Embryonic Signaling Pathways Shape Colorectal Cancer Subtypes: Linking Gut Development to Tumor Biology
Abstract
1. Introduction
2. Embryologic Development
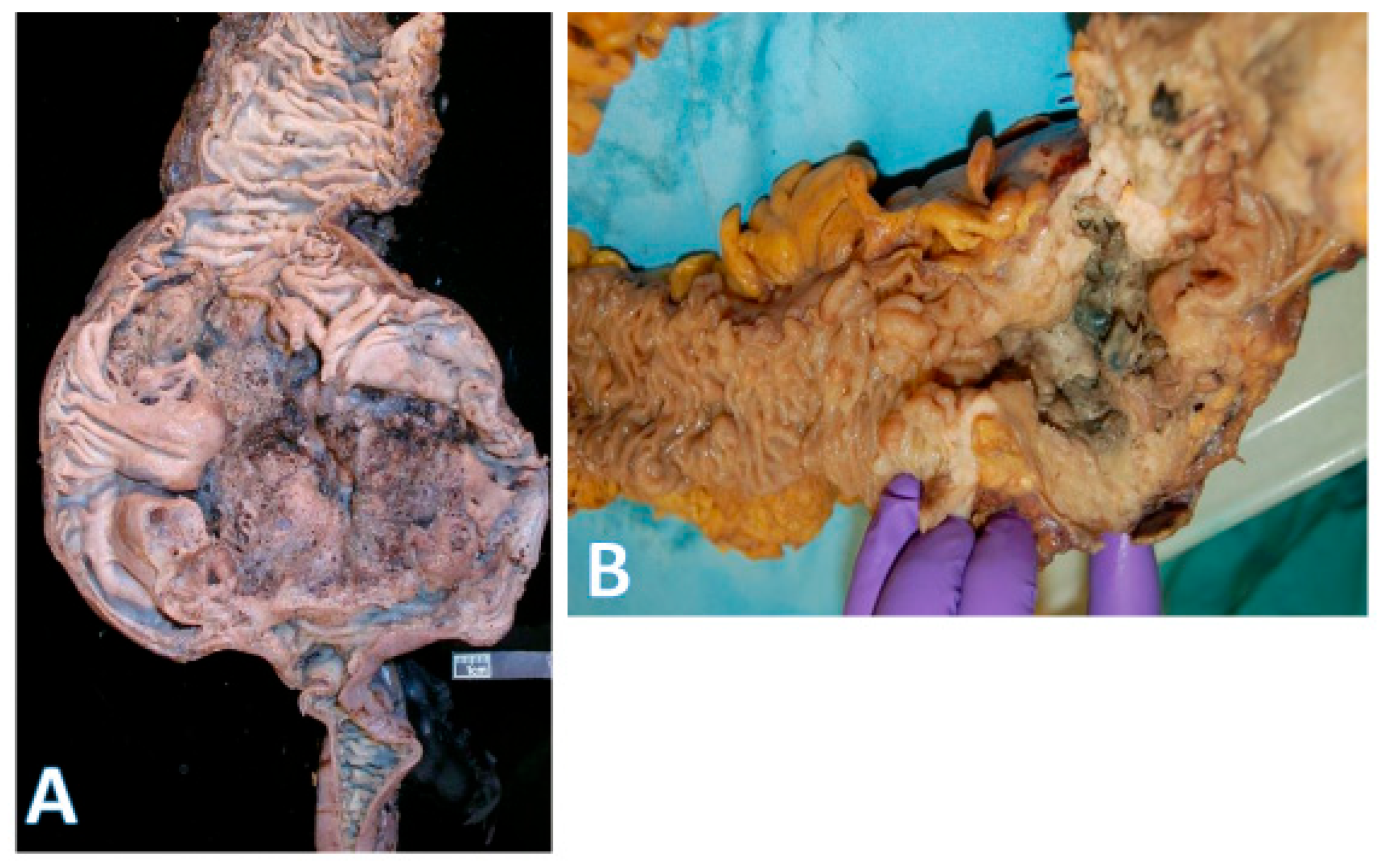
3. Right-Versus Left-Sided Cancers
4. Comparisons of Epithelial Cancers of the Midgut
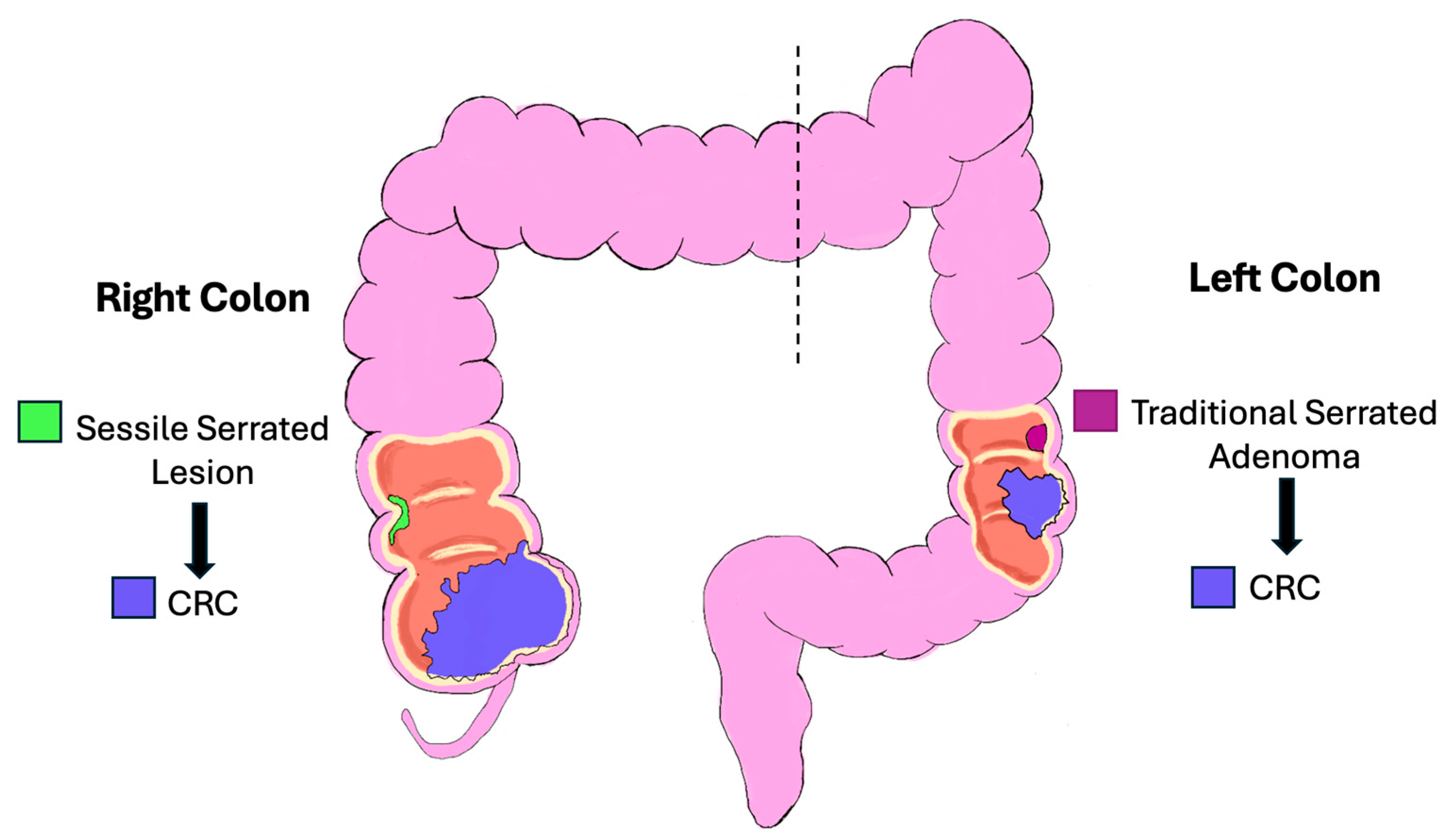
5. Precursor Lesions for CRC
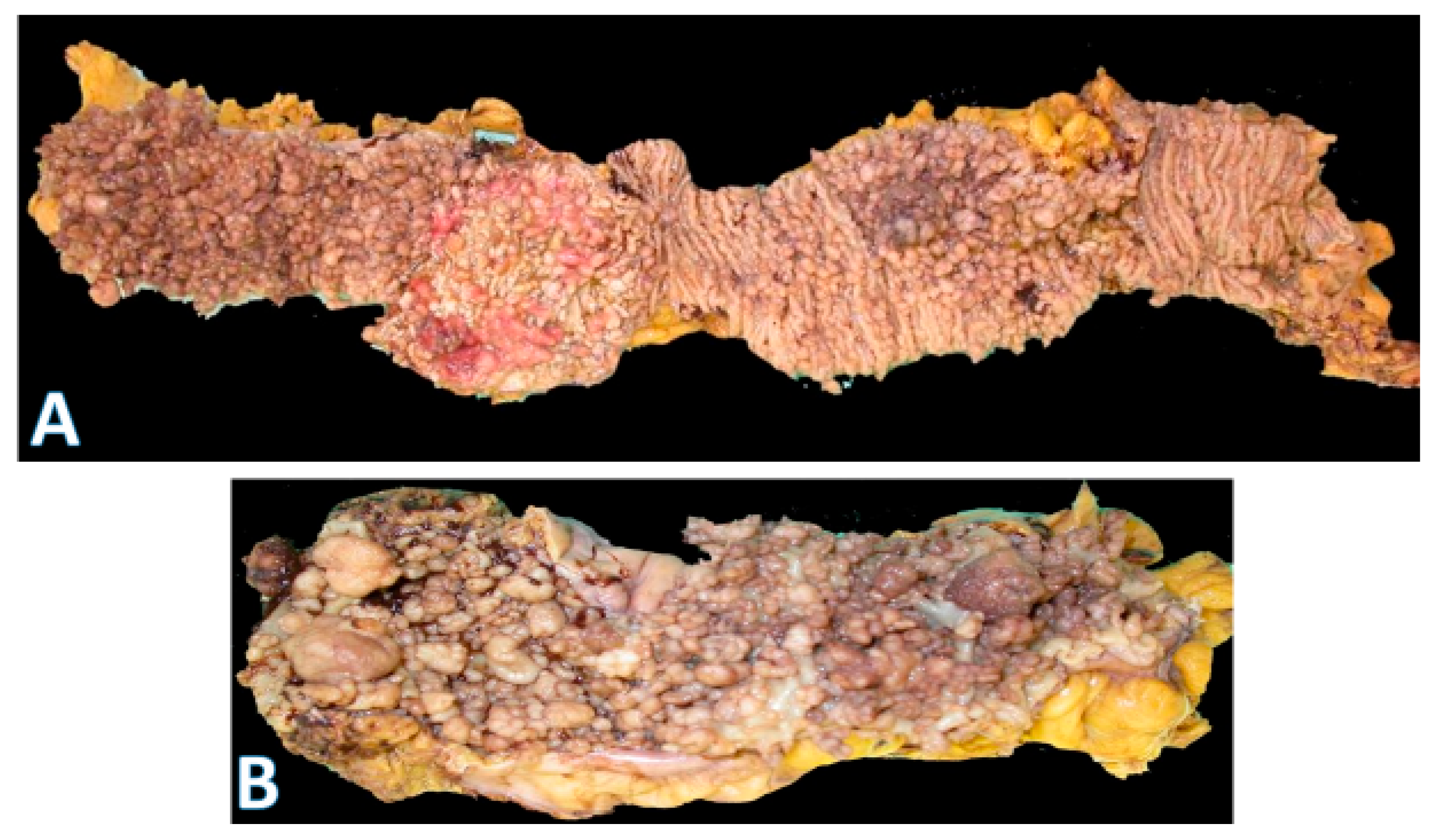
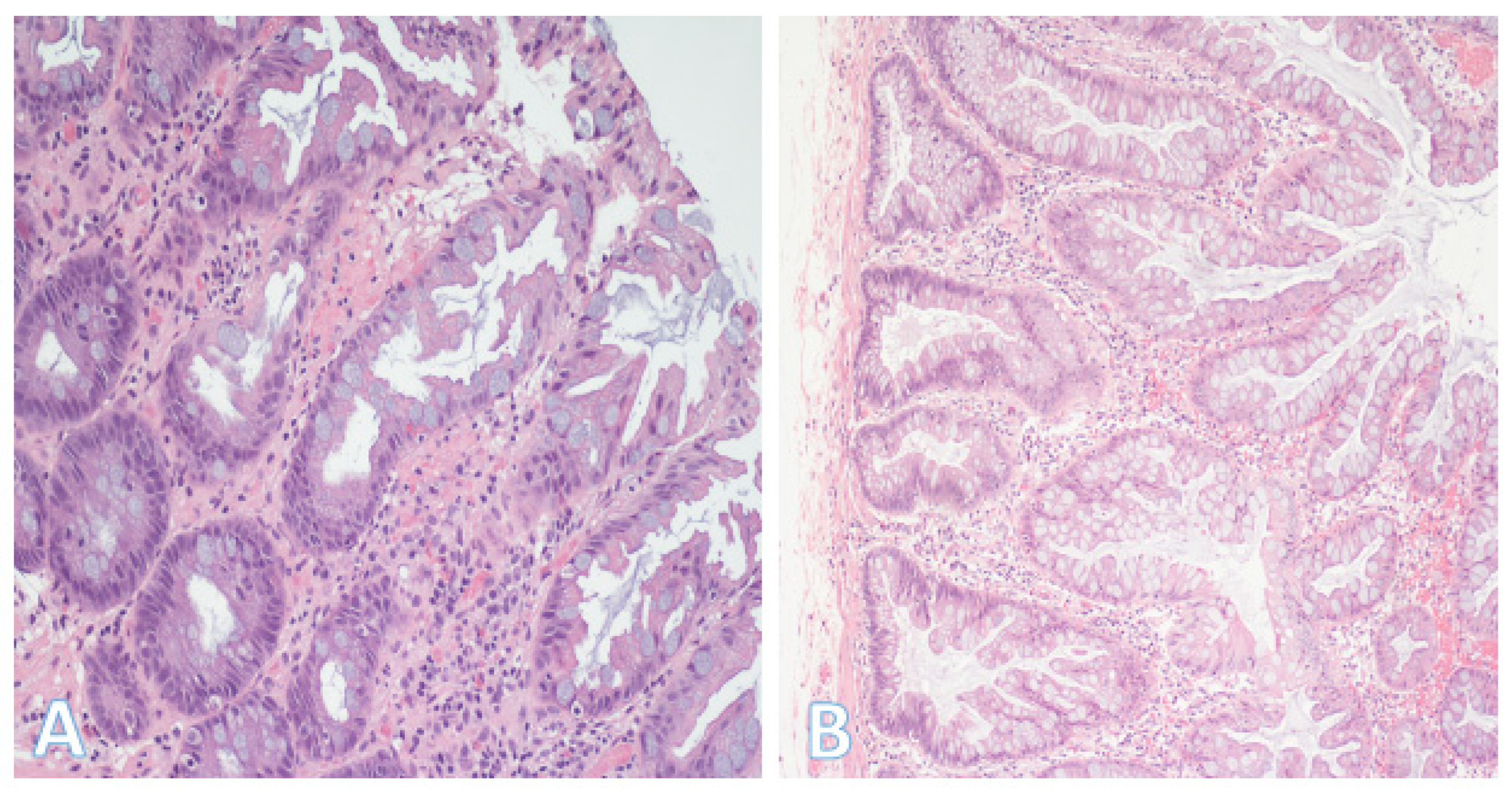
5.1. Conventional Adenomas
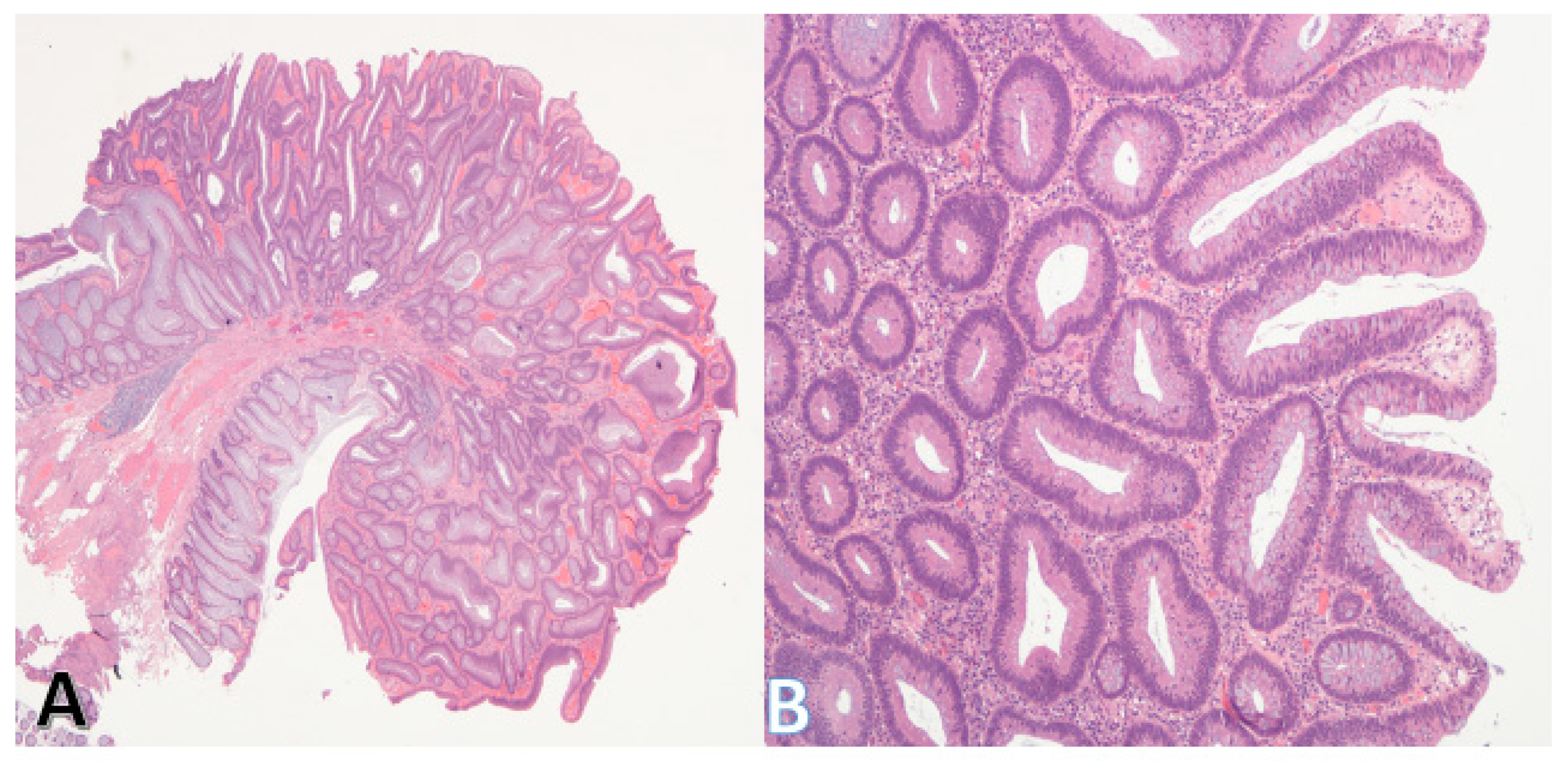
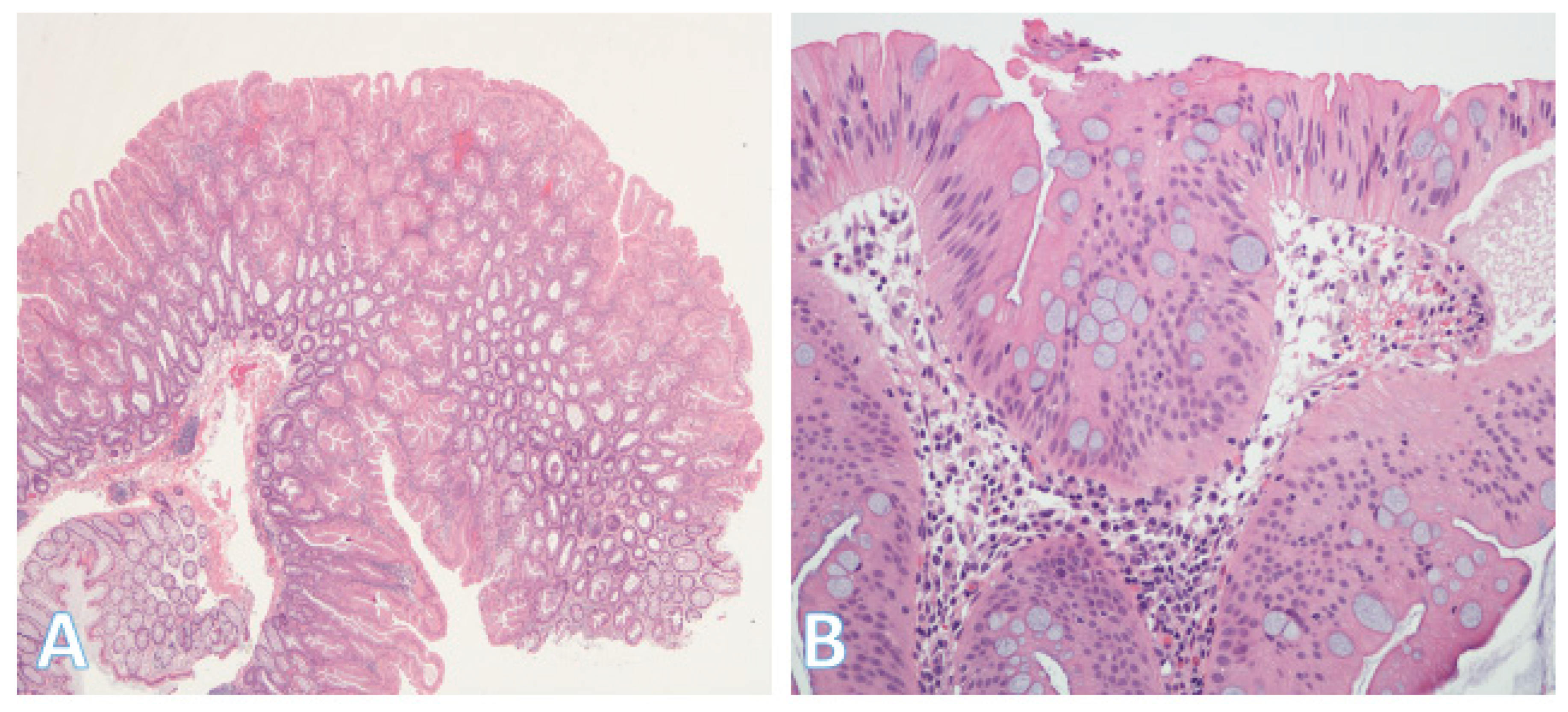
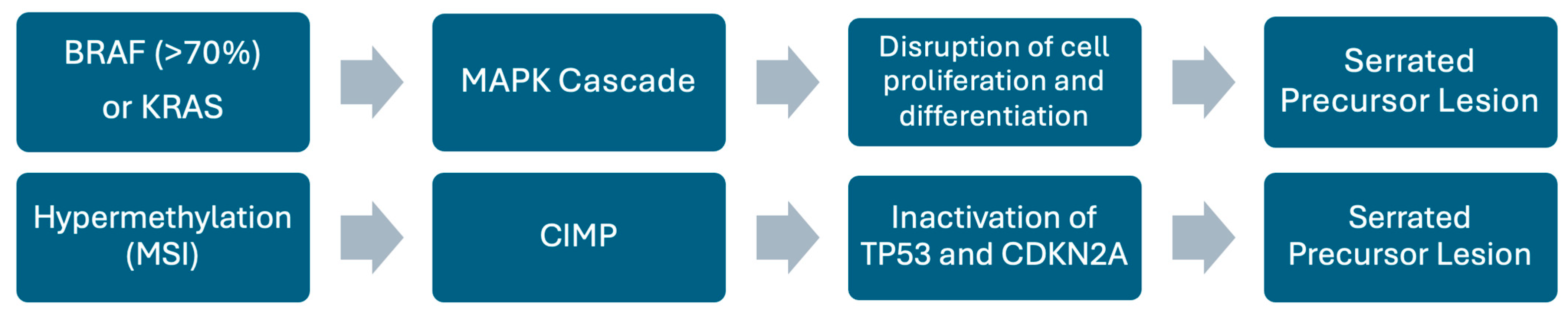
5.2. Serrated Pathway
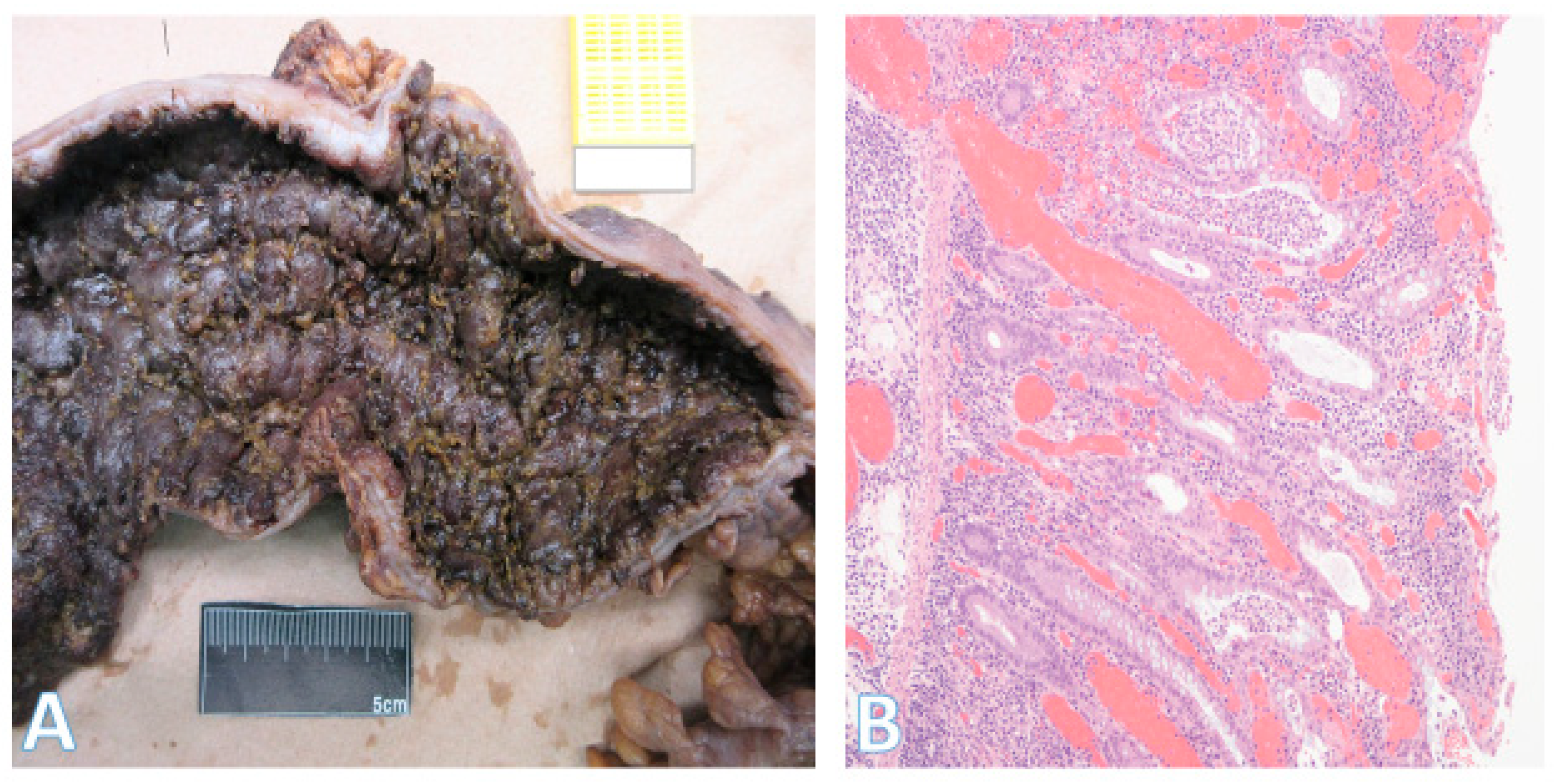
5.3. Inflammatory Bowel Disease-Associated Dysplasia
6. Histologic Subtypes of CRC
7. Consensus Molecular Subtypes (CMS)
7.1. Prevalence of CMSs
7.2. Limitations of CMS
7.2.1. Overlap
7.2.2. Treatment and Drug Resistance
7.2.3. Cost/Benefit Analysis
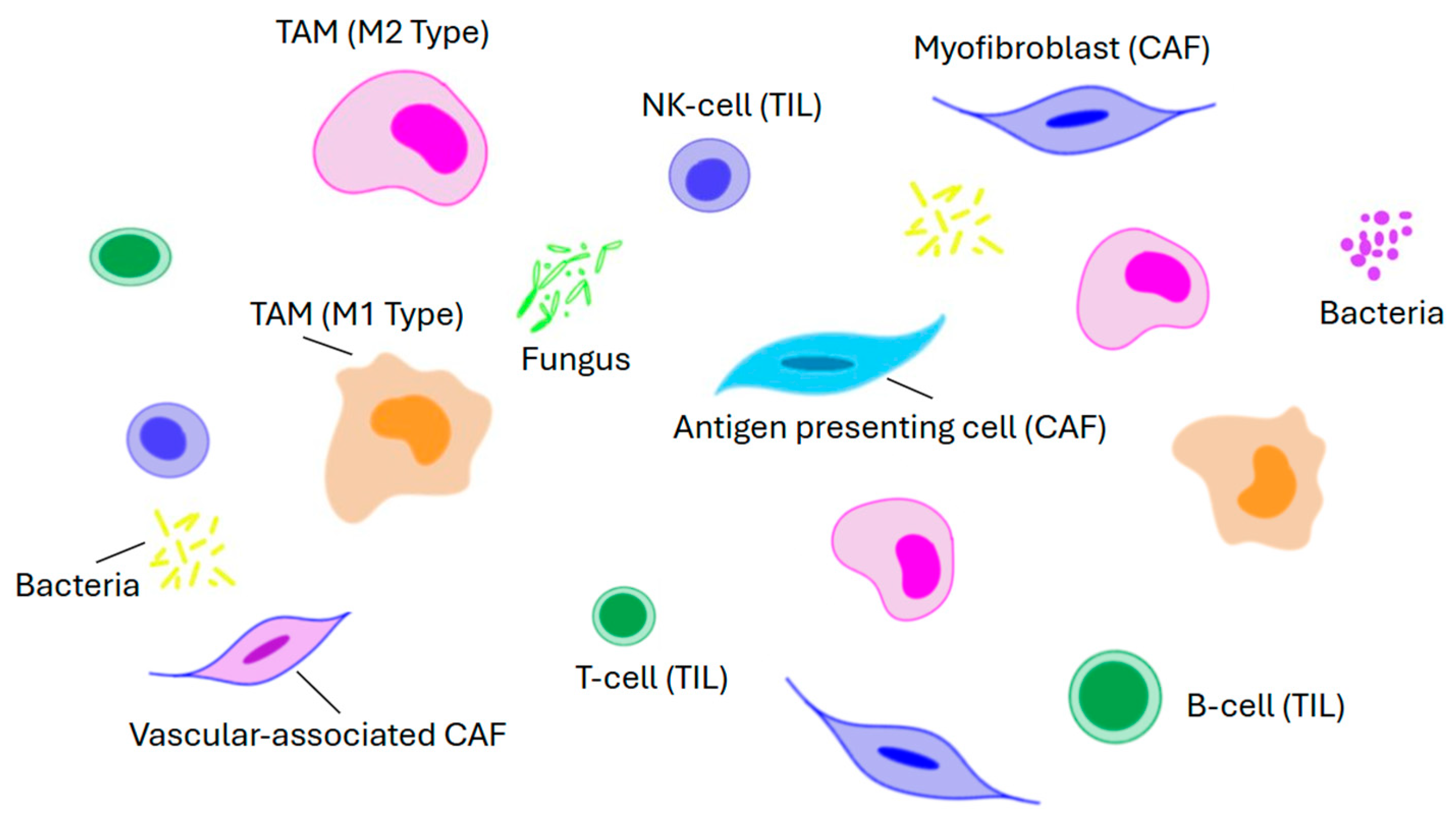
8. The Tumor Micro-Environment
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CIMP | CpG island methylator phenotype |
| CMS | Consensus Molecular Subtype |
| CRC | Colorectal Carcinoma |
| D3/D4 | Third/Fourth Part of the Duodenum |
| EMT | Epithelial Mesenchymal Transition |
| FAP | Familial Adenomatous Polyposis |
| GI | Gastrointestinal |
| IBD | Inflammatory Bowel Disease |
| JI | Jejunoileal |
| LAMN | Low-grade Appendiceal Mucinous Neoplasm |
| MSI | Microsatellite Instability |
| NET | Neuroendocrine Tumor |
| NOS | Not Otherwise Specified |
| SSL | Sessile Serrated Lesion |
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal cancer: Epidemiology, risk factors, and prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; O’Connell, K.; Jeon, J.; Song, M.; Hunter, D.; Hoffmeister, M.; Lin, Y.; Berndt, S.; Brenner, H.; Chan, A.T.; et al. Combined effect of modifiable and non-modifiable risk factors for colorectal cancer risk in a pooled analysis of 11 population-based studies. BMJ Open Gastroenterol. 2019, 6, e000339. [Google Scholar] [CrossRef] [PubMed]
- Filho, A.M.; Laversanne, M.; Ferlay, J.; Colombet, M.; Piñeros, M.; Znaor, A.; Parkin, D.M.; Soerjomataram, I.; Bray, F. The GLOBOCAN 2022 cancer estimates: Data sources, methods, and a snapshot of the cancer burden worldwide. Int. J. Cancer 2025, 156, 1336–1346. [Google Scholar] [CrossRef]
- Wang, W.; Kandimalla, R.; Huang, H.; Zhu, L.; Li, Y.; Gao, F.; Goel, A.; Wang, X. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin. Cancer Biol. 2019, 55, 37–52. [Google Scholar] [CrossRef]
- Bufill, J.A. Colorectal cancer: Evidence for distinct genetic categories based on proximal or distal tumor location. Ann. Intern. Med. 1990, 113, 779–788. [Google Scholar] [CrossRef]
- Salem, M.E.; Weinberg, B.A.; Xiu, J.; El-Deiry, W.S.; Hwang, J.J.; Gatalica, Z.; Philip, P.A.; Shields, A.F.; Lenz, H.J.; Marshall, J.L. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget 2017, 8, 86356–86368. [Google Scholar] [CrossRef]
- Kalantzis, I.; Nonni, A.; Pavlakis, K.; Delicha, E.M.; Miltiadou, K.; Kosmas, C.; Ziras, N.; Gkoumas, K.; Gakiopoulou, H. Clinicopathological differences and correlations between right and left colon cancer. World J. Clin. Cases 2020, 8, 1424–1443. [Google Scholar] [CrossRef]
- Noah, T.K.; Donahue, B.; Shroyer, N.F. Intestinal development and differentiation. Exp. Cell Res. 2011, 317, 2702–2710. [Google Scholar] [CrossRef]
