ITIH4 in Rheumatoid Arthritis Pathogenesis: Network Pharmacology and Molecular Docking Analysis Identify CXCR4 as a Potential Receptor
Abstract
1. Introduction
2. Materials and Methods
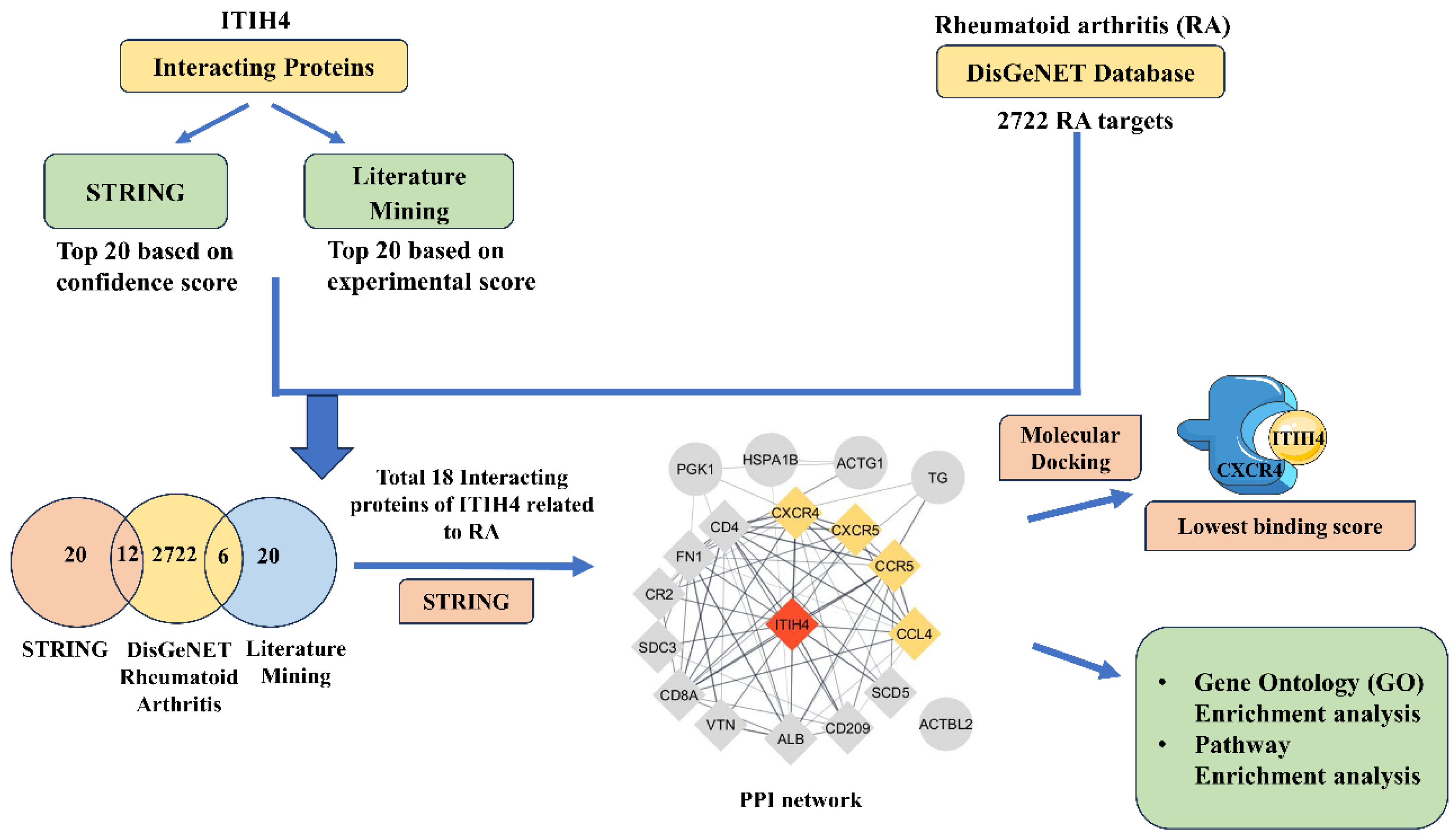
2.1. Workflow
2.2. In Silico Analysis
Screening of Interacting Proteins of ITIH4 and Their Association with RA
2.3. Gene Ontology (GO) and Pathway Enrichment Analysis of Common Proteins
2.4. Protein–Protein Interaction (PPI) Network Construction of Common Proteins
2.5. Preparation of Ligand Protein for Molecular Docking
2.6. Preparation of Receptor Protein
2.7. Active Binding Site Prediction
2.8. Molecular Docking
2.9. Cell Culture and Treatment
Collection and Isolation of Primary Cells from Biopsy Synovium
2.10. In Vitro Knockdown of ITIH4
2.11. Western Blot
2.12. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.13. Statistical Analysis
3. Results
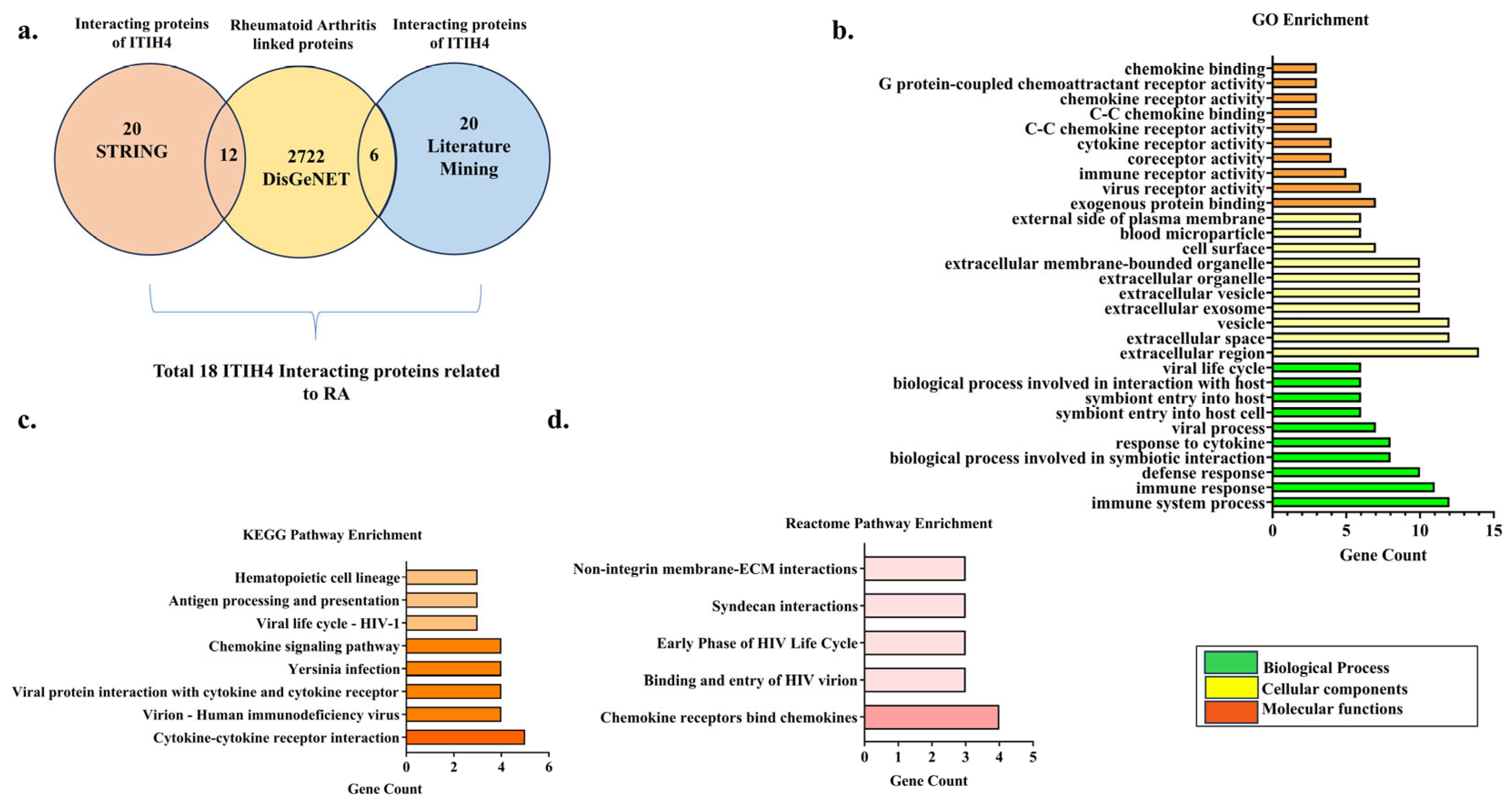
3.1. Retrieval of Overlapping Targets of ITIH4 Associated with RA
3.2. Gene Ontology (GO) Enrichment Analysis
3.3. Pathway Enrichment Analysis
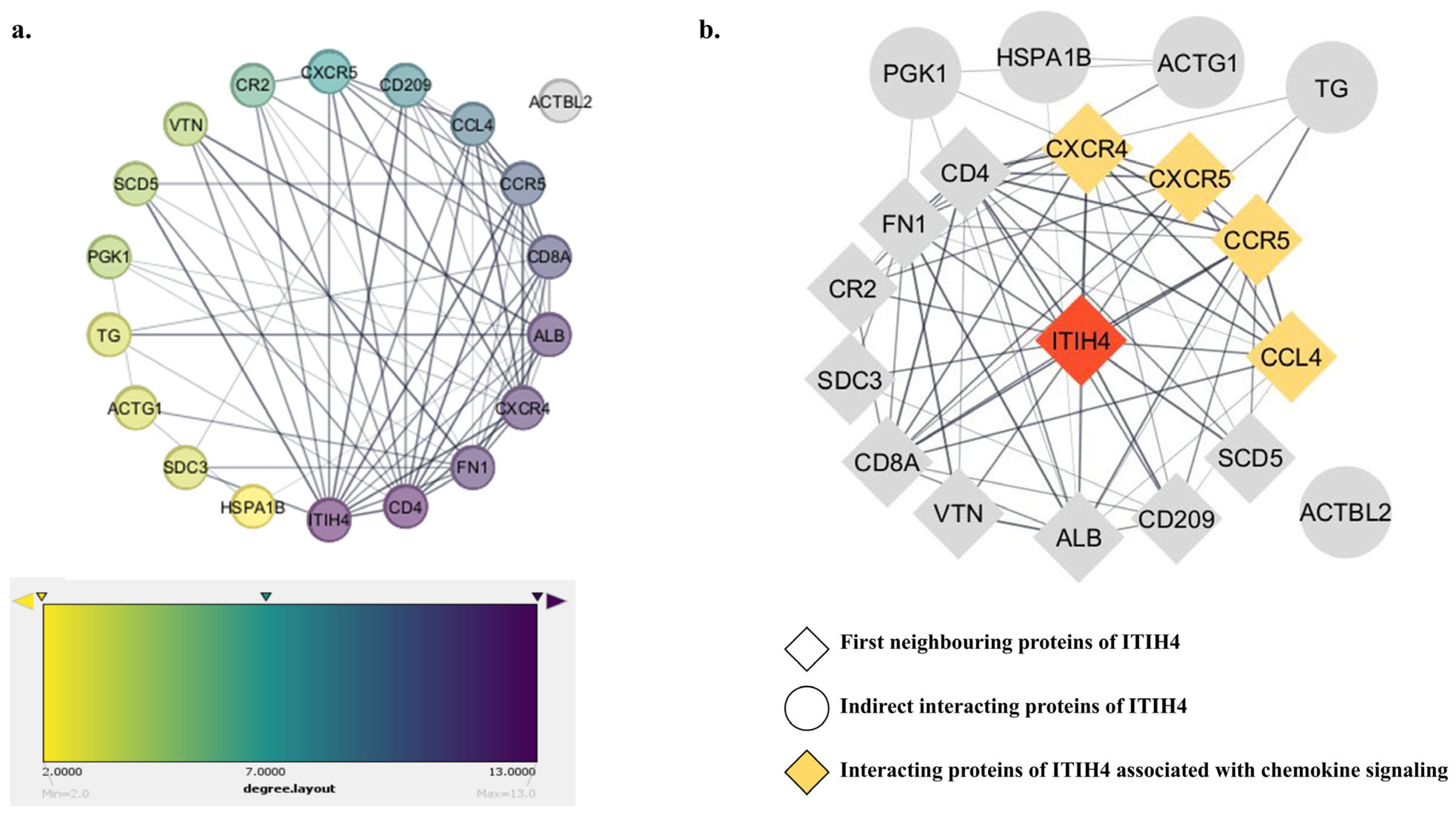
3.4. PPI Network of the Common Interacting Proteins
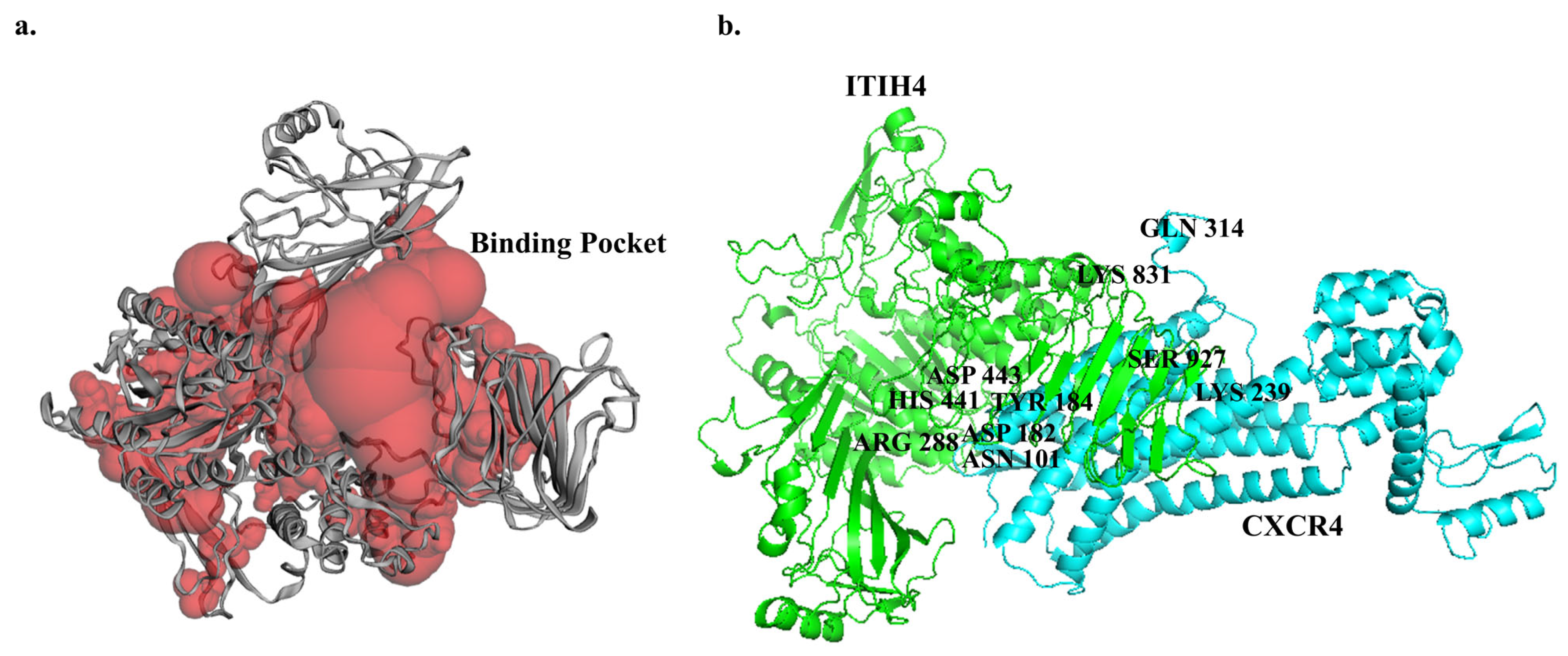
3.5. Predicted Binding Sites of ITIH4
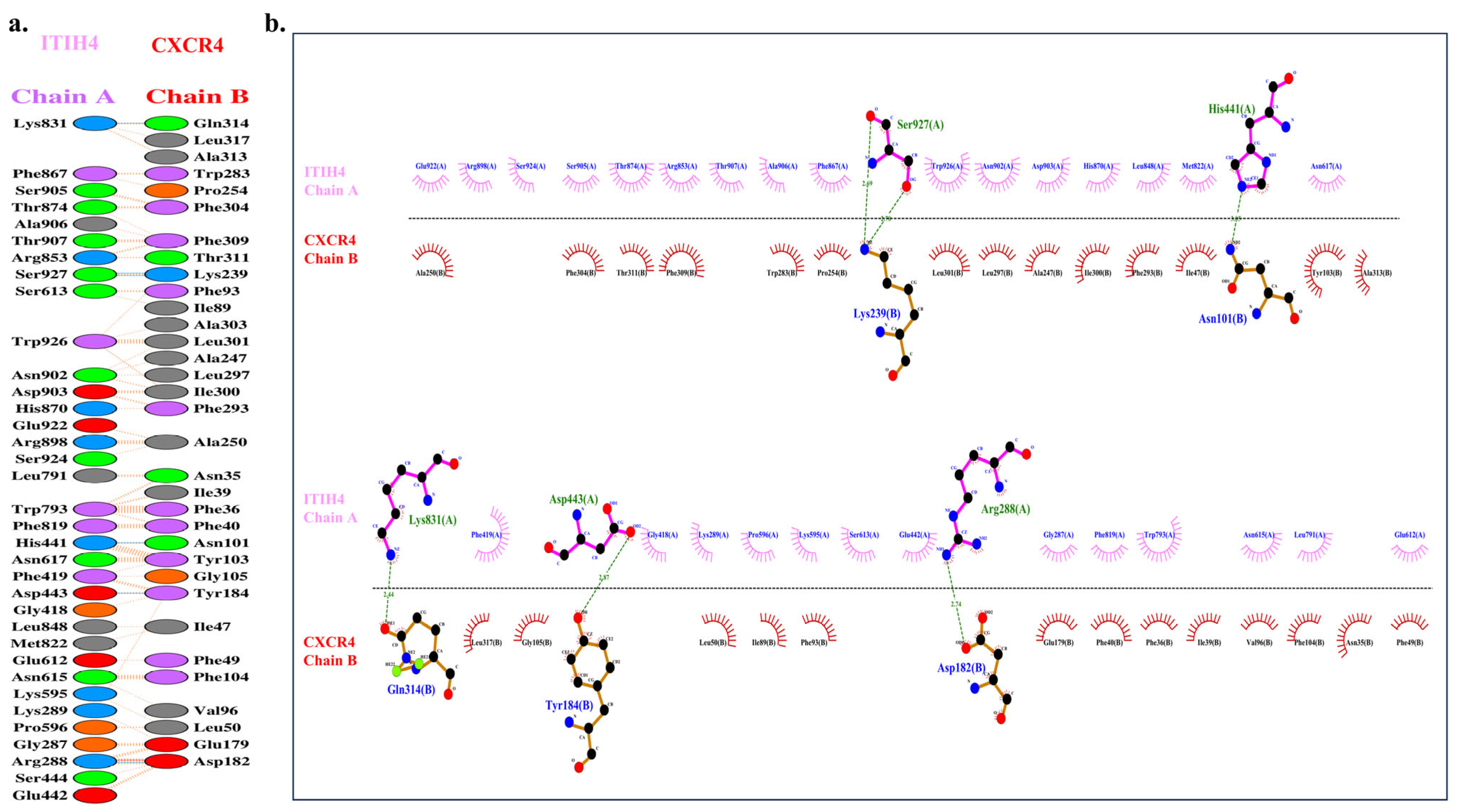
3.6. Molecular Docking of ITIH4 with Common Proteins
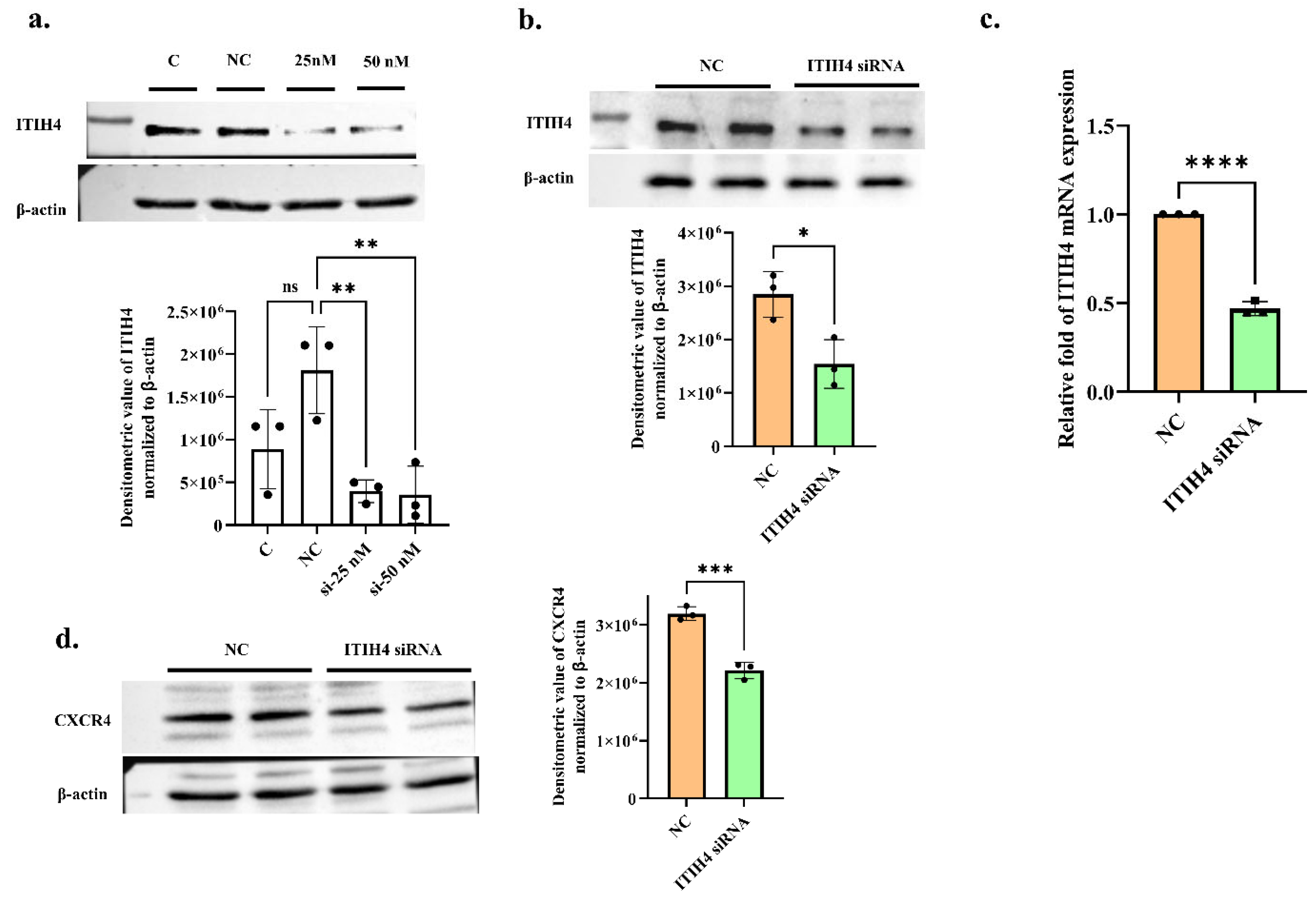
3.7. ITIH4 Knockdown Reduces CXCR4 Expression in RA-FLS
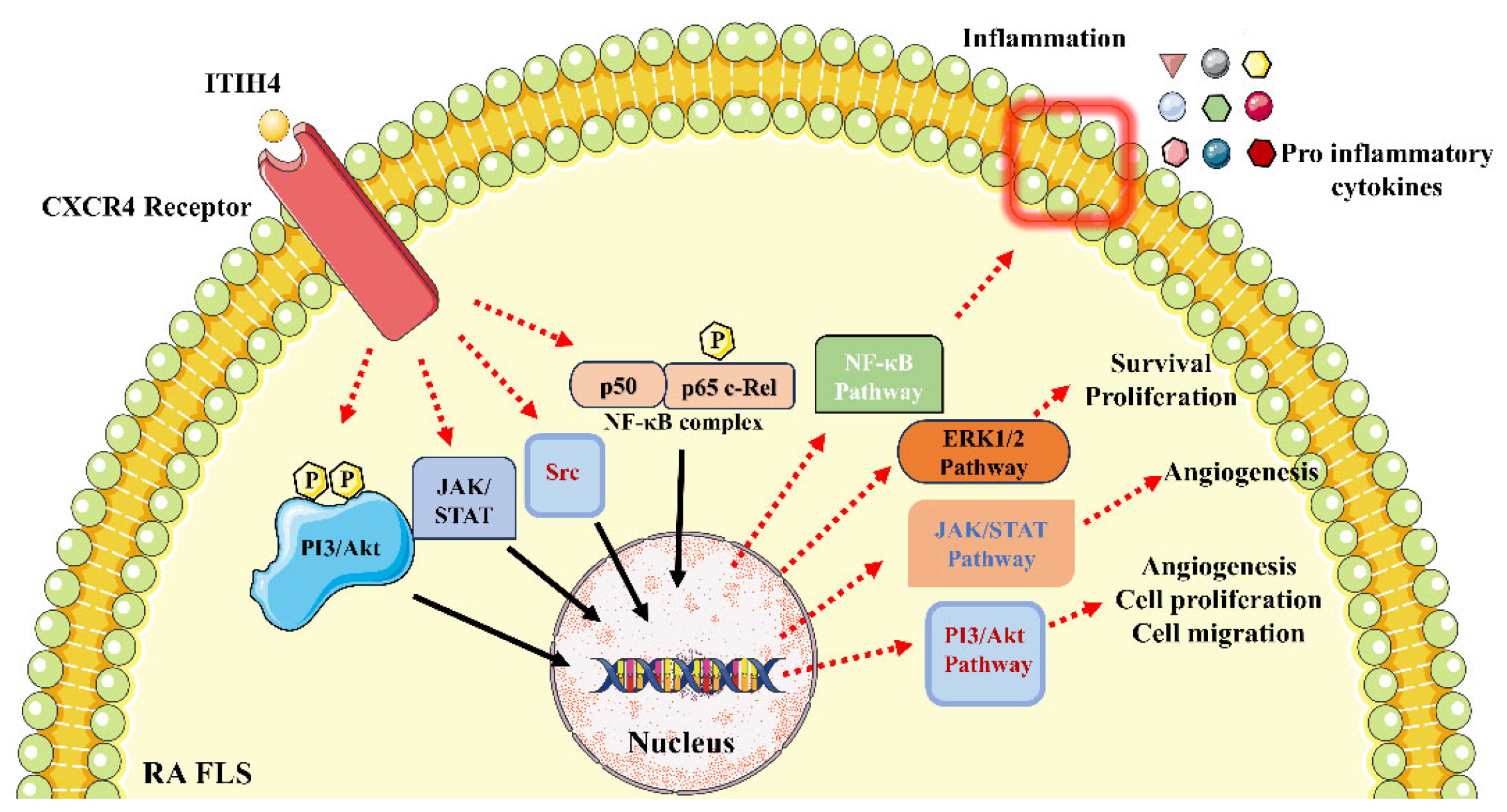
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, Q.; Zhou, C.; Nandakumar, K.S. Molecular and Cellular Pathways Contributing to Joint Damage in Rheumatoid Arthritis. Mediat. Inflamm. 2020, 2020, 3830212. [Google Scholar] [CrossRef] [PubMed]
- Finckh, A.; Gilbert, B.; Hodkinson, B.; Bae, S.-C.; Thomas, R.; Deane, K.D.; Alpizar-Rodriguez, D.; Lauper, K. Global epidemiology of rheumatoid arthritis. Nat. Rev. Rheumatol. 2022, 18, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Alamanos, Y.; Drosos, A.A. Epidemiology of adult rheumatoid arthritis. Autoimmun. Rev. 2022, 4, 130–136. [Google Scholar] [CrossRef]
- Jang, S.; Kwon, E.-J.; Lee, J.J. Rheumatoid arthritis: Pathogenic roles of diverse immune cells. Int. J. Mol. Sci. 2022, 23, 905. [Google Scholar] [CrossRef]
- Chen, Z.; Bozec, A.; Ramming, A.; Schett, G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat. Rev. Rheumatol. 2019, 15, 9–17. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Anzaghe, M.; Schülke, S. Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells 2020, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N.; Schiller, J.; Stern, R.; Soltes, L. Role, metabolism, chemical modifications and applications of hyaluronan. Curr. Med. Chem. 2009, 16, 1718–1745. [Google Scholar] [CrossRef] [PubMed]
