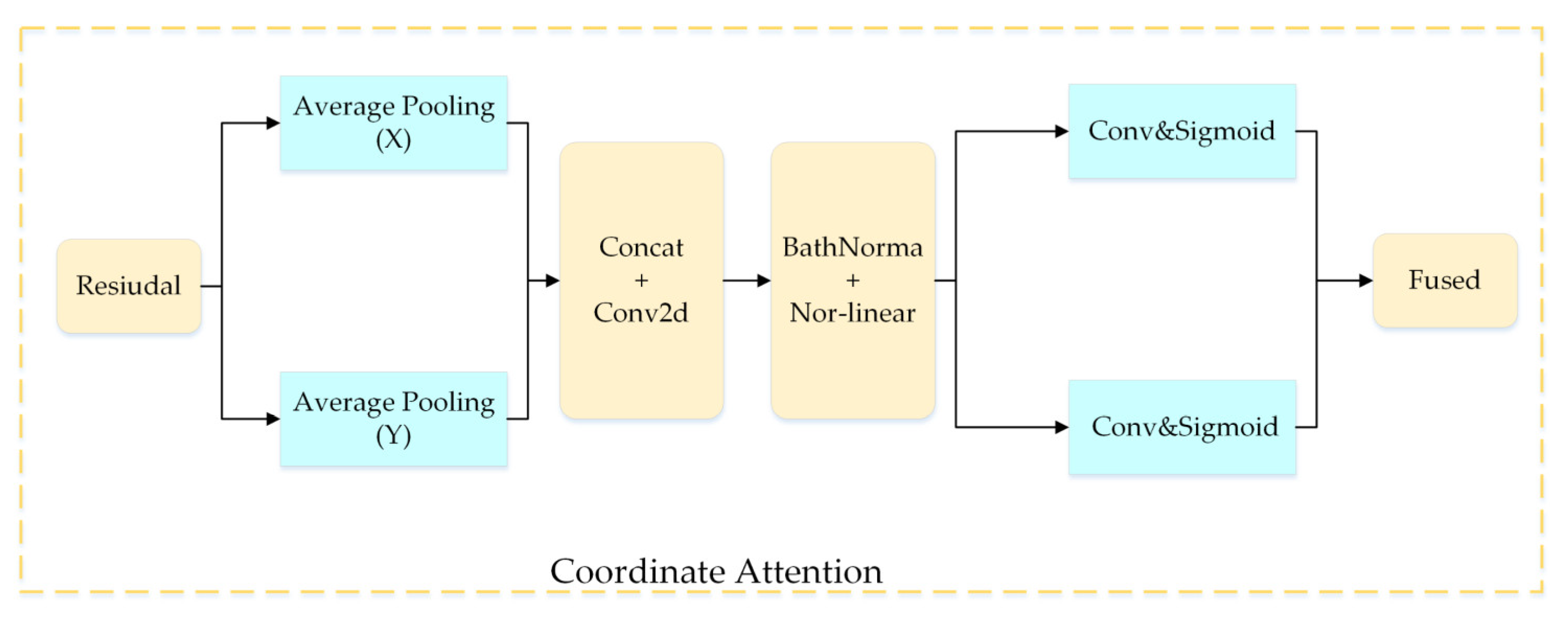
2.2.5. Coordinate Attention Module
Figure 9 shows the details of the CA module.
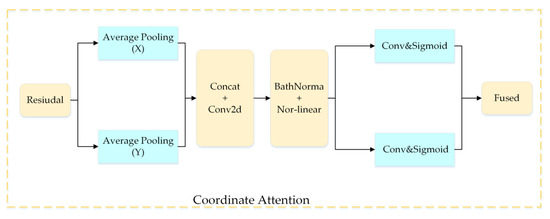
Figure 9.
Coordinate attention module.
Specifically, the coordinate attention module is decomposed in the vertical and horizontal directions and is transformed into a pair of one-dimensional feature codes as shown in the equation below:
where denotes the input of the CA module. and denote the height and width of the pooling kernel, respectively. represents the c-th channel’s output.
The global pooling of the above formula can encode spatial information globally, but it retains location information with difficulty. Thus, the pooling along in both directions is decomposed. After the horizontal decomposition, the output of the first channel with a height is as follows:
Equally, after applying the vertical decomposition, the output of the c-th channel with a width of is as follows:
The transformation of the attention module, described above, captures long-term dependencies along one spatial direction while saving position information in the other spatial direction. Finally, the convolution is fed after being spliced together by the feature diagram of the aggregation above:
where represents a feature map encoded in the horizontal and vertical directions, denotes a nonlinear activation function, and represents the convolution layer.
Then, we used the convolution to compress the channels. After that, we used the sigmoid function for normalization, from which we can obtain two outputs:
where and denote the attention weights of the two spatial directions.
The final output can be expressed as:
As shown in Figure 6, the CA module is added after the inverse bottleneck in the DFCA block. It not only ensures that the network is lightweight, but also makes the allocation of resources more reasonable, and the coordinate attention module can quickly find the area of interest in the disease image, ignoring the background and the noise information. Specifically, the coordinate module can use the sigmoid activation function, described above, to weight the characteristic map of the convolutional network (mainly learning to weight the coefficient) in order to obtain a new salient feature map. This new map is integrated with the original feature map, which can effectively emphasize the disease area and suppress noise and background information, heightening the learning ability of the network.