- Kolev, H.M.; Kaestner, K.H. Mammalian intestinal development and differentiation—The state of the art. Cell. Mol. Gastroenterol. Hepatol. 2023, 16, 809–821. [Google Scholar] [CrossRef]
- Roberts, D.J. Molecular mechanisms of development of the gastrointestinal tract. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2000, 219, 109–120. [Google Scholar] [CrossRef]
- Smith, R.J.; Zhang, H.; Hu, S.S.; Yung, T.; Francis, R.; Lee, L.; Onaitis, M.W.; Dirks, P.B.; Zang, C.; Kim, T.H. Single-cell chromatin profiling of the primitive gut tube reveals regulatory dynamics underlying lineage fate decisions. Nat. Commun. 2022, 13, 2965. [Google Scholar] [CrossRef]
- Kiefer, J.C. Molecular mechanisms of early gut organogenesis: A primer on development of the digestive tract. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2003, 228, 287–291. [Google Scholar] [CrossRef]
- Sylvestre, M.; Di Carlo, S.E.; Peduto, L. Stromal regulation of the intestinal barrier. Mucosal Immunol. 2023, 16, 221–231. [Google Scholar] [CrossRef]
- Gasnier, M.; Lim, H.Y.; Barker, N. Role of Wnt signaling in the maintenance and regeneration of the intestinal epithelium. Curr. Top. Dev. Biol. 2023, 153, 281–326. [Google Scholar]
- Burclaff, J. Transcriptional regulation of metabolism in the intestinal epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 2023, 325, G501–G507. [Google Scholar] [CrossRef]
- Zhao, L.; Song, W.; Chen, Y.G. Mesenchymal-epithelial interaction regulates gastrointestinal tract development in mouse embryos. Cell Rep. 2022, 40, 111053. [Google Scholar] [CrossRef]
- Fink, M.; Njah, K.; Patel, S.J.; Cook, D.P.; Man, V.; Ruso, F.; Rajan, A.; Narimatsu, M.; Obersterescu, A.; Pye, M.J.; et al. Chromatin remodelling in damaged intestinal crypts orchestrates redundant TGFβ and Hippo signalling to drive regeneration. Nat. Cell Biol. 2024, 26, 2084–2098. [Google Scholar] [CrossRef] [PubMed]
- Kazakevych, J.; Sayols, S.; Messner, B.; Krienke, C.; Soshnikova, N. Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 2017, 45, 5770–5784. [Google Scholar] [CrossRef] [PubMed]
- Aiderus, A.; Barker, N.; Tergaonkar, V. Serrated colorectal cancer: Preclinical models and molecular pathways. Trends Cancer 2024, 10, 76–91. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.J.; Zhao, Q.; Yang, S. Colorectal serrated pathway cancers and precursors. Histopathology 2015, 66, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Najdi, R.; Holcombe, R.F.; Waterman, M.L. Wnt signaling and colon carcinogenesis: Beyond APC. J. Carcinog. 2011, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Menter, D.G.; Kopetz, S. Right versus left colon cancer biology: Integrating the consensus molecular subtypes. J. Natl. Compr. Cancer Netw. 2017, 15, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zeng, J.H.; Qin, X.G.; Chen, J.Q.; Luo, D.Z.; Chen, G. Distinguishable prognostic signatures of left-and right-sided colon cancer: A study based on sequencing data. Cell. Physiol. Biochem. 2018, 48, 475–490. [Google Scholar] [CrossRef]
- Abdel Hamid, M.; Pammer, L.M.; Oberparleiter, S.; Günther, M.; Amann, A.; Gruber, R.A.; Mair, A.; Nocera, F.I.; Ormanns, S.; Zimmer, K.; et al. Multidimensional differences of right-and left-sided colorectal cancer and their impact on targeted therapies. npj Precis. Oncol. 2025, 9, 116. [Google Scholar] [CrossRef]