- Petrey, A.C.; de la Motte, C.A. Hyaluronan, a Crucial Regulator of Inflammation. Front. Immunol. 2014, 5, 101. Available online: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2014.00101 (accessed on 11 March 2014). [CrossRef]
- Ragan, C.; Meyer, K. The hyaluronic acid of synovial fluid in rheumatoid arthritis. J. Clin. Investig. 1949, 28, 56–59. [Google Scholar] [CrossRef]
- Kida, D.; Yoneda, M.; Miyaura, S.; Ishimaru, T.; Yoshida, Y.; Ito, T.; Ishiguro, N.; Iwata, H.; Kimata, K. The SHAP-HA complex in sera from patients with rheumatoid arthritis and osteoarthritis. J. Rheumatol. 1999, 26, 1230–1238. Available online: http://europepmc.org/abstract/MED/10381035 (accessed on 1 June 1999).
- Zhao, M.; Yoneda, M.; Ohashi, Y.; Kurono, S.; Iwata, H.; Ohnuki, Y.; Kimata, K. Evidence for the covalent binding of SHAP, heavy chains of inter-α-trypsin inhibitor, to hyaluronan. J. Biol. Chem. 1995, 270, 26657–26663. [Google Scholar] [CrossRef]
- Saroha, A.; Kumar, S.; Chatterjee, B.P.; Das, H.R. Jacalin bound plasma O-glycoproteome and reduced sialylation of alpha 2-HS glycoprotein (A2HSG) in rheumatoid arthritis patients. PLoS ONE 2012, 7, e46374. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Yu, P.; Monga, S.P.S.; Mishra, B.; Mishra, L. Identification of mouse itih-4 encoding a glycoprotein with two EF-hand motifs from early embryonic liver. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 1998, 1398, 32–37. [Google Scholar] [CrossRef]
- Piñeiro, M.; Alava, M.A.; González-Ramón, N.; Osada, J.; Lasierra, P.; Larrad, L.; Piñeiro, A.; Lampreave, F. ITIH4 Serum Concentration Increases during Acute-Phase Processes in Human Patients and Is Up-Regulated by Interleukin-6 in Hepatocarcinoma HepG2 Cells. Biochem. Biophys. Res. Commun. 1999, 263, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Bhanumathy, C.D.; Tang, Y.; Monga, S.P.S.; Katuri, V.; Cox, J.A.; Mishra, B.; Mishra, L. Itih-4, a serine protease inhibitor regulated in interleukin-6–dependent liver formation: Role in liver development and regeneration. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2002, 223, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Monu, A.P.; Saquib, M.; Sarkar, A.; Chakraborty, D.; Kumar, U.; Biswas, S. Transthyretin and receptor for advanced glycation end product’s differential levels associated with the pathogenesis of rheumatoid arthritis. J. Inflamm. Res. 2021, 14, 5581–5596. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Matsumoto, I.; Osada, A.; Kurata, I.; Ebe, H.; Tanaka, Y.; Inoue, A.; Umeda, N.; Kondo, Y.; Tsuboi, H.; et al. Identification of novel biomarker as citrullinated inter-alpha-trypsin inhibitor heavy chain 4, specifically increased in sera with experimental and rheumatoid arthritis. Arthritis Res. Ther. 2018, 20, 1–13. [Google Scholar] [CrossRef]
- Khakha, S.; Sharma, A.; Kumari, P.; Sahi, S.; Biswas, S. In-silico molecular characterization and mutational analysis of inter-alpha-trypsin inhibitor heavy chain 4 in rheumatoid arthritis. J. Proteins Proteom. 2019, 10, 313–323. [Google Scholar] [CrossRef]
- Sharma, A.; Kumari, P.; Rajan, G.S.; Malhotra, R.; Biswas, S. In-Silico Structure Prediction and Domain Characterization of Inter Alpha Trypsin Inhibitor Heavy Chain 4. In Proceedings of the 2016 International Conference on Bioinformatics and Systems Biology (BSB), Allahabad, India, 4–6 March 2016; pp. 1–5. [Google Scholar]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Park, H.-B.; Choi, B.-C.; Baek, K.-H. PGK1 modulates balance between pro-and anti-inflammatory cytokines by interacting with ITI-H4. Biomed. Pharmacother. 2023, 161, 114437. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.-C.; Duan, W.-G.; Chen, S.-Y.; Fang, J.-K. Analysis of the Composition and Anti-Rheumatoid Arthritis Mechanism of Qintengtongbi Decoction Based on Network Pharmacology. Nat. Prod. Commun. 2021, 16, 1934578X211041421. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chakraborty, D.; Gupta, K.; Biswas, S. Potential role of Bavachin in Rheumatoid arthritis: Informatics approach for rational based selection of phytoestrogen. J. Herb. Med. 2023, 38, 100640. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Alekseenko, A.; Ignatov, M.; Jones, G.; Sabitova, M.; Kozakov, D. Protein–protein and protein–peptide docking with ClusPro server. Protein Struct. Predict. 2020, 2165, 157–174. [Google Scholar]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., III; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Saquib, M.; Agnihotri, P.; Sarkar, A.; Malik, S.; Mann, S.; Chakraborty, D.; Joshi, L.; Malhotra, R.; Biswas, S. Functional Significance of miR-4693-5p in Targeting HIF1α and Its Link to Rheumatoid Arthritis Pathogenesis. Non-Coding RNA 2024, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Suto, T.; Tosevska, A.; Dalwigk, K.; Kugler, M.; Dellinger, M.; Stanic, I.; Platzer, A.; Niederreiter, B.; Sevelda, F.; Bonelli, M.; et al. TNFR2 is critical for TNF-induced rheumatoid arthritis fibroblast-like synoviocyte inflammation. Rheumatology 2022, 61, 4535–4546. [Google Scholar] [CrossRef]