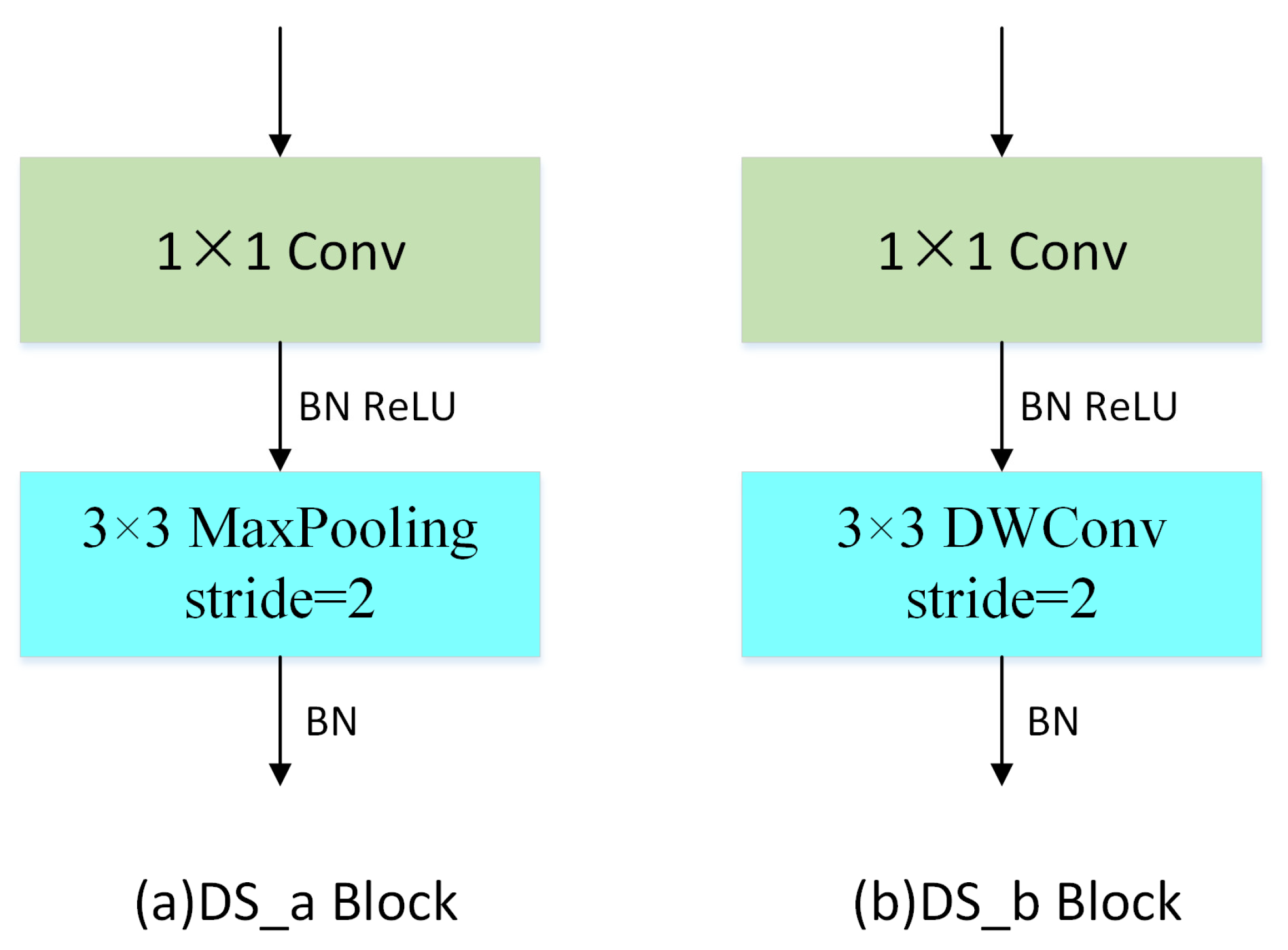
2.2.6. DS Block
The down-sampling operation, by reducing the size of the feature map, can not only increase the receptive field but also reduce the amount of computation. Usually, the down-sampling operation leads to the loss of some feature information; so, in this paper, the DS block used a 1 × 1 pointwise convolution to extend the channel dimension to reduce the loss of information during the down-sampling process. Two different DS modules were used in this work. Disease images acquired in real scenarios often contain complex backgrounds and noise; thus, they affect the recognition accuracy of CNN. In response to this problem, the Max-Pooling operation remains an important feature to remove distracting information in the initial stages of DFCANet. Depthwise convolution is used to down-sample the feature map, which preserves more useful information than pooling in the later stages. Simultaneously, the characteristics of the disease can be further extracted, and the network performance can be enhanced. Figure 10 illustrates the structure of the two proposed DS blocks.
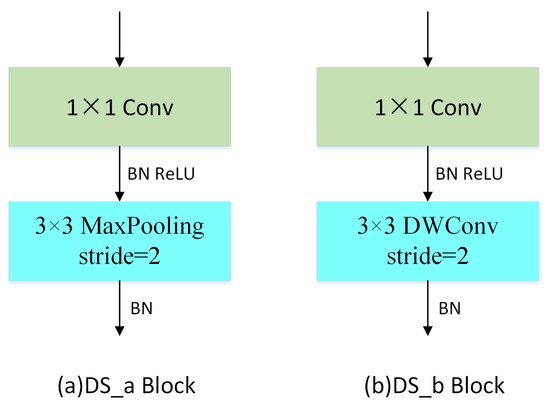
Figure 10.
Down-sampling module with (a) max-pooling and (b) depthwise convolution.
2.3. Experimental Environment and Hyperparameter Setting
The experimental hardware in this study was the Windows 10 Operation System and the Intel(R) Xeon(R) W-2235. The GPU model was NVIDIA GeForce RTX 2080Ti and the software environment adopted was Python 3.8.8 and the Pytorch 1.11.0 framework. The hyperparameters were set as follows: The cross-entropy loss function (CE) was used as the loss function and the Adam optimizer [41] was used to optimize the model. The initial learning rate and batch size during training were set to 0.002 and 32, respectively. The number of iterations was set to 100.
2.4. Evaluation Indexes
In this study, the accuracy, precision, recall, and F1 score were utilized to perform the evaluation metrics to measure the model’s performance.