- Vlachou, E.; Koffas, A.; Toumpanakis, C.; Keuchel, M. Updates in the diagnosis and management of small-bowel tumors. Best. Pract. Res. Clin. Gastroenterol. 2023, 64–65, 101860. [Google Scholar] [CrossRef]
- Puccini, A.; Battaglin, F.; Lenz, H.J. Management of advanced small bowel cancer. Curr. Treat. Options Oncol. 2018, 19, 69. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Overman, M.J.; Hu, C.Y.; Kopetz, S.; Abbruzzese, J.L.; Wolff, R.A.; Chang, G.J. A population-based comparison of adenocarcinoma of the large and small intestine: Insights into a rare disease. Ann. Surg. Oncol. 2012, 19, 1439–1445. [Google Scholar] [CrossRef]
- Pedersen, K.S.; Raghav, K.; Overman, M.J. Small bowel adenocarcinoma: Etiology, presentation, and molecular alterations. J. Natl. Compr. Cancer Netw. 2019, 17, 1135–1141. [Google Scholar] [CrossRef]
- Barsouk, A.; Rawla, P.; Barsouk, A.; Thandra, K.C. Epidemiology of cancers of the small intestine: Trends, risk factors, and prevention. Med. Sci. 2019, 17, 46. [Google Scholar] [CrossRef]
- Moyana, T.N.; Satkunam, N. A comparative immunohistochemical study of jejunoileal and appendiceal carcinoids. Implications for histogenesis and pathogenesis. Cancer 1992, 70, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Ranot, J.M.; Hamid, J.S.; Montazeri, A.; Harper, K.; McCudden, C.; Moyana, T.N. Well-Differentiated Jejunoileal Neuroendocrine Tumors and Corresponding Liver Metastases: Mesenteric Fibrogenesis and Extramural Vascular Invasion in Tumor Progression. Cancers 2025, 17, 1486. [Google Scholar] [CrossRef] [PubMed]
- Nagtegaal, I.D.; Arends, M.J.; Odze, R.D. Tumours of the colon and rectum. In World Health Organization (WHO) Classification of Tumours: Digestive System Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2020; Volume 4. [Google Scholar]
- Zhu, Y.; Li, X. Advances of Wnt signalling pathway in colorectal cancer. Cells 2023, 12, 447. [Google Scholar] [CrossRef] [PubMed]
- Voloshanenko, O.; Erdmann, G.; Dubash, T.D.; Augustin, I.; Metzig, M.; Moffa, G.; Hundsrucker, C.; Kerr, G.; Sandmann, T.; Anchang, B.; et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat. Commun. 2013, 4, 2610. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Antas, P.; Li, V.S. Targeting Wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am. J. Physiol. Cell Physiol. 2015, 309, C511–C521. [Google Scholar] [CrossRef]
- Mezzapesa, M.; Losurdo, G.; Celiberto, F.; Rizzi, S.; d’Amati, A.; Piscitelli, D.; Ierardi, E.; Di Leo, A. Serrated colorectal lesions: An up-to-date review from histological pattern to molecular pathogenesis. Int. J. Mol. Sci. 2022, 23, 4461. [Google Scholar] [CrossRef]
- De Palma, F.D.; D’argenio, V.; Pol, J.; Kroemer, G.; Maiuri, M.C.; Salvatore, F. The molecular hallmarks of the serrated pathway in colorectal cancer. Cancers 2019, 11, 1017. [Google Scholar] [CrossRef]
- Wang, J.D.; Xu, G.S.; Hu, X.L.; Li, W.Q.; Yao, N.; Han, F.Z.; Zhang, Y.; Qu, J. The histologic features, molecular features, detection and management of serrated polyps: A review. Front. Oncol. 2024, 14, 1356250. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kitamoto, S.; Kamada, N. Inflammatory bowel disease and carcinogenesis. Cancer Metastasis Rev. 2022, 41, 301–316. [Google Scholar] [CrossRef]