- Sarkar, A.; Sharma, S.; Agnihotri, P.; Sarkar, T.; Kumari, P.; Malhotra, R.; Datta, B.; Kumar, V.; Biswas, S. Synovial Fluid Cell Proteomic Analysis Identifies Upregulation of Alpha-Taxilin Proteins in Rheumatoid Arthritis: A Potential Prognostic Marker. J. Immunol. Res. 2020, 2020, 4897983. [Google Scholar] [CrossRef]
- Sarkar, A.; Chakraborty, D.; Kumar, V.; Malhotra, R.; Biswas, S. Upregulation of leucine-rich alpha-2 glycoprotein: A key regulator of inflammation and joint fibrosis in patients with severe knee osteoarthritis. Front. Immunol. 2022, 13, 1028994. [Google Scholar] [CrossRef]
- Shree, P.; Srivastava, S.; Chaube, R.; Tripathi, Y.B. Protein-Protein docking interaction of Nephrin protein with the expression of MMP-9: An anti-inflammatory agent. Res. J. Biotechnol. 2022, 17, 6. [Google Scholar] [CrossRef]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. CXCR4 signaling in health and disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef]
- Kondo, N.; Kuroda, T.; Kobayashi, D. Cytokine Networks in the Pathogenesis of Rheumatoid Arthritis. Int. J. Mol. Sci. 2021, 22, 10922. [Google Scholar] [CrossRef]
- Ma, Y.; Li, R.; Wang, J.; Jiang, W.; Yuan, X.; Cui, J.; Wang, C. ITIH4, as an inflammation biomarker, mainly increases in bacterial bloodstream infection. Cytokine 2021, 138, 155377. [Google Scholar] [CrossRef] [PubMed]
- Laursen, T.L.; Bossen, L.; Pihl, R.; Troldborg, A.; Sandahl, T.D.; Hansen, A.G.; Folserass, T.; Vesterhus, M.; Grønbæk, H.; Thiel, S. Highly increased levels of inter-α-inhibitor heavy chain 4 (ITIH4) in autoimmune cholestatic liver diseases. J. Clin. Transl. Hepatol. 2022, 10, 796. [Google Scholar] [CrossRef] [PubMed]
- Elemam, N.M.; Hannawi, S.; Maghazachi, A.A. Role of Chemokines and Chemokine Receptors in Rheumatoid Arthritis. Immuno Targets Ther. 2020, 9, 43–56. [Google Scholar] [CrossRef]
- Haringman, J.J.; Smeets, T.J.M.; Reinders-Blankert, P.; Tak, P.P. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann. Rheum. Dis. 2006, 65, 294–300. [Google Scholar] [CrossRef]
- García-Vicuña, R.; Gómez-Gaviro, M.V.; Domínguez-Luis, M.J.; Pec, M.K.; González-Alvaro, I.; Alvaro-Gracia, J.M.; Díaz-González, F. CC and CXC chemokine receptors mediate migration, proliferation, and matrix metalloproteinase production by fibroblast-like synoviocytes from rheumatoid arthritis patients. Arthritis Rheum. 2004, 50, 3866–3877. [Google Scholar] [CrossRef] [PubMed]