3. Results
The experiment consisted of three parts: The first was to explore the effects of different data enhancement methods on corn disease identification; the second was to compare different network models; and the third was the ablation experiment of DFCA.
3.1. Impact of Different Data Augmentation Methods on the Model
To explore the online data augmentation methods applicable to corn leaf disease under real environmental conditions, we conducted the experiments as shown in Table 3. The experimental results indicate that most online augmentation methods can increase the diversity of the data, enhance the generalization ability of the DFCANet, and improve the recognition accuracy of the model. The data augmentation strategy using the KeepAugment method, while training DFCANet, eventually led the DFCANet model to reach, respectively, the values of 0.9785, 0.9817, 0.9792, and 0.9803 when computing the average accuracy, precision, recall, and F1 score. Thus, using the KeepAugment data augmentation method leads to an increase in the accuracy by 2.44% compared to the same simulation launched without the use of the data augmentation method. While using the GridMask data augmentation method, we found that the recognition accuracy of the model was reduced by approximately 0.61% because the feature areas of corn diseases might be masked during the application of this data augmentation algorithm.

Table 3.
Results of different data augmentation methods when applying the DFCAnet algorithm.
3.2. Comparative Experiment of Different Network Models
The above results demonstrate the effectiveness of the KeepAugment online augmentation strategy on the corn leaf disease dataset, and the strategy was used by default in all the subsequent experiments.
The proposed DFCANet model was compared with the classical CNN models. As shown in Table 4, DFCANet’s average classification accuracy, precision, recall, and F1 score reached 0.9785, 0.9817, 0.9792, and 0.9803, respectively, which are all superior to other CNN methods’ performances. The DFCANet model’s Params and Flops (Floating points of operations) have far lower values than heavyweight CNN models (VGG16, ResNet50, EffcientNetV2_b0 and ConvNeXt-base). The accuracy of the DFCANet model is 5.19% higher than that of ResNet50. EffcientNet-B0 performs better because its network structure is based on the Neural Architecture Search (NAS) technique to obtain the optimal set of parameters. In addition, EffcientNet has higher accuracy while having lower Flops. The number of model parameters (Params) of DenseNet121 is only 6.96M, but, as this method connects all channels to each other for feature reusing, it helps the model to retain the background noise information in the complex environment of the dataset easily and affects the classification accuracy of the model. Due to the redundant connection mechanism of DenseNet121, the model has 2.88G Flops. ShuffleNet V2 reduces the complexity of the model by channel splitting and channel disruption, so the Params and Flops of ShuffleNet V2 are lower than those of DFCANet, but the evaluation metrics, such as the accuracy and recall of the model, are lower than those of DFCANet. MobileNet-V2 and MobileNet-V3 have similar network structures, but MobileNet-V3′s structure is derived from NAS and has lower Flops. DFCANet’s Params are lower than those of MobileNet-V2 and MobileNetV3-large. In general, lightweight network models are better suited to smaller datasets than heavy models. This indicates that lighter models are more effective in small samples.

Table 4.
Results of the comparative experiment of corn leaf disease classification performance using the proposed and classic CNN models.
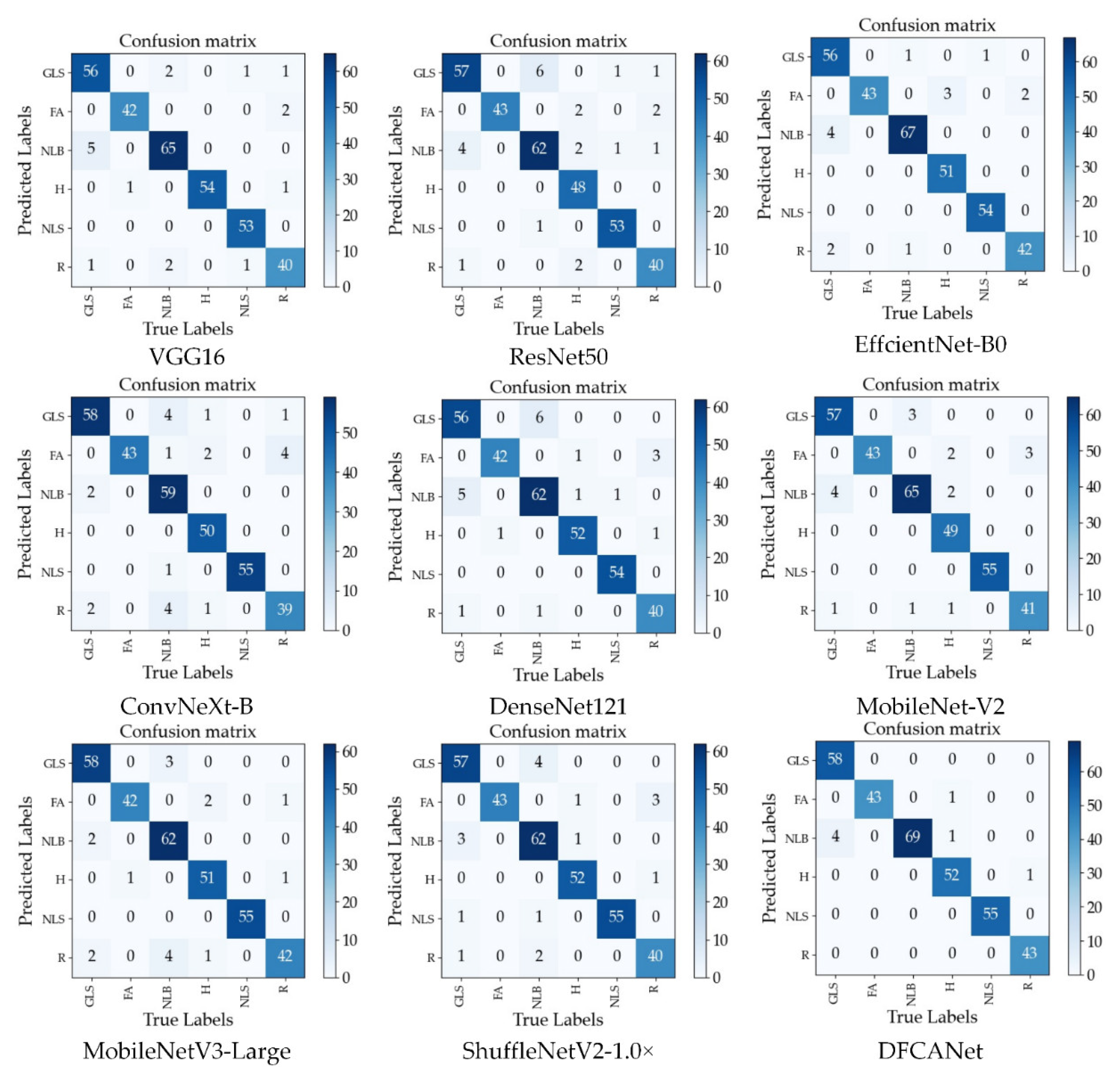
GLS, FA, NLB, H, NLS, and R represent, respectively, corn gray leaf spot, corn leaf infected by fall armyworm, corn northern leaf blight, corn healthy leaf, corn northern leaf spot, and corn rust leaf. Figure 11 shows the confusion matrix for the nine models. DFCANet leads other network models in the number of true positive samples in each category of the test set. This model achieves all correct predictions in three categories (FA, NLB, and NLS). In the test set of NLB, other network models predicted the highest number of errors, e.g., ConvNeXt-B predicted 10 errors, and most of the models incorrectly predicted NLB as GLS, because NLB and GLS have very similar disease characteristics (the symptoms of GLS include multiple greyish-brown and narrow rectangular lesions and the symptoms of NLS include multiple brown spots with circular concentric lesions). In the test set of FA, many models were able to complete all the correct predictions due to the fact that, unlike the complex characteristics of the disease, the insect pest caused the corn leaves to have more distinctive characteristics of mutilation. All obtained results demonstrate the effectiveness of our proposed network model for corn disease identification when dealing with complex backgrounds.
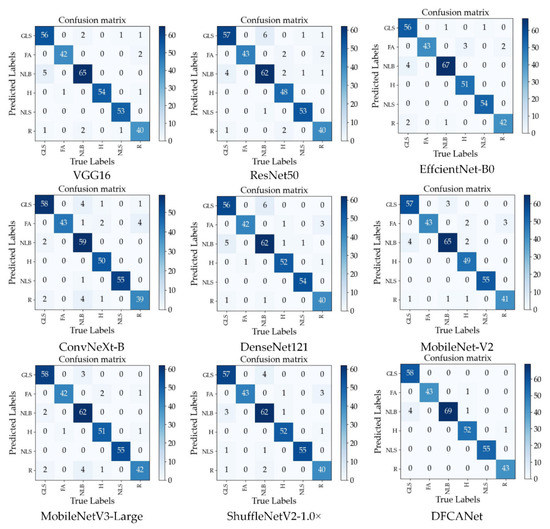
Figure 11.
Confusion matrix for the nine models.
3.3. Ablation Experiments
Table 5 shows a comparison of the results of adding different attention modules (the Squeeze-and-Excitation (SE) module, the CBAM module, and the Coordinate Attention (CA) module). All three attention mechanisms are lightweight, so they do not significantly increase the number of parameters of the model. The accuracy of DFCANet with the CA module is 1.52% and 2.75% higher than that of the SE and CBAM modules, respectively. This is due to the fact that the CA module can capture not only cross-channel information but also direction perception and location perception information. This combination helped the model to accurately locate and identify disease areas and disease features in corn leaves.

Table 5.
Comparison of the results of different attention mechanisms.
As shown in Figure 12, the class activation maps of the Grad-cam [49] visualization model vividly demonstrate the regions on which the model focused with different attention mechanisms. It is easy to find that all three attention mechanisms can effectively ignore the background and focus on the lesion area in the images of the real scene; however, compared to the SE and CBAM modules, the CA module can more accurately locate the lesion region of the corn leaves and focus on the disease characteristics more precisely.

Figure 12.
Grad-cam visualization results obtained using different attention mechanisms.
To better illustrate the effect of different modules on the model, we designed ablation experiments. First, we simplified DFCANet to a baseline. Specifically, the CA module and the double feature fusion branch were removed from DFCANet, and the DS block was replaced by a common down-sampling module. As shown in Table 6, the accuracy of the model increased by 1.22% after adding the CA module to the baseline network, indicating that the CA attention mechanism can effectively improve the recognition ability of the model. After adding the double-feature fusion branch, the model recognition accuracy increased by 1.53%, indicating that the second double-feature fusion can effectively fuse both the shallow feature information and the deep feature information. Finally, the DS module designed in this paper improved the model recognition accuracy by 2.44%, which proves that the DS Block can retain effective information and filter interference information at the same time. Compared to the baseline, the subsequent modules do not increase the Params too much, and the increase in Flops is within limits.

Table 6.
Results of the ablation experiments.
3.4. Simulation of Real Weather Data Augmentation Experiments
To simulate more realistic scenarios, we need to consider the weather conditions of the real environment, which are ignored in the existing datasets. Therefore, we algorithmically added rain, fog, and stronger sunlight effects to the images. As shown in Figure 13, the data augmentation of simulated real weather introduced noise and interference, which has higher requirements on the feature extraction ability of the model and the ability to suppress noise. Data augmentation expanded the number of images to twice the original. In the above experiments, we demonstrated the effectiveness of KeepAugment, so the augmentation of KeepAugment was performed after simulating the data augmentation of real conditions.

Figure 13.
Data augmentation by simulating real weather.
Simulating data augmentation under real conditions inevitably causes the image to introduce considerable noise and interference. These disturbances are also commonly encountered in images taken in real environments, so this affords good robustness to the model. As shown in Table 7, the accuracy, recall, and other evaluation metrics of each model improved after the data enhancement of the simulated real environment. It is worth noting that MobileNetv3-Large performs well in data enhancement with noise interference due to the addition of the SE attention module in MobileNetv3-Large. This shows the effectiveness of the attention module for noise suppression.

Table 7.
Comparisons of the recognition accuracy of different models.
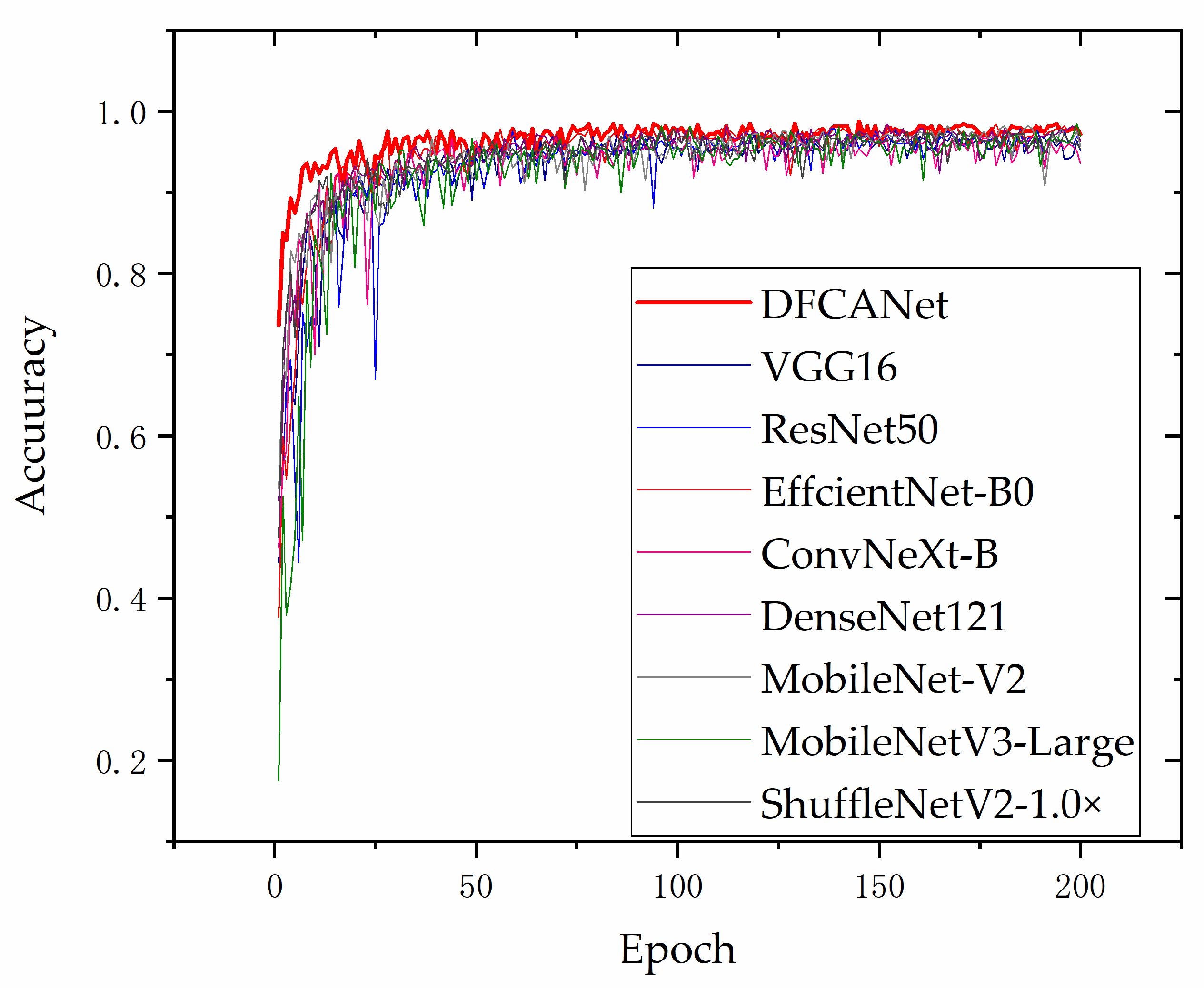
Figure 14 shows the validation curves in the validation set for different models. As the data augmentation of simulating real environment poses challenges to model training, in order to achieve a better convergence, we increased the training epoch to 200. It can be found that our proposed DCFCANet achieved the most advanced performance compared to other network models. DFCANet’s accuracy after curve smoothing averaged approximately 97%, ahead of other networks throughout the training period. Noise and disturbances were introduced in the data enhancement for simulating severe weather, which tested the robustness of the model. From the perspective of the magnitude of the curve fluctuations, the proposed model DFCANet has strong robustness. In addition, DFCANet is substantially ahead of the other models in the initial stage of training, at approximately 73% accuracy (other models are around 45%), which indicates that DFCANet has excellent fitting ability.
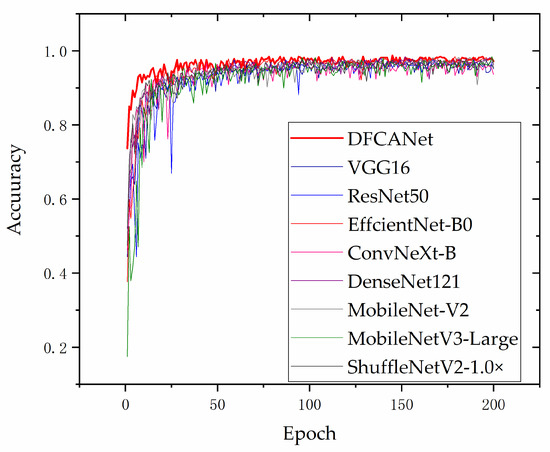
Figure 14.
Accuracy variation curves of the different models on the validation set.
3.5. Comparative Experiments of the Public Datasets
To further verify the superiority of the model in this paper, we conducted the experiments in the public dataset Plantvillage (https://github.com/spMohanty/PlantVillage-Dataset, accessed on 10 August 2022), as shown in Table 8. Plantvillage provides the same training and validation sets, so the experimental results in the literature are highly comparable. In recent years, numerous researchers have made significant contributions to the identification of plant diseases. In the task of maize disease identification, Aravind and Budiarianto et al. used machine learning algorithms for feature extraction and classification, so the recognition accuracy was lower than that of CNN models. Ramar and Panigrahi et al. obtained a better performance by improving a CNN model. The CNN model designed by Mishra et al. performed excellently in hardware. The DFCANet designed in this paper achieved the highest recognition accuracy of 99.47% in maize disease identification. In the task of identifying all diseases in Plantvillage, Mohanty, Mohameth, and Huang et al. all achieved high accuracy after fine-tuning by transfer learning. The CNN model designed in this paper was not pre-trained by ImageNet, yet it obtained the highest accuracy, which reflects that DFCANet has excellent feature extraction ability and fitting ability.

Table 8.
Comparisons of the recognition accuracy of different models on PlantVillage.
The images in the Plantvillage dataset were collected under laboratory conditions with simple background images. To further validate the accuracy of DFCANet in field conditions, we performed comparison experiments in the CD&S dataset. The CD&S dataset was the main source of data for this paper and includes images of three types of maize diseases (NLS, GLS, and LB). As with Plantvillage, CD&S provides a fixed training and validation set, which has high comparability. As shown in Table 9, numerous classical models achieved excellent accuracy due to the balanced data distribution of the CD&S dataset. DFCANet achieved the highest accuracy, further verifying its superiority.

Table 9.
Comparisons of the recognition accuracy of different models on CD&S dataset.
4. Discussion
Crop disease recognition is a challenging task in fine-grained classification, mainly because of the small number of crop disease samples, the difficulty of recognizing the type of diseases in complex scenes, and the large intra-class variation and small inter-class variation of crop disease features [52]. In addition, in crop recognition tasks, traditional CNN models with a large number of parameters require more computational resources and are difficult to scale up widely [6,53]. Hence, in this study, we designed a novel model structure to meet the challenges encountered in the real environment when capturing corn leaf photos in order to detect and classify diseases.
To address the problem of a large number of parameters in the traditional CNN model, we replaced ordinary convolution with deep separable convolution. It was demonstrated in Equation (1) that the computation of deep convolution and point convolution is times that of ordinary convolution. The authors of [16] used the same approach to simplify DenseNet, but our custom-designed CNN structure is more specialized and lightweight. Generally, attention mechanisms allow for better resource allocation and effective focus on crop disease areas. The authors of [20,54] introduced CBAM and SE modules to improve the model by 3% and 4.26%, respectively. In this study, we introduced the CA module, and the above-presented ablation experimental results (see Table 5) show that adding the CA modules improved the accuracy of the baseline by 1.22%. Additionally, the visualization results (see Figure 12) show that the CA module focuses better on the lesion area than the SE and CBAM modules. As the complexity of the disease situation increases, a bottleneck emerges in the role of attention mechanisms, which requires models with stronger feature extraction capabilities. The fusion of features is an effective way to enhance the model’s ability to extract features. Inspired by the different feature fusion methods of ResNet [39] and DenseNet [44], we discarded the redundant connection method of DenseNet and adopted a deeper fusion method than ResNet. The low-level features were fused to the middle-level and high-level layers, respectively. The experimental results show that our DFCA improved the accuracy of ResNet and DenseNet by 5.17% and 4.28%, respectively. Feature fusion enriches the diversity of features, but the way to retain information in down-sampling has been neglected by researchers [55,56]. Therefore, we designed a DS block to reduce the loss of feature information, and the model accuracy was improved by 2.44% in the ablation experiment. However, most previous studies use traditional offline data augmentation methods or simple online augmentation methods [19,20,28,29,30]. The experimental results obtained in this study show that the wrong data augmentation method can mask the disease area and reduce the accuracy of the model. For example, the accuracy of using GridMask is 0.61% lower than that of not using data augmentation. Our study validated an online data augmentation method named KeepAugment for crop diseases with a 4.2% higher accuracy than without augmentation. These findings provide implications for future studies on crop disease identification. In addition, despite pixel-based data enhancement methods such as random erasure and masking, we also took full account of what may happen in real environments and performed data enhancement by simulating severe weather. After this data augmentation, DFCANet model was improved to 98.47% accuracy. The experimental results show that data enhancement by simulating real weather not only improves the accuracy of the model but also improves the robustness of the model.
In addition to performing extensive experiments to verify the superiority of each module in DFCANet, we also compared it to the other literature and models in public datasets. In the public dataset Plantvillage, for disease identification of corn leaves, DFCANet achieved 99.74% classification accuracy, and the highest accuracy of 98.78% in [4,5,6,7,9], which is 0.96% lower than this study. Our method achieved the highest accuracy of 99.58% in the identification of all categories in Plantvillage. The accuracy of our method was 0.23%, 1.76%, and 0.43% higher than that of Mohanty et al. [8], Mohameth et al. [50], and Huang et al. [51], respectively. In addition, DFCANet achieves 99.12% accuracy in CD&S (public dataset with complex background), surpassing other classical CNN models.
Overall, the DFCANet proposed in this study is lightweight and effective for crop disease identification in complex backgrounds. Moreover, our study provided an insightful exploration of the effectiveness and robustness of data enhancement methods for crop disease datasets, which provided a reference for future crop disease data enhancement.
5. Future Work
In the future, our research will focus on the following aspects: (1) Collecting datasets and solving data imbalance problems; (2) exploring more ways to augment data for agricultural imagery; (3) changing the input size of the model; and (4) deploying models to mobile or other edge devices.
The datasets collected in the field environment are critical. However, due to the differences in seasons and regions, the collected datasets showed a long-tail distribution. In this study, we balanced the data volume of each category by artificially supplementing other data sources. In future work, we will focus on solving the problem of the long-tail distribution of data.
For agricultural images, we will explore more data augmentation approaches, such as GAN.
The input image size in this paper was 224 × 224, which can be applied to the general CNN model and lightweight model. However, such an input size ignores some information, so in the follow-up research, we will adopt a larger input size or slice the image into a path. Further, we will adapt the learnable Resizer Model to learn a more appropriate resizing method.
We also need to deploy the model on mobile phones or other edge devices for users to identify disease types.
6. Conclusions
In this study, a lightweight CNN model, called DFCANet, was designed based on the DFCA block and the DS block. DFCA blocks are used to pinpoint disease areas on corn leaves and extract subtle differences to identify and classify different diseases. DS blocks are designed to reduce the loss of disease signature information. The experimental results showed that the accuracy, precision, recall, and F1 score of the proposed model, when used in the classification of corn leaf disease images in real environments, reached 0.9847, 0.9853, 0.9853, and 0.9853, respectively. The accuracy was 1.84%, 5.20%, 1.23%, 4.19%, 2.76%, 3.06%, 2.14%, and 3.67% higher than the VGG16, ResNet50, EffcientNet-B0, Con-vNeXt-B, DenseNet121, MobileNet-V2, Mo-bileNetv3-Large, and ShuffleNetV2-1.0× methods. Furthermore, the Params and Flops of DFCANet were 1.91M and 309.1M, respectively, which are more lightweight than those of other CNN models. We validated the effectiveness of KeepAugment and simulated real-weather data augmentation approaches in crop disease identification. In summary, DFCANet has the advantage of being lightweight and efficient for corn disease identification.
Author Contributions
Conceptualization, X.Z. and Y.C.; methodology, X.Z. and X.C.; software, R.P.; validation, Y.C., X.Z., J.L. and R.P.; formal analysis, J.L. and T.C. (Tengbao Cao); investigation, J.C.; data curation, D.Y. and T.C. (Tengbao Cao); writing—original draft preparation, J.L., R.P. and T.C. (Tomislav Cernava); writing—review and editing, X.Z., T.C. (Tomislav Cernava) and X.C.; visualization, R.P.; supervision, X.Z. and X.C.; project administration, X.Z. and X.C.; funding acquisition, X.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key Research and Development Plan Key Special Projects, grant number 2021YFE0107700, National Nature Science Foundation of China, grant numbers 61865002 and 31960555, Guizhou Science and Technology Program, grant number 2019-1410, and Outstanding Young Scientist Program of Guizhou Province, grant number KY2021-026. In addition, the study received support by the Program for Introducing Talents to Chinese Universities, 111Program, grant number D20023.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Qiqi, W.; Yinjun, C. Advantages Analysis of Corn Planting in China. J. Agric. Sci. Technol. 2018, 20, 1. [Google Scholar]
- Li, X.; Dong, Y.; Huang, H.; You, H. Effects of fungicides on disease control and yield and quality of silage corn. Mod. Anim. Husb. Technol. 2020, 4, 7–9. [Google Scholar]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by corn in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Aravind, K.R.; Raja, P.; Mukesh, K.V.; Aniirudh, R.; Ashiwin, R.; Szczepanski, C. Disease Classification in Corn Crop Using Bag of Features and Multiclass Support Vector Machine. In Proceedings of the 2018 2nd International Conference on Inventive Systems and Control (ICISC), Coimbatore, India, 19–20 January 2018; pp. 1191–1196. [Google Scholar]
- Kusumo, B.S.; Heryana, A.; Mahendra, O.; Pardede, H.F. Machine Learning-Based for Automatic Detection of Corn-Plant Diseases Using Image Processing. In Proceedings of the International conference on computer, control, informatics and its applications (IC3INA), IEEE, Tangerang, Indonesia, 1–2 November 2018; pp. 93–97. [Google Scholar]
- Ahila Priyadharshini, R.; Arivazhagan, S.; Arun, M.; Mirnalini, A. Maize leaf disease classification using deep convolutional neural networks. Neural Comput. Appl. 2019, 31, 8887–8895. [Google Scholar] [CrossRef]
- Panigrahi, K.P.; Sahoo, A.K.; Das, H. A cnn approach for corn leaves disease detection to support digital agricultural system. In Proceedings of the 2020 4th International Conference on Trends in Electronics and Informatics (ICOEI) (48184), Tirunelveli, India, 15–17 June 2020; pp. 678–683. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Mishra, S.; Sachan, R.; Rajpal, D. Deep convolutional neural network based detection system for real-time corn plant disease recognition. Procedia Comput. Sci. 2020, 167, 2003–2010. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Lv, M.; Zhou, G.; He, M.; Chen, A.; Zhang, W.; Hu, Y. Corn leaf disease identification based on feature enhancement and DMS-robust alexnet. IEEE Access. 2020, 8, 57952–57966. [Google Scholar] [CrossRef]
- Zeng, W.; Li, H.; Hu, G.; Liang, D. Identification of corn leaf diseases by using the SKPSNet-50 convolutional neural network model. Sustain. Comput. Inform. Syst. 2020, 35, 100695. [Google Scholar]