- Yin, Y.; Wan, J.; Yu, J.; Wu, K. Molecular pathogenesis of colitis-associated colorectal cancer: Immunity, genetics, and intestinal microecology. Inflamm. Bowel Dis. 2023, 29, 1648–1657. [Google Scholar] [CrossRef]
- Zhou, R.W.; Harpaz, N.; Itzkowitz, S.H.; Parsons, R.E. Molecular mechanisms in colitis-associated colorectal cancer. Oncogenesis 2023, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Itzkowitz, S.H. Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology 2022, 162, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Chatila, W.K.; Walch, H.; Hechtman, J.F.; Moyer, S.M.; Sgambati, V.; Faleck, D.M.; Srivastava, A.; Tang, L.; Benhamida, J.; Ismailgeci, D.; et al. Integrated clinical and genomic analysis identifies driver events and molecular evolution of colitis-associated cancers. Nat. Commun. 2023, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Riddell, R.H.; Petras, R.E.; Williams, G.T. Atlas of Tumor Pathology, Fascicle 32: Tumors of the Intestines; Armed Forces Institute of Pathology: Washington, DC, USA, 2002; p. 327. [Google Scholar]
- Jass, J. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef]
- Auld, F.M.; Moyana, T.N. Invasive stratified mucin-producing carcinoma of the colorectum: Expanding the morphologic spectrum of large bowel cancer. Diagn. Pathol. 2023, 18, 113. [Google Scholar] [CrossRef]
- Palvio, D.H.; Sørensen, F.B.; Kløve-Mogensen, M. Stem cell carcinoma of the colon and rectum: Report of two cases and review of the literature. Dis. Colon. Rectum 1985, 28, 440–445. [Google Scholar] [CrossRef]
- Lipka, S.; Hurtado-Cordovi, J.; Avezbakiyev, B.; Freedman, L.; Clark, T.; Rizvon, K.; Mustacchia, P. Synchronous small cell neuroendocrine carcinoma and adenocarcinoma of the colon: A link for common stem cell origin? ACG Case Rep. J. 2014, 1, 96–99. [Google Scholar] [CrossRef]
- Onishi, R.; Sano, T.; Nakamura, Y.; Ihara, C.; Shimatsu, A.; Namiuchi, S.; Sawada, S. Ectopic adrenocorticotropin syndrome associated with undifferentiated carcinoma of the colon showing multidirectional neuroendocrine, exocrine, and squamous differentiation. Virchows Arch. 1996, 427, 537–541. [Google Scholar] [CrossRef]
- Petrelli, M.; Tetangco, E.; Reid, J.D. Carcinoma of the colon with undifferentiated, carcinoid, and squamous cell features. Am. J. Clin. Pathol. 1981, 75, 581–584. [Google Scholar] [CrossRef]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Göktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Miyagi, Y.; Sekiyama, A.; Yoshida, S.; Nakao, S.; Okamoto, N.; Koganei, K.; Sugita, A. Colorectal small cell carcinoma in ulcerative colitis with identical rare p53 gene mutation to associated adenocarcinoma and dysplasia. J. Crohn’s Colitis 2012, 6, 112–115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mouillet-Richard, S.; Cazelles, A.; Sroussi, M.; Gallois, C.; Taieb, J.; Laurent-Puig, P. Clinical challenges of consensus molecular subtype CMS4 colon cancer in the era of precision medicine. Clin. Cancer Res. 2024, 30, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Torang, A.; van de Weerd, S.; Lammers, V.; van Hooff, S.; van den Berg, I.; van den Bergh, S.; Koopman, M.; IJzermans, J.N.; Roodhart, J.M.; Koster, J.; et al. NanoCMSer: A consensus molecular subtype stratification tool for fresh-frozen and paraffin-embedded colorectal cancer samples. Mol. Oncol. 2025, 19, 1332–1346. [Google Scholar] [CrossRef]
- Ten Hoorn, S.; de Back, T.R.; Sommeijer, D.W.; Vermeulen, L. Clinical value of consensus molecular subtypes in colorectal cancer: A systematic review and meta-analysis. JNCI J. Natl. Cancer Inst. 2022, 114, 503–516. [Google Scholar] [CrossRef]
- Valdeolivas, A.; Amberg, B.; Giroud, N.; Richardson, M.; Gálvez, E.J.; Badillo, S.; Julien-Laferrière, A.; Túrós, D.; Voith von Voithenberg, L.; Wells, I.; et al. Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics. npj Precis. Oncol. 2024, 8, 10. [Google Scholar] [CrossRef]
- Trinh, A.; Lädrach, C.; Dawson, H.E.; Ten Hoorn, S.; Kuppen, P.J.; Reimers, M.S.; Koopman, M.; Punt, C.J.; Lugli, A.; Vermeulen, L.; et al. Tumour budding is associated with the mesenchymal colon cancer subtype and RAS/RAF mutations: A study of 1320 colorectal cancers with Consensus Molecular Subgroup (CMS) data. Br. J. Cancer 2018, 119, 1244–1251. [Google Scholar] [CrossRef]
- Rejali, L.; Seifollahi Asl, R.; Sanjabi, F.; Fatemi, N.; Asadzadeh Aghdaei, H.; Saeedi Niasar, M.; Ketabi Moghadam, P.; Nazemalhosseini Mojarad, E.; Mini, E.; Nobili, S. Principles of molecular utility for CMS classification in colorectal cancer management. Cancers 2023, 15, 2746. [Google Scholar] [CrossRef]
- Yamauchi, M.; Lochhead, P.; Morikawa, T.; Huttenhower, C.; Chan, A.T.; Giovannucci, E.; Fuchs, C.; Ogino, S. Colorectal cancer: A tale of two sides or a continuum? Gut 2012, 61, 794–797. [Google Scholar] [CrossRef]
- Han, T.; Goswami, S.; Hu, Y.; Tang, F.; Zafra, M.P.; Murphy, C.; Cao, Z.; Poirier, J.T.; Khurana, E.; Elemento, O.; et al. Lineage reversion drives WNT independence in intestinal cancer. Cancer Discov. 2020, 10, 1590–1609. [Google Scholar] [CrossRef]
- He, X.; Xie, T.; Shi, L.; Kuang, X.; Li, L.; Shang, X.; Fu, B. Research hotspots and frontiers in the tumor microenvironment of colorectal cancer: A bibliometric study from 2014 to 2024. Front. Oncol. 2025, 15, 1525280. [Google Scholar] [CrossRef]
- Xun, J.; Hu, Z.; Wang, M.; Jiang, X.; Liu, B.; Han, Y.; Gao, R.; Wu, X.; Zhang, A.; Yang, S.; et al. Hydroxygenkwanin suppresses peritoneal metastasis in colorectal cancer by modulating tumor-associated macrophages polarization. Chem. Biol. Interact. 2024, 396, 111038. [Google Scholar] [CrossRef]
- Li, Y.; Liao, W.; Huang, W.; Liu, F.; Ma, L.; Qian, X. Mechanism of gambogic acid repressing invasion and metastasis of colorectal cancer by regulating macrophage polarization via tumor cell-derived extracellular vesicle-shuttled miR-21. Drug Dev. Res. 2024, 85, e22141. [Google Scholar] [CrossRef]
- Qayoom, H.; Sofi, S.; Mir, M.A. Targeting tumor microenvironment using tumor-infiltrating lymphocytes as therapeutics against tumorigenesis. Immunol. Res. 2023, 71, 588–599. [Google Scholar] [CrossRef]
- Liu, C.; Zou, H.; Ruan, Y.; Fang, L.; Wang, B.; Cui, L.; Wu, T.; Chen, Z.; Dang, T.; Lan, Y.; et al. Multiomics reveals the immunologic features and the immune checkpoint blockade potential of colorectal medullary carcinoma. Clin. Cancer Res. 2025, 31, 773–786. [Google Scholar] [CrossRef]
- Ding, K.; Mou, P.; Wang, Z.; Liu, S.; Liu, J.; Lu, H.; Yu, G. The next bastion to be conquered in immunotherapy: Microsatellite stable colorectal cancer. Front. Immunol. 2023, 14, 1298524. [Google Scholar] [CrossRef]
- Zhang, R.; Lan, S.; Jia, M.; Liu, F.; Wang, M.; Jin, Q.; Su, L.; Liu, G. Theranostic Applications of Taurine-Derived Carbon Dots in Colorectal Cancer: Ferroptosis Induction and Multifaceted Antitumor Mechanisms. Int. J. Nanomed. 2025, 20, 7613–7635. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Wang, S.; Ding, J.; Qu, W.; Zhang, C.; Luan, F.; Wang, N.; Hou, Y.; Suo, M.; Liu, H.; et al. The allogenic non-proteinogenic amino acid BMAA-based vaccine breaks up the immune tolerance against colorectal cancer. Theranostics 2025, 15, 3143–3158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, X.; Jin, H. Oxidized Low-Density Lipoprotein as a Potential Target for Enhancing Immune Checkpoint Inhibitor Therapy in Microsatellite-Stable Colorectal Cancer. Antioxidants 2025, 14, 726. [Google Scholar] [CrossRef] [PubMed]
- Pandey, H.; Tang, D.W.; Wong, S.H.; Lal, D. Gut microbiota in colorectal cancer: Biological role and therapeutic opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar]
- García Menéndez, G.; Sichel, L.; López, M.D.; Hernández, Y.; Arteaga, E.; Rodríguez, M.; Fleites, V.; Fernández, L.T.; Cano, R.D. From colon wall to tumor niche: Unraveling the microbiome’s role in colorectal cancer progression. PLoS ONE 2024, 19, e0311233. [Google Scholar] [CrossRef]
- Yadav, D.; Sainatham, C.; Filippov, E.; Kanagala, S.G.; Ishaq, S.M.; Jayakrishnan, T. Gut microbiome–colorectal cancer relationship. Microorganisms 2024, 12, 484. [Google Scholar] [CrossRef]



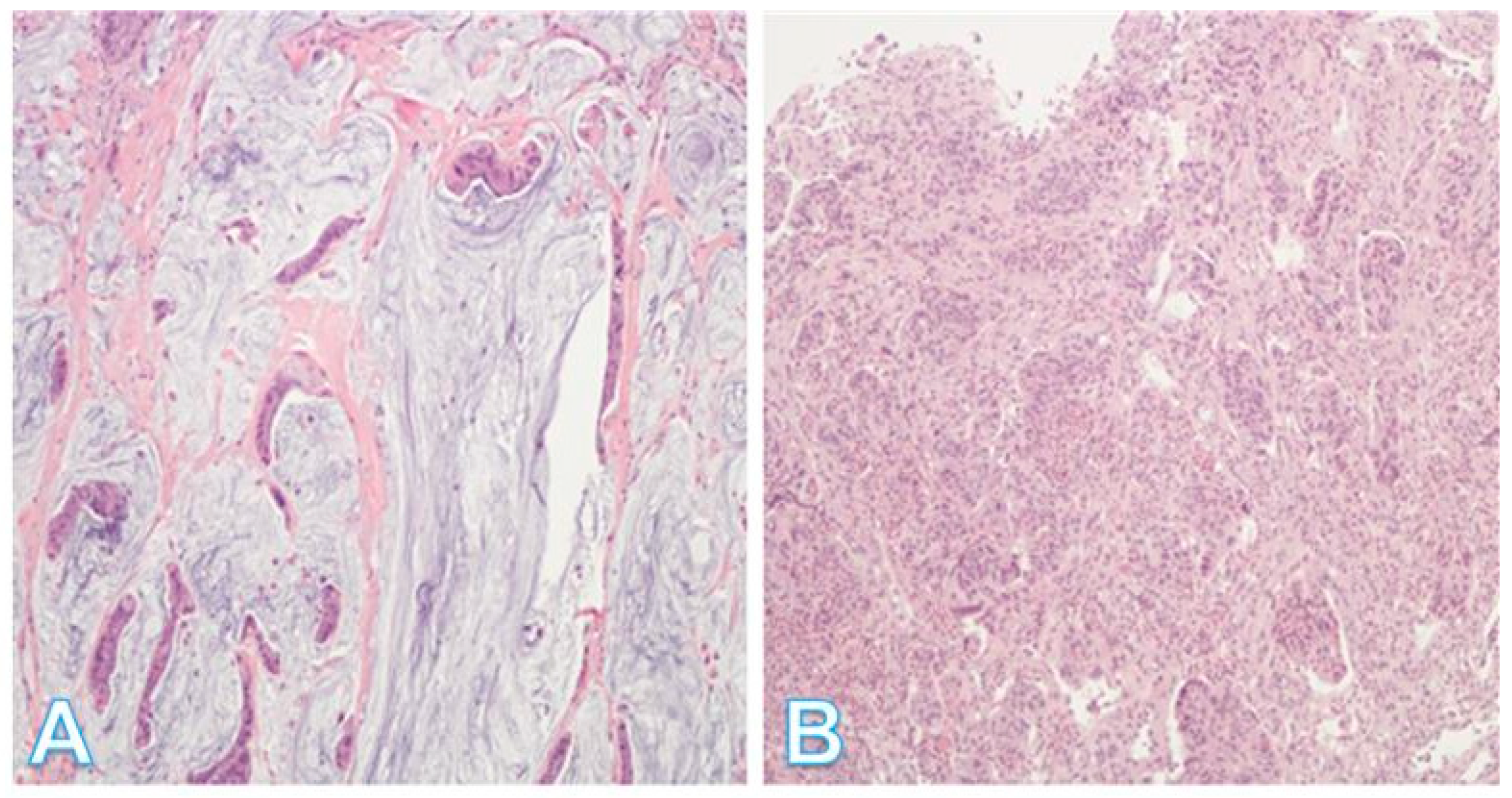

| Process/Region | Key Signals and Regulators | General Functional Role | General Clinical/Tumor Biology Relevance |
|---|---|---|---|
| Germ layer contributions | Endoderm, mesoderm, ectoderm | Establish formation of gut epithelium, mesenchyme, and enteric nervous system | Early developmental programs may be reactivated in cancer biology |
| Regional identity (foregut, midgut, hindgut) | WNT, FGF, CDX transcription factors | Pattern craniocaudal axis and segmental identity | Regional differences in signaling may contribute to site-specific CRC biology |
| Ileocecal/appendix formation | Morphogenetic processes (herniation, rotation, cecal budding) | Shape midgut structures including cecum and appendix | Developmental variations may underlie anatomic and biological differences in tumors |
| Crypt–villus axis formation | Hedgehog, BMP, PDGFR-α, Sox9 | Guide villus emergence and crypt–villus organization | Maintenance of epithelial turnover; dysregulation associated with polyp formation |
| Crypt stem cell niche | Notch, WNT, BMP | Balance self-renewal and differentiation of intestinal stem cells | Disturbances predispose to adenomas and other precursor lesions |
| Posterior axis patterning | WNT, FGF gradients | Promote distal intestinal identity | May influence molecular subtypes of distal CRC |
| Hippo pathway | YAP, TAZ | Regulate growth, polarity, and stem cell behavior | Overactivation associated with aggressive CRC features |
| Lineage plasticity and regeneration | Mesenchymal–epithelial signals; TGF-β; stem cell factors | Allow cell fate flexibility and epithelial regeneration after injury | Reactivation of plasticity pathways implicated in progression and therapy resistance |
| Precursor Lesion | Morphology/Pathology | Key Molecular Events | Clinical Notes/Risk |
|---|---|---|---|