- Murayama, M.A.; Shimizu, J.; Miyabe, C.; Yudo, K.; Miyabe, Y. Chemokines and chemokine receptors as promising targets in rheumatoid arthritis. Front. Immunol. 2023, 14, 1100869. Available online: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1100869 (accessed on 13 February 2023). [CrossRef]
- Peng, L.; Zhu, N.; Mao, J.; Huang, L.; Yang, Y.; Zhou, Z.; Wang, L.; Wu, B. Expression levels of CXCR4 and CXCl12 in patients with rheumatoid arthritis and its correlation with disease activity. Exp. Ther. Med. 2020, 20, 1925–1934. [Google Scholar] [CrossRef]
- Kanbe, K.; Chiba, J.; Inoue, Y.; Taguchi, M.; Yabuki, A. SDF-1 and CXCR4 in synovium are associated with disease activity and bone and joint destruction in patients with rheumatoid arthritis treated with golimumab. Mod. Rheumatol. 2016, 26, 46–50. [Google Scholar] [CrossRef]
- Shadidi, K.R.; Aarvak, T.; Henriksen, J.E.; Natvig, J.B.; Thompson, K.M. The Chemokines CCL5, CCL2 and CXCL12 Play Significant Roles in the Migration of Th1 Cells into Rheumatoid Synovial Tissue. Scand. J. Immunol. 2003, 57, 192–198. [Google Scholar] [CrossRef]
- Nanki, T.; Hayashida, K.; El-Gabalawy, H.S.; Suson, S.; Shi, K.; Girschick, H.J.; Yavuz, S.; Lipsky, P.E. Stromal cell-derived factor-1-CXC chemokine receptor 4 interactions play a central role in CD4+ T cell accumulation in rheumatoid arthritis synovium. J. Immunol. 2000, 165, 6590–6598. [Google Scholar] [CrossRef]
- Buckley, C.D.; Amft, N.; Bradfield, P.F.; Pilling, D.; Ross, E.; Arenzana-Seisdedos, F.; Amara, A.; Curnow, S.J.; Lord, J.M.; Scheel-Toellner, D.; et al. Persistent induction of the chemokine receptor CXCR4 by TGF-β1 on synovial T cells contributes to their accumulation within the rheumatoid synovium. J. Immunol. 2000, 165, 3423–3429. [Google Scholar] [CrossRef] [PubMed]
- Blades, M.C.; Ingegnoli, F.; Wheller, S.K.; Manzo, A.; Wahid, S.; Panayi, G.S.; Perretti, M.; Pitzalis, C. Stromal cell–derived factor 1 (CXCL12) induces monocyte migration into human synovium transplanted onto SCID Mice. Arthritis Rheum. 2002, 46, 824–836. [Google Scholar] [CrossRef]
- Kanbe, K.; Takemura, T.; Takeuchi, K.; Chen, Q.; Takagishi, K.; Inoue, K. Synovectomy reduces stromal-cell-derived factor-1 (SDF-1) which is involved in the destruction of cartilage in osteoarthritis and rheumatoid arthritis. J. Bone Jt. Surg. Br. Vol. 2004, 86, 296–300. [Google Scholar] [CrossRef]
