RhoG-Binding Domain of Elmo1 Ameliorates Excessive Process Elongation Induced by Autism Spectrum Disorder-Associated Sema5A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Key Antibodies, Chemicals, and Plasmids
2.2. Generation of Plasmid-Encoding Mutated Proteins and Other Constructs
2.3. Cell Line Culture, Stable Clone, and Differentiation
2.4. Plasmid Transfection
2.5. Preparation of Cell Extracts or Lysates, Fractionation, and Denatured Polyacrylamide Electrophoresis and Immunoblotting
2.6. Affinity Precipitation Assay to Monitor Guanosine Triphosphate (GTP)-Bound Rac1 Protein
2.7. Fluorescent Images
2.8. Statistical Analyses
2.9. Ethics Statement
3. Results
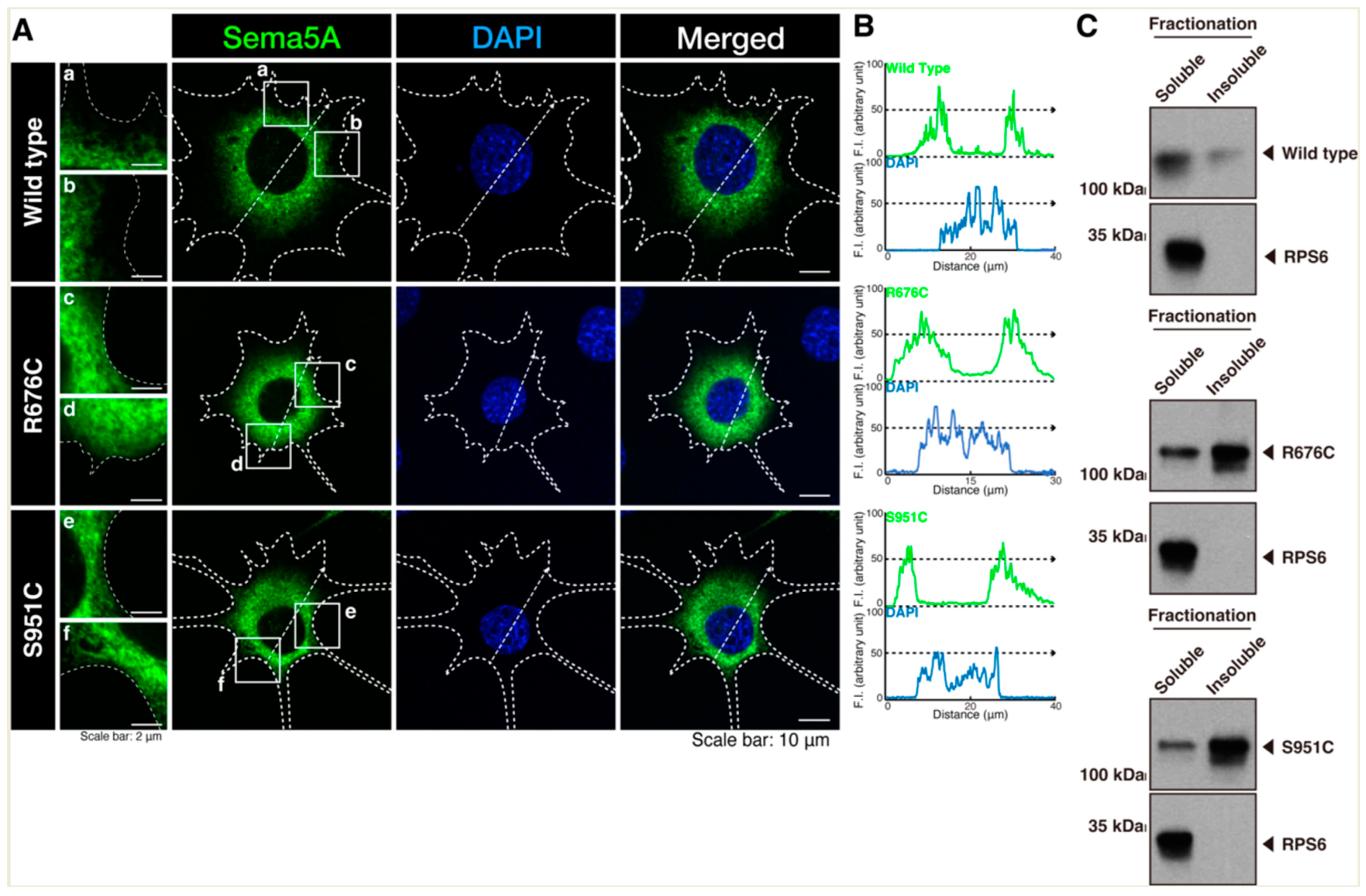
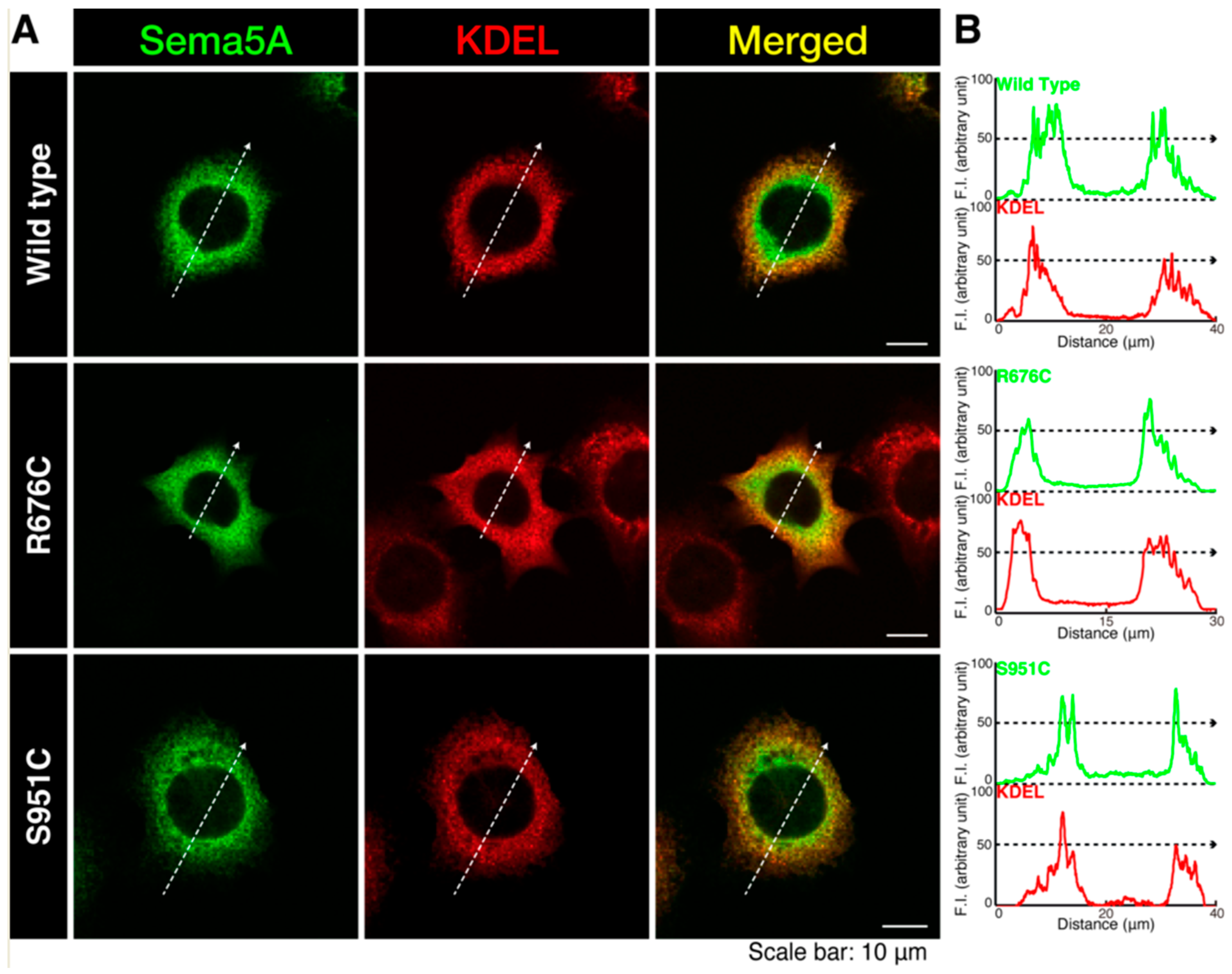
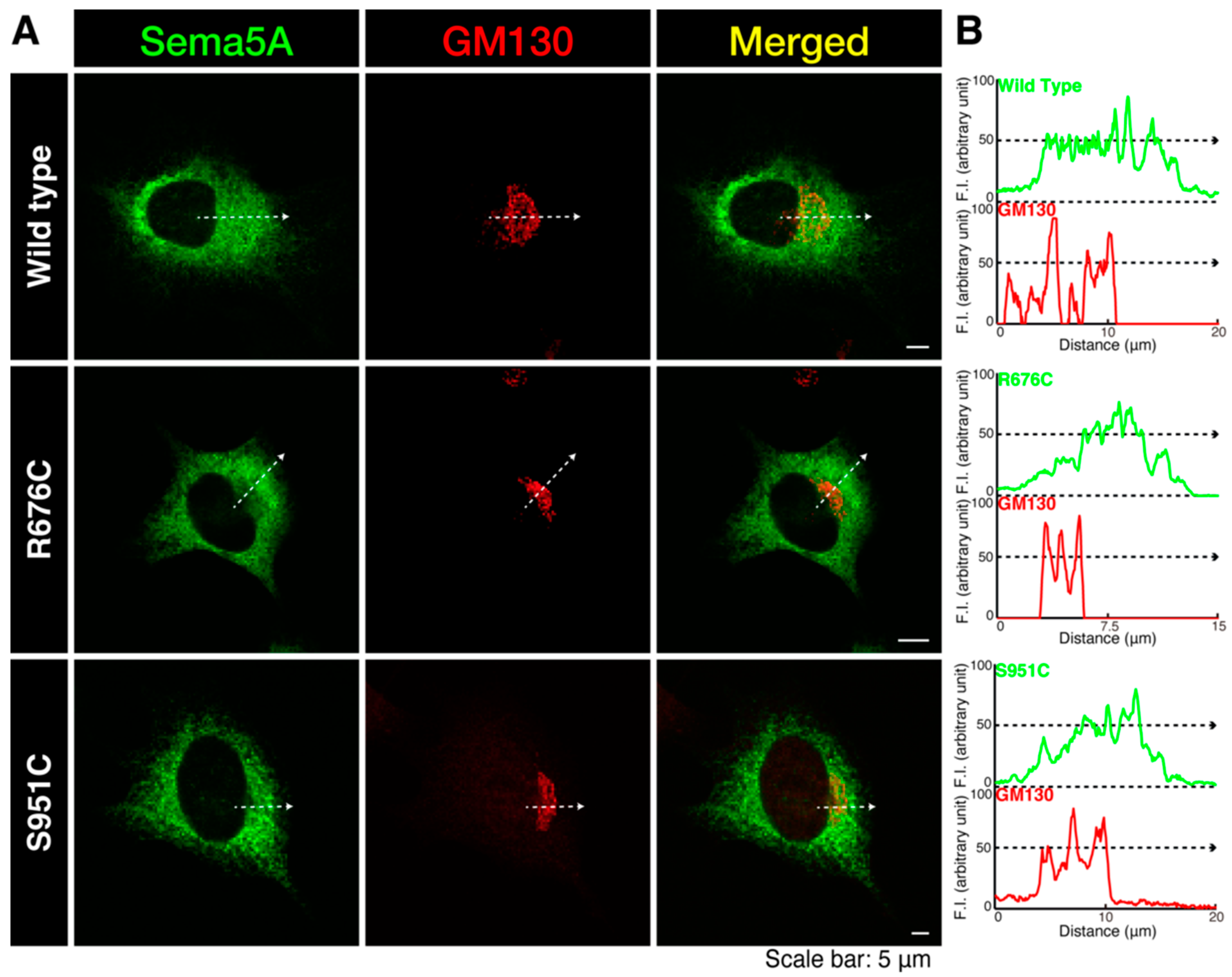
3.1. Mutated Sema5A Preferentially Localizes around the Plasma Membrane
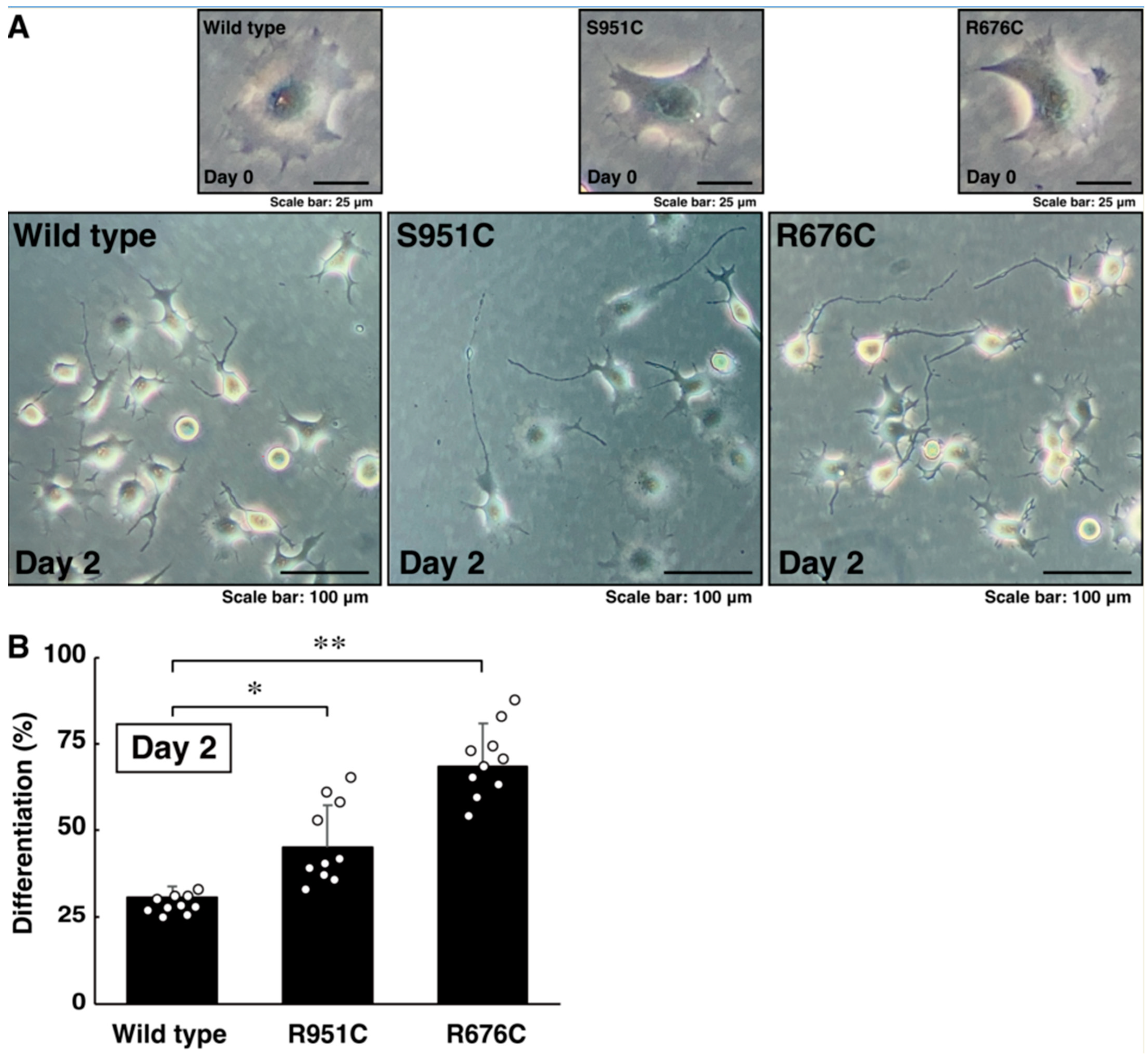
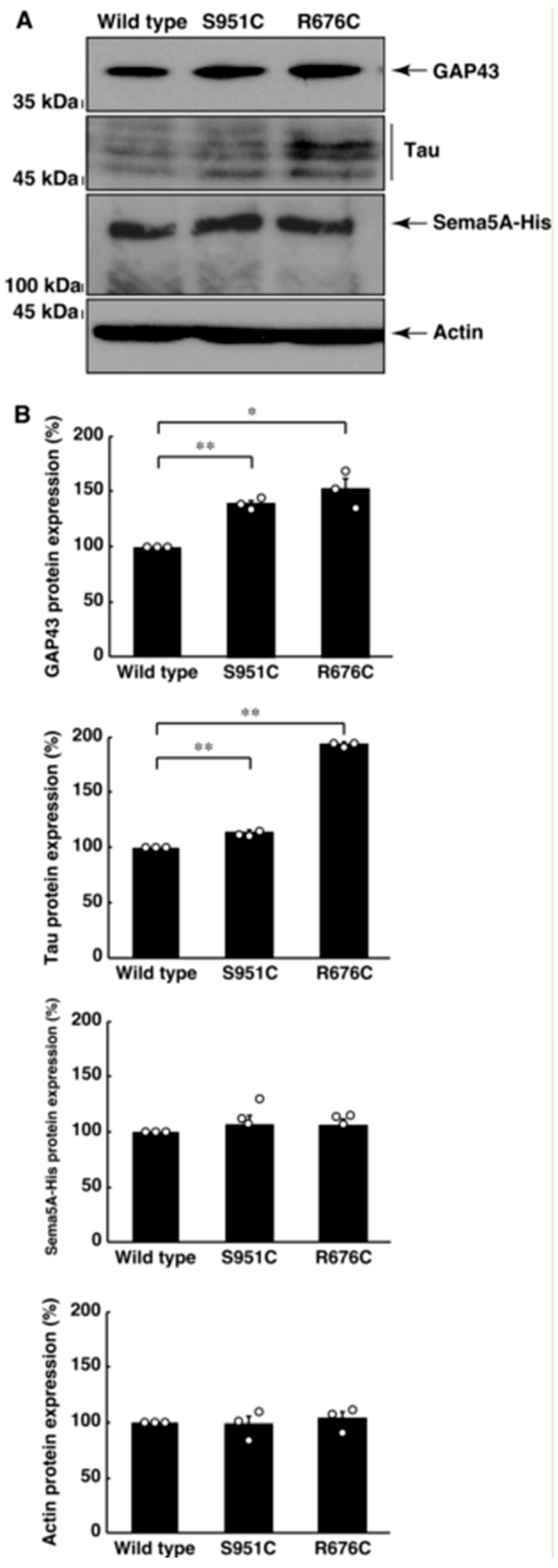