| Adenomas (conventional pathway) | Tubular, tubulovillous, or villous architecture; progressive dysplasia | Early: APC inactivation (60–80%) → β-catenin accumulation, Wnt pathway activation; Later: KRAS, SMAD4, PTEN, PIK3CA, TP53 mutations; neo-angiogenesis and stromal activation | Most common precursor; driver of sporadic CRC and FAP (multiple adenomas); progression risk increases with size, dysplasia, and villous component |
| Sessile Serrated Lesions (SSLs)/Serrated pathway | Mucin-vacuolated hyperplastic polyps and SSLs; serrated/“saw-tooth” crypts | Early: BRAF mutations (≥70% of SSLs), MAPK activation; Hypermethylation → CIMP, MSI; Later: silencing of tumor suppressors (TP53, CDKN2A); KRAS mutations rare (<10%), mutually exclusive with BRAF | Accounts for up to 30% of sporadic CRC; precursor lesions often in proximal colon; serrated adenocarcinomas have worse prognosis than conventional adenocarcinoma |
| IBD-associated dysplasia (colitis-associated CRC) | Flat or raised dysplasia; often multifocal due to ‘field cancerization’ | TP53 mutations: early and frequent; also KRAS, SMAD4; APC mutations uncommon; oxidative stress and microbiota dysbiosis drive clonal expansion | Risk of CRC is 2–3× higher in IBD patients; however, ≤1% of all CRC arise from IBD; cancer often synchronous/metachronous; pathogenesis differs from sporadic MSS CRC |
| Histologic Subtype | Morphology/Defining Features | Molecular/Pathobiologic Features | Prognosis/Clinical Notes |
|---|---|---|---|
| Adenocarcinoma NOS | Conventional malignant glands; no special features | Heterogeneous; baseline reference category | Standard comparator; variable prognosis depending on stage |
| Medullary carcinoma | Solid growth; sheets of cells with vesicular nuclei, prominent nucleoli, intraepithelial lymphocytes | MSI-H; can be sporadic or Lynch syndrome–associated | Relatively favorable prognosis compared to adenocarcinoma NOS |
| Mucinous adenocarcinoma | Extracellular mucin pools ≥50% of tumor volume | Heterogeneous; overlaps with adenocarcinoma NOS, signet-ring, and invasive stratified mucinous carcinomas | Prognosis variable; can behave more aggressively, depending on coexisting subtype |
| Signet-ring cell carcinoma | Intracytoplasmic mucin displacing nucleus peripherally; diffuse infiltrative pattern | Often associated with MSI-L/MSS; overlaps with mucinous subtypes | Poor prognosis, independent of stage; highly infiltrative |
| Micropapillary carcinoma | Small tight clusters in cleft-like spaces; inverted polarity | Reverse cell polarity with ‘inside-out’ MUC1/EMA staining; frequent lymphovascular invasion | Aggressive course; early nodal/vascular spread |
| Serrated adenocarcinoma | Serrated/’sawtooth’ glandular architecture; often eosinophilic cytoplasm | Associated with serrated adenomas; frequently CIMP-high, MSI variable | Worse prognosis compared to adenocarcinoma NOS |
| Adenoma-like carcinoma | Morphology mimicking conventional adenomas with malignant cytology | Overlaps with adenocarcinoma NOS pathways | Clinical relevance limited; prognosis similar to adenocarcinoma NOS |
| Adenosquamous carcinoma | Combined glandular and squamous differentiation | Variable; may show p53 mutations | Rare, aggressive; worse prognosis than adenocarcinoma NOS |