- Pandey, A.; Jain, K. A robust deep attention dense convolutional neural network for plant leaf disease identification and classification from smart phone captured real world images. Ecol. Inform. 2022, 70, 101725. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, J.; Wang, S.; Shen, J.; Yang, K.; Zhou, X. Identifying Field Crop Diseases Using Transformer-Embedded Convolutional Neural Network. Agriculture 2022, 12, 1083. [Google Scholar] [CrossRef]
- Chen, W.; Chen, J.; Duan, R.; Fang, Y.; Ruan, Q.; Zhang, D. MS-DNet: A mobile neural network for plant disease identification. Comput. Electron. Agric. 2020, 199, 107175. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Identification of rice plant diseases using lightweight attention networks. Expert Syst. Appl. 2021, 169, 114514. [Google Scholar] [CrossRef]
- Yin, C.; Zeng, T.; Zhang, H.; Fu, W.; Wang, L.; Yao, S. Maize small leave spot classification based on improved deep convolutional neural network with multi-scale attention mechanism. Agronomy 2022, 12, 906. [Google Scholar] [CrossRef]
- Zeng, W.; Li, H.; Hu, G.; Liang, D. Lightweight dense-scale network (LDSNet) for corn leaf disease identification. Comput. Electron. Agri. 2022, 197, 106943. [Google Scholar] [CrossRef]
- Lin, J.; Chen, X.; Pan, R.; Cao, T.; Cai, J.; Chen, Y.; Peng, X.; Cernava, T.; Zhang, X. GrapeNet: A Lightweight Convolutional Neural Network Model for Identification of Grape Leaf Diseases. Agriculture 2022, 12, 887. [Google Scholar] [CrossRef]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft Coco: Common objects in context. In Proceedings of the European Conference on Computer Vision (ECCV), Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Pan, S.-Q.; Qiao, J.-F.; Wang, R.; Yu, H.-L.; Wang, C.; Taylor, K.; Pan, H.-Y. Intelligent diagnosis of northern corn leaf blight with deep learning model. J. Integr. Agric. 2022, 21, 1094–1105. [Google Scholar] [CrossRef]
- Richey, B.; Shirvaikar, M.V. Deep learning based real-time detection of northern corn leaf blight crop disease using YoloV4. In Proceedings of the Real-Time Image Processing and Deep Learning 2021, Electric Network, 12–16 April 2021; Volume 11736, pp. 39–45. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27, 139–144. [Google Scholar]
- Chen, J.; Wang, W.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Attention embedded lightweight network for corn disease recognition. Plant Pathol. 2021, 70, 630–642. [Google Scholar] [CrossRef]
- Albarrak, K.; Gulzar, Y.; Hamid, Y.; Mehmood, A.; Soomro, A.B. A Deep Learning-Based Model for Date Fruit Classification. Sustainability 2022, 14, 6339. [Google Scholar] [CrossRef]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A convolution neural network-based seed classification system. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Hamid, Y.; Wani, S.; Soomro, A.B.; Alwan, A.A.; Gulzar, Y. Smart seed classification system based on MobileNetV2 architecture. In Proceedings of the 2022 2nd International Conference on Computing and Information Technology (ICCIT), Tabuk, Saudi Arabia, 25–27 January 2022; pp. 217–222. [Google Scholar]
- Ahmad, A.; Saraswat, D.; Gamal, A.E.; Johal, G. CD&S Dataset: Handheld Imagery Dataset Acquired Under Field Conditions for Corn Disease Identification and Severity Estimation. arXiv 2021, arXiv:2110.12084. [Google Scholar]
- Singh, D.; Jain, N.; Jain, P.; Kayal, P.; Kumawat, S.; Batra, N. PlantDoc: A dataset for visual plant disease detection. In Proceedings of the 7th ACM IKDD CoDS and 25th COMAD, ACM KDD, Hyderabad, India, 5–7 January 2020; pp. 249–253. [Google Scholar]
- Gong, C.; Wang, D.; Li, M.; Chandra, V.; Liu, Q. Keepaugment: A simple information-preserving data augmentation approach. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Electr Network, 19–25 June 2021; pp. 1055–1064. [Google Scholar]
- DeVries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random erasing data augmentation. Proc. AAAI Conf. Artif. Intell. 2020, 34, 13001–13008. [Google Scholar] [CrossRef]
- Chen, P.; Liu, S.; Zhao, H.; Jia, J. Gridmask data augmentation. arXiv 2020, arXiv:2001.04086. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate Attention for Efficient Mobile Network Design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Electr Network, 19–25 June 2021; pp. 13713–13722. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A Convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 21–24 June 2022; pp. 11976–11986. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning (ICML) PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for Mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. Shufflenet v2: Practical guidelines for efficient Cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-And-Excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-Cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Mohameth, F.; Chen, B.C.; Kane, A.S. Plant disease detection with deep learning and feature extraction using plant village. J. Computer. Commun. 2020, 8, 10–22. [Google Scholar] [CrossRef]
- Huang, J.P.; Chen, J.; Li, K.X.; Li, J.Y.; Liu, H. Identification of multiple plant leaf diseases using neural architecture search. Trans. Chin. Soc. Agric. Eng. 2020, 36, 166–173. [Google Scholar]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Subetha, T.; Khilar, R.; Christo, M.S. A comparative analysis on plant pathology classification using deep learning architecture–Resnet and VGG19. Mater. Today Proc. 2021, in press. [Google Scholar]
- Zhao, S.; Peng, Y.; Liu, J.; Wu, S. Tomato Leaf Disease Diagnosis Based on Improved Convolution Neural Network by Attention Module. Agriculture 2021, 11, 651. [Google Scholar] [CrossRef]
- Lin, J.; Chen, Y.; Pan, R.; Cao, T.; Cai, J.; Yu, D.; Chi, X.; Cernava, T.; Zhang, X.; Chen, X. CAMFFNet: A novel convolutional neural network model for tobacco disease image recognition. Comput. Electron. Agric. 2022, 202, 107390. [Google Scholar] [CrossRef]
- Gao, R.; Wang, R.; Feng, L.; Li, Q.; Wu, H. Dual-branch, efficient, channel attention-based crop disease identification. Comput. Electron. Agric. 2021, 190, 106410. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).