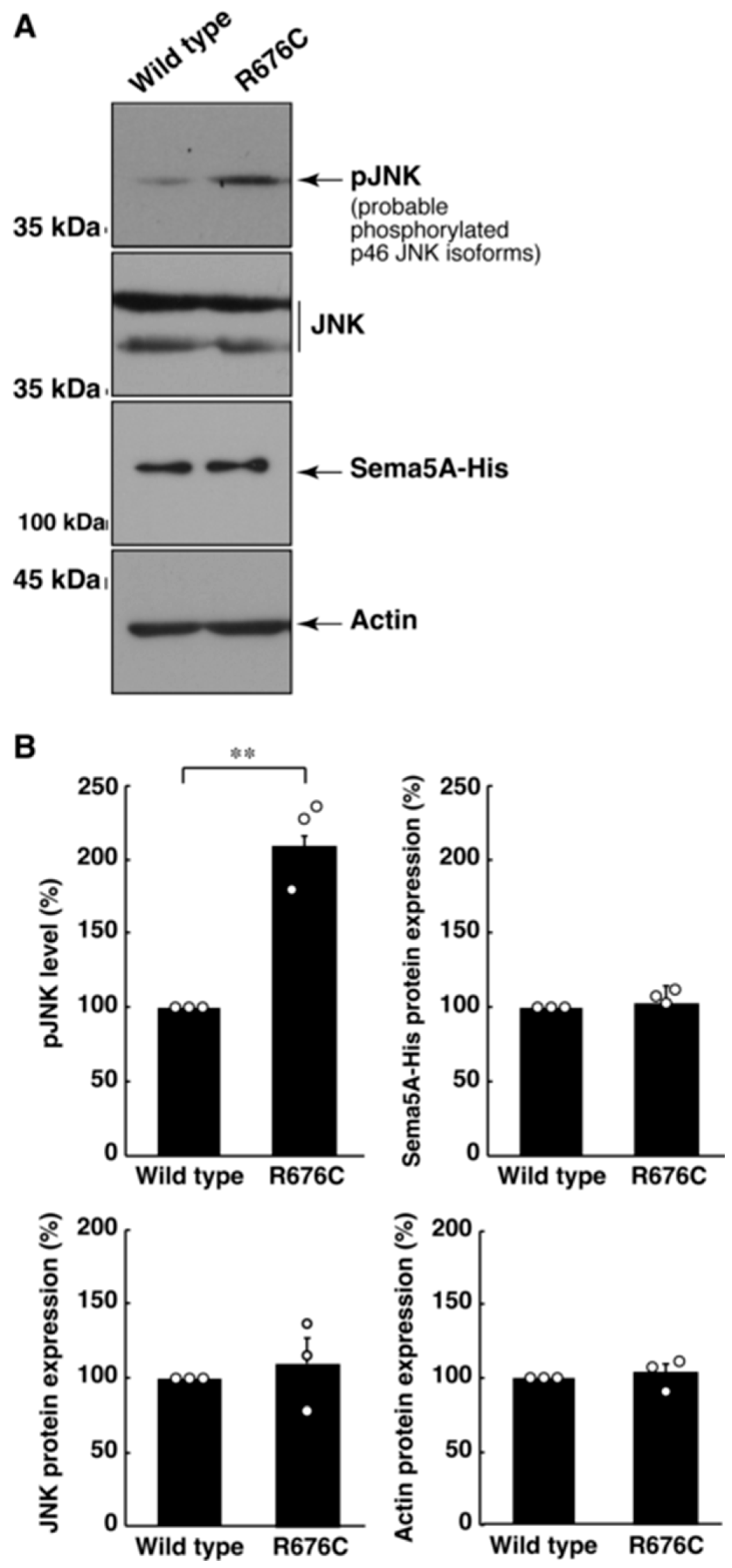
3.2. Mutated Sema5A Excessively Leads to Morphological Differentiation
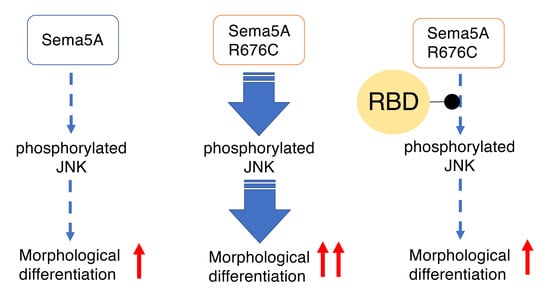
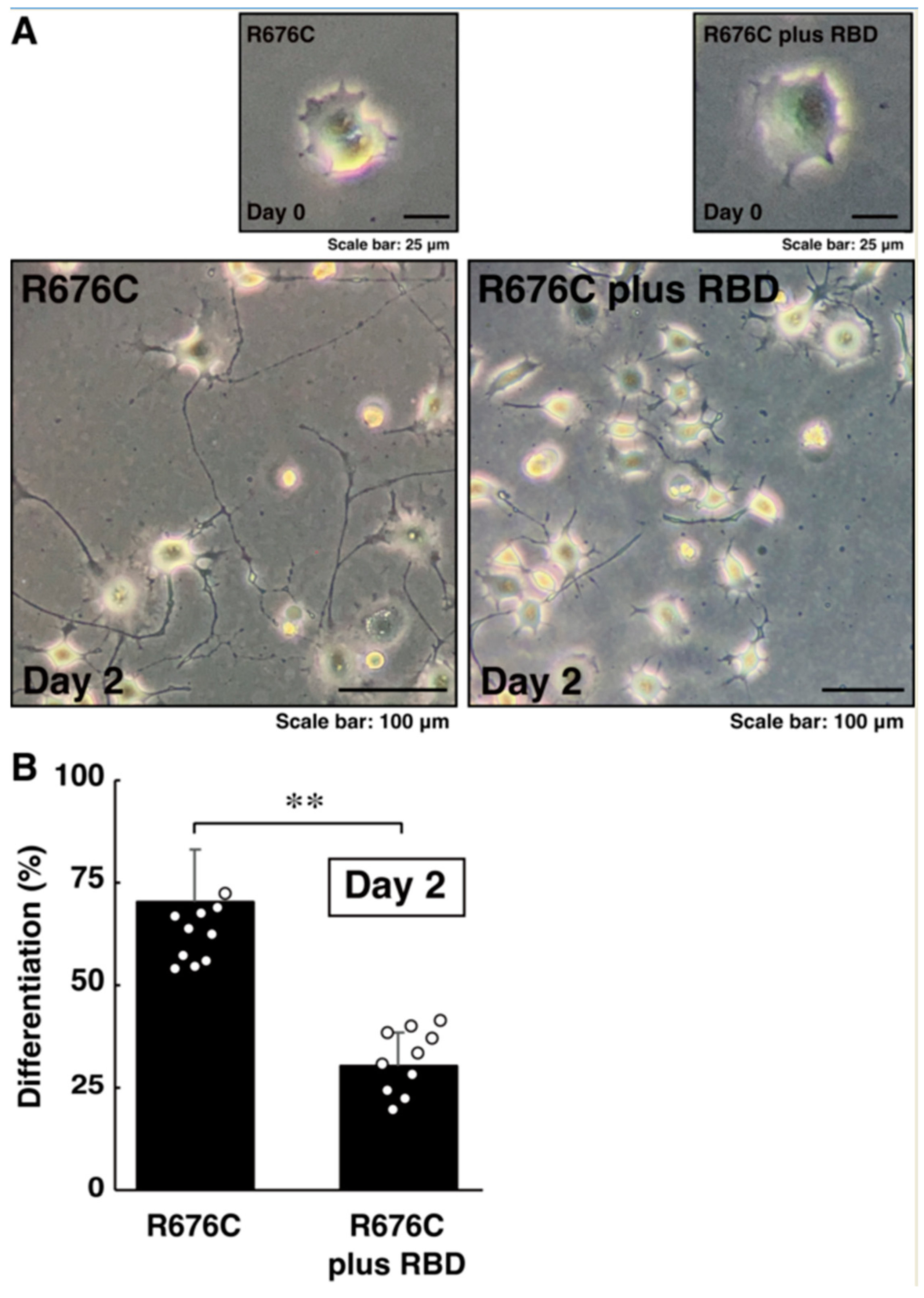
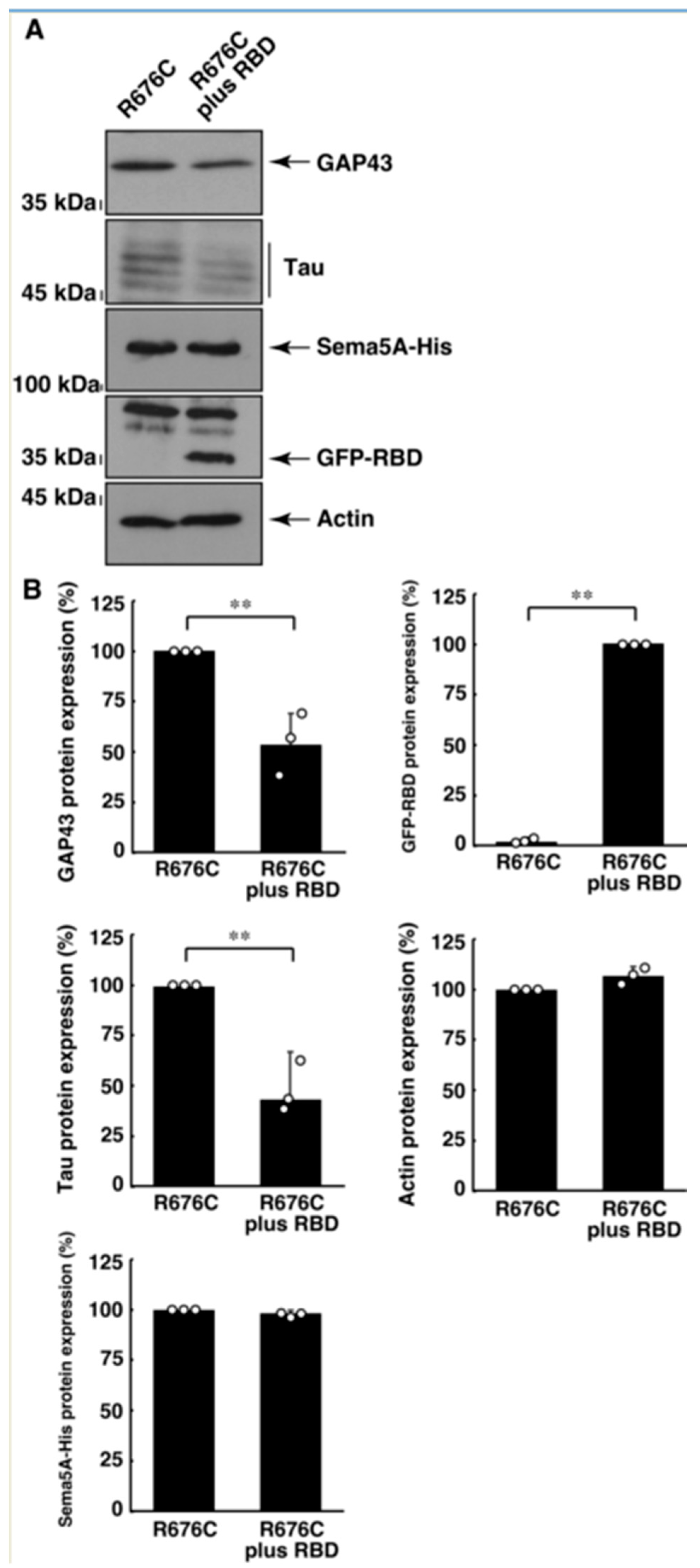
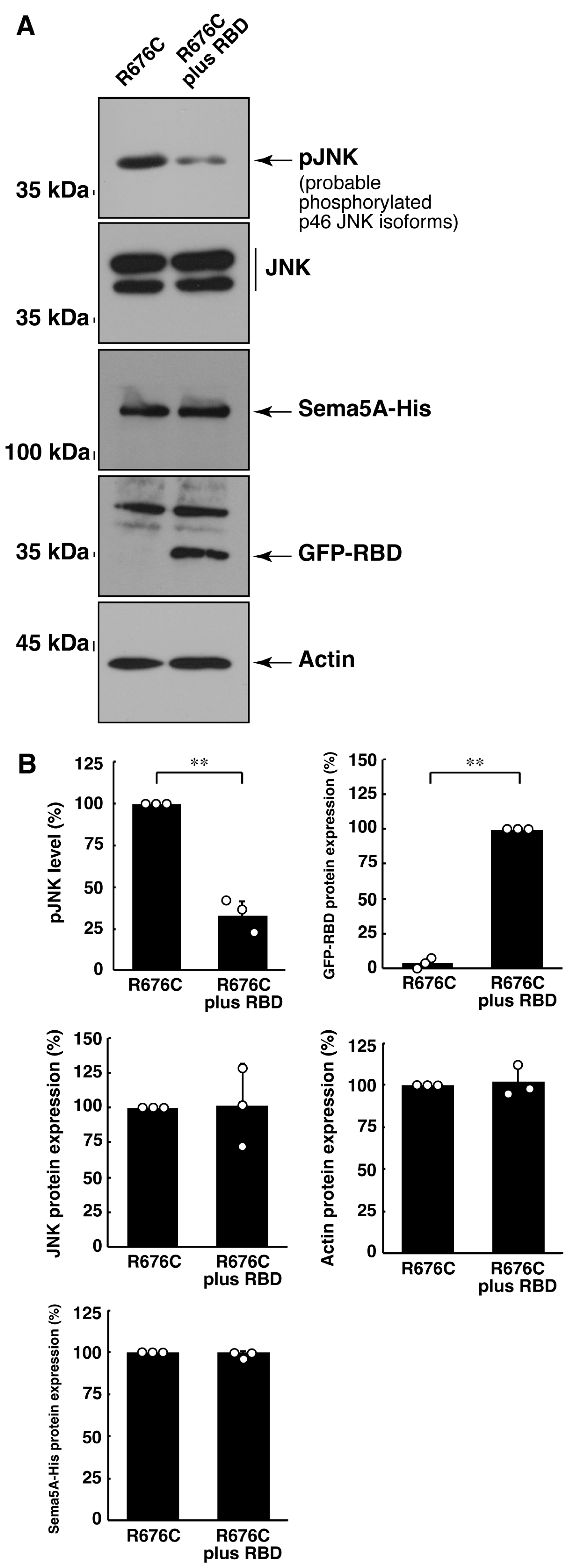
3.3. RhoG-Binding Domain of Elmo1 Can Recover the Excessive