| Sarcomatoid/undifferentiated carcinoma | Spindle-cell or pleomorphic morphology with minimal gland formation | EMT signatures; variable molecular profiles | Highly aggressive; poor outcomes |
| Neuroendocrine carcinoma (small/large cell) | Small-cell: scant cytoplasm, high N:C ratio. Large-cell: organoid growth, necrosis | Expression of neuroendocrine markers (synaptophysin, chromogranin, INSM1); TP53/RB1 alterations common; high Ki-67 | Poor prognosis; aggressive, often metastatic at diagnosis |
| Invasive stratified mucinous carcinoma | Stratified epithelium with mucin production; overlaps with mucinous subtype | Heterogeneous; may share features with MSI or KRAS-mutated tumors | Prognosis variable; emerging recognition |
| Stem-cell/multidirectional carcinoma | Bidirectional/multilineage differentiation (glandular, squamous, neuroendocrine elements) | Suggests stem-cell origin; lineage plastic-ity; dedifferentiation pathways | Rare, aggressive; reflects tumor heterogeneity |
| Subtype | Key Features | Molecular Profile | Prevalence/Stage | Prognosis and Therapy | Limitations |
|---|---|---|---|---|---|
| CMS1 (Immunogenic) | Right-sided; SSL association; immune-rich TME; CIMP-high | MSI-H, BRAF mut, immune activation | ~15%; stable across stages | Early stage: good; Stage IV: worst OS; potential benefit from ICIs | Overlap with other CMS; right-sided tumors not exclusive; genotype not definitive |
| CMS2 (Canonical) | Left-sided; epithelial morphology; CIN phenotype | High SCNA; WNT, MYC, SRC/EGFR activation | ~35%; stable across stages | Most favorable OS; best response to adjuvant chemo | Mixed molecular features common |
| CMS3 (Metabolic) | Mixed sidedness; epithelial; metabolic dysregulation | Low SCNA/CIMP; mixed MSI; KRAS mut; metabolic pathway activation | ~15%; can transition to CMS4 | Intermediate OS; adjuvant chemo benefit (like CMS2) | Not exclusive; overlap with CMS1/4 |
| CMS4 (Mesenchymal) | Stromal-rich; CAF infiltration; EMT; angiogenesis; tumor budding | EMT, TGF-β, angiogenesis, YAP/TAZ | ~25%; 10% in Stage I → 40% in Stage IV | Worst OS; poor therapy response; high metastatic risk | Therapy can shift profile (e.g., WNT resistance → YAP/TAZ reprogramming) |
| Mixed/Unclassified | Weighted combinations of CMS (‘continuum’ model) | Variable | ~10% overall | Prognosis variable | Reflects heterogeneity and technical limits |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toews, K.P.; Auld, F.M.; Moyana, T.N. Embryonic Signaling Pathways Shape Colorectal Cancer Subtypes: Linking Gut Development to Tumor Biology. Pathophysiology 2025, 32, 52. https://doi.org/10.3390/pathophysiology32040052
Toews KP, Auld FM, Moyana TN. Embryonic Signaling Pathways Shape Colorectal Cancer Subtypes: Linking Gut Development to Tumor Biology. Pathophysiology. 2025; 32(4):52. https://doi.org/10.3390/pathophysiology32040052
Chicago/Turabian StyleToews, Kitty P., Finn Morgan Auld, and Terence N. Moyana. 2025. "Embryonic Signaling Pathways Shape Colorectal Cancer Subtypes: Linking Gut Development to Tumor Biology" Pathophysiology 32, no. 4: 52. https://doi.org/10.3390/pathophysiology32040052
APA StyleToews, K. P., Auld, F. M., & Moyana, T. N. (2025). Embryonic Signaling Pathways Shape Colorectal Cancer Subtypes: Linking Gut Development to Tumor Biology. Pathophysiology, 32(4), 52. https://doi.org/10.3390/pathophysiology32040052