- Nanki, T.; Takada, K.; Komano, Y.; Morio, T.; Kanegane, H.; Nakajima, A.; E Lipsky, P.; Miyasaka, N. Chemokine receptor expression and functional effects of chemokines on B cells: Implication in the pathogenesis of rheumatoid arthritis. Arthritis Res. Ther. 2009, 11, R149. [Google Scholar] [CrossRef] [PubMed]
- Pablos, J.L.; Santiago, B.; Galindo, M.; Torres, C.; Brehmer, M.T.; Blanco, F.J.; García-Lázaro, F.J. Synoviocyte-derived CXCL12 is displayed on endothelium and induces angiogenesis in rheumatoid arthritis. J. Immunol. 2003, 170, 2147–2152. [Google Scholar] [CrossRef] [PubMed]
- Tamamura, H.; Fujisawa, M.; Hiramatsu, K.; Mizumoto, M.; Nakashima, H.; Yamamoto, N.; Otaka, A.; Fujii, N. Identification of a CXCR4 antagonist, a T140 analog, as an anti-rheumatoid arthritis agent. FEBS Lett. 2004, 569, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.-H.; Seki, K.; Choi, B.-I.; Kimura, K.B.; Ito, A.; Fujikado, N.; Saijo, S.; Iwakura, Y. CXC chemokine receptor 4 expressed in T cells plays an important role in the development of collagen-induced arthritis. Arthritis Res. Ther. 2010, 12, 1–14. [Google Scholar] [CrossRef]
- Choi, W.-T.; Duggineni, S.; Xu, Y.; Huang, Z.; An, J. Drug Discovery Research Targeting the CXC Chemokine Receptor 4 (CXCR4). J. Med. Chem. 2012, 55, 977–994. [Google Scholar] [CrossRef]
| S. No. | Patient’s Characteristics | RA (n = 3) |
|---|---|---|
| 1 | Age (yrs.) | 50 ± 5 |
| 2 | Sex (Female, Male) | F (2), M (1) |
| 3 | ESR (mm/h) | 35 ± 5 |
| 4 | RF (+ve/−ve) | +ve |
| 5 | CRP (mg/L) | 80 ±15 |
| 6 | Tender Joint | 20 ± 6 |
| 7 | Swollen joints | 10 ± 4 |
| 8 | DAS-28 score | 6 ± 0.5 |
| 9 | Disease duration yrs. | 10 ± 5 |
| 10 | Medication (Yes/No) | Yes |
| Chain | Binding Sites |
|---|---|
| A | 36TYR, 37SER, 39THR, 41ASP, 42SER, 43ARG, 44VAL, 45SER, 46SER, 47ARG, 48PHE, 50HIS, 52VAL, 54THR, 56ARG, 78PHE, 80THR, 81ASN, 85ILE, 87ASP, 88GLY, 141THR, 143GLU, 145VAL, 147GLU, 149LEU, 150LEU, 151LYS, 152ARG, 153ARG, 154LEU, 155GLY, 169VAL, 170LYS, 171HIS, 173GLN, 174MET, 175ASP, 177HIS, 179PHE, 180GLU, 181PRO, 182GLN, 183GLY, 184ILE, 185SER, 187LEU, 190GLU, 205TRP, 206GLN, 207ASN, 208LYS, 209THR, 210LYS, 211ALA, 212HIS, 214ARG, 215PHE, 216LYS, 217PRO, 218THR, 219LEU, 221GLN, 222GLN, 225SER, 239ILE, 241TYR, 242ASP, 243VAL, 244ASP, 245ARG, 246ALA, 247ILE, 248SER, 249GLY, 250GLY, 251SER, 252ILE, 253GLN, 254ILE, 255GLU, 256ASN, 257GLY, 265PRO, 266GLU, 268LEU, 269THR, 270THR, 271MET, 272PRO, 273LYS, 274ASN, 275VAL, 288ARG, 292GLN, 294ARG, 295GLU, 298ILE, 299LYS, 301LEU, 302ASP, 303ASP, 304LEU, 305SER, 306PRO, 307ARG, 308ASP, 309GLN, 326LEU, 329ALA, 331ALA, 334VAL, 338ARG, 342ALA, 363LEU, 366SER, 367ASN, 370GLU, 371ARG, 372LEU, 373PRO, 374GLU, 375GLY, 376SER, 377VAL, 378SER, 379LEU, 380ILE, 381ILE, 405ALA, 406VAL, 408GLY, 409ARG, 410TYR, 411SER, 413PHE, 431LEU, 432ASP, 433ASN, 434GLY, 435GLY, 436LEU, 438ARG, 439ARG, 440ILE, 441HIS, 442GLU, 443ASP, 444SER, 445ASP, 447ALA, 448LEU, 449GLN, 451GLN, 452ASP, 453PHE, 454TYR, 455GLN, 456GLU, 457VAL, 458ALA, 459ASN, 460PRO, 461LEU, 462LEU, 463THR, 464ALA, 465VAL, 467PHE, 469TYR, 470PRO, 471SER, 472ASN, 473ALA, 476GLU, 478THR, 479GLN, 480ASN, 482PHE, 483ARG, 484LEU, 485LEU, 486PHE, 487LYS, 488GLY, 489SER, 490GLU, 491MET, 492VAL, 493VAL, 494ALA, 495GLY, 496LYS, 497LEU, 498GLN, 500ARG, 502PRO, 504VAL, 505LEU, 506THR, 507ALA, 508THR, 509VAL, 513LEU, 514PRO, 515THR, 516GLN, 517ASN, 518ILE, 519THR, 520PHE, 521GLN, 522THR, 524SER, 525SER, 526VAL, 537LYS, 538TYR, 539ILE, 540PHE, 542ASN, 545GLU, 546ARG, 573ASN, 576LEU, 577ASN, 578LEU, 580LEU, 581ALA, 582TYR, 583SER, 584PHE, 585VAL, 586THR, 587PRO, 588LEU, 590SER, 591MET, 593VAL, 594THR, 595LYS, 596PRO, 597ASP, 599GLN, 600GLU, 601GLN, 604VAL, 615ASN, 617ASN, 618VAL, 621GLY, 624PHE, 625PHE, 627TYR, 628TYR, 629LEU, 630GLN, 631GLY, 632ALA, 633LYS, 634ILE, 635PRO, 636LYS, 647TRP, 649ARG, 650GLN, 651ALA, 652GLY, 653ALA, 654ALA, 655GLY, 656SER, 657ARG, 658MET, 659ASN, 660PHE, 661ARG, 662PRO, 663GLY, 664VAL, 665LEU, 666SER, 667SER, 668ARG, 669GLN, 670LEU, 671GLY, 672LEU, 673PRO, 674GLY, 675PRO, 677ASP, 678VAL, 679PRO, 681HIS, 682ALA, 683ALA, 684TYR, 685HIS, 686PRO, 687PHE, 688ARG, 689ARG, 690LEU, 691ALA, 692ILE, 693LEU, 694PRO, 695ALA, 696SER, 697ALA, 698PRO, 699PRO, 703ASN, 705ASP, 707ALA, 708VAL, 710ARG, 711VAL, 712MET, 714MET, 715LYS, 716ILE, 717GLU, 718GLU, 719THR, 720THR, 721MET, 722THR, 723THR, 724GLN, 725THR, 726PRO, 727ALA, 728PRO, 729ILE, 730GLN, 731ALA, 732PRO, 733SER, 734ALA, 735ILE, 736LEU, 737PRO, 738LEU, 739PRO, 741GLN, 742SER, 744GLU, 745ARG, 746LEU, 749ASP, 750PRO, 751ARG, 754GLN, 755GLY, 756PRO, 757VAL, 758ASN, 759LEU, 760LEU, 761SER, 762ASP, 763PRO, 764GLU, 765GLN, 766GLY, 767VAL, 768GLU, 769VAL, 770THR, 771GLY, 772GLN, 773TYR, 774GLU, 775ARG, 776GLU, 777LYS, 778ALA, 779GLY, 780PHE, 781SER, 782TRP, 783ILE, 784GLU, 785VAL, 786THR, 787PHE, 788LYS, 789ASN, 790PRO, 791LEU, 792VAL, 793TRP, 794VAL, 795HIS, 796ALA, 797SER, 798PRO, 799GLU, 800HIS, 801VAL, 802VAL, 803VAL, 807ARG, 810SER, 811ALA, 812TYR, 814TRP, 815LYS, 820SER, 821VAL, 822MET, 823PRO, 824GLY, 825LEU, 826LYS, 837LEU, 845ILE, 846GLY, 847LEU, 849PHE |
| S.No. | Ligand Protein–Receptor Protein | Weighted Scores of Lowest Energy (kcal/mol) | No. of H-Bonds |
|---|---|---|---|
| 1. | ITIH4-SCD5 (Chain A: Chain A) | −1814.8 | None |
| 2. | ITIH4-CXCR5 (Chain A: Chain A) | −1733.3 | None |
| 3. | ITIH4-CXCR4 (Chain A: Chain B) | −1705.7 | 6 (A: B) |
| 4. | ITIH4-TG (Chain A: Chain B) | −1514.8 | 19 (A:B) |
| 5. | ITIH4-CCR5 (Chain A: Chain B) | −1440.6 | 5 (A: B) |
| 6. | ITIH4-SDC3 (Chain A: Chain A) | −1300.6 | None |
| 7. | ITIH4-FN1 (Chain A: Chain A) | −1271 | None |
| 8. | ITIH4-CD209 | −1184.9 | 20 (A: B) |
| 9. | ITIH4-ACTG1 (Chain A: Chain B) | −1148 | 10 (A: B) |
| 10. | ITIH4-CD8A (Chain A: Chain A) | −1139.7 | None |
| 11. | ITIH4-ALB (Chain A: Chain B) | −1008.6 | 10 (A: B) |
| 12. | ITIH4-ACTBL2 (Chain A: Chain A) | −980.9 | None |
| 13. | ITIH4-PGK1 (Chain A: Chain B) | −968.6 | 16 (A: B) |
| 14. | ITIH4-CCL4 (Chain A: Chain B) | −934.6 | 23 (A: B) |
| 15. | ITIH4-HSPA1B (Chain A: Chain A) | −933 | None |
| 16. | ITIH4-CR2 (Chain A: Chain B) | −873.9 | 8 (A: B) |
| 17. | ITIH4-CD4 (Chain A: Chain B) | −863.3 | 19 (A: B) |
| 18. | ITIH4-VTN (Chain A: Chain B) | −855.1 | 11 (A: B) |
| AA Residues of Chain A ITIH4 | AA Residues of Chain B CXCR4 | Bond Length (Å) |
|---|---|---|
| HIS 441 | ASN 101 | 3.05 |
| ARG 288 | ASP 182 | 2.74 |
| ASP 443 | TYR 184 | 2.87 |
| SER 927 | LYS 239 | 2.69 |
| SER 927 | LYS 239 | 2.70 |
| LYS 831 | GLN 314 | 2.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, L.; Chakraborty, D.; Kumar, V.; Biswas, S. ITIH4 in Rheumatoid Arthritis Pathogenesis: Network Pharmacology and Molecular Docking Analysis Identify CXCR4 as a Potential Receptor. Pathophysiology 2024, 31, 514-530. https://doi.org/10.3390/pathophysiology31030038
Joshi L, Chakraborty D, Kumar V, Biswas S. ITIH4 in Rheumatoid Arthritis Pathogenesis: Network Pharmacology and Molecular Docking Analysis Identify CXCR4 as a Potential Receptor. Pathophysiology. 2024; 31(3):514-530. https://doi.org/10.3390/pathophysiology31030038
Chicago/Turabian StyleJoshi, Lovely, Debolina Chakraborty, Vijay Kumar, and Sagarika Biswas. 2024. "ITIH4 in Rheumatoid Arthritis Pathogenesis: Network Pharmacology and Molecular Docking Analysis Identify CXCR4 as a Potential Receptor" Pathophysiology 31, no. 3: 514-530. https://doi.org/10.3390/pathophysiology31030038
APA StyleJoshi, L., Chakraborty, D., Kumar, V., & Biswas, S. (2024). ITIH4 in Rheumatoid Arthritis Pathogenesis: Network Pharmacology and Molecular Docking Analysis Identify CXCR4 as a Potential Receptor. Pathophysiology, 31(3), 514-530. https://doi.org/10.3390/pathophysiology31030038