Differentiation Phenotypes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernier, R.; Mao, A.; Yen, J. Psychopathology, families, and culture: Autism. Child Adolesc. Psychiatr. Clin. N. Am. 2010, 19, 855–867. [Google Scholar]
- Mazza, M.; Pino, M.C.; Mariano, M.; Tempesta, D.; Ferrana, M.; Berardis, D.D.; Masedu, F.; Valenti, M. Affective and cognitive empathy in adolescents with autism spectrum disorder. Front. Hum. Neurosci. 2014, 8, 791. [Google Scholar] [CrossRef] [PubMed]
- Arnett, A.B.; Trinh, S.; Bernier, R.A. The state of research on the genetics of autism spectrum disorder: Methodological, clinical and conceptual progress. Curr. Opin. Psychol. 2019, 27, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.L.; Earl, R.; Fox, E.A.; Ma, R.; Haidar, G.; Pepper, M.; Berliner, L.; Wallace, A.; Bernier, R. Trauma and autism spectrum disorder: Review, proposed treatment adaptations and future directions. J. Child. Adolesc. Trauma 2019, 12, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Modabbernia, A.; Velthorst, E.; Reichenberg, A. Environmental risk factors for autism: An evidence-based review of systematic reviews and meta-analyses. Mol. Autism. 2017, 8, 13. [Google Scholar] [CrossRef]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism spectrum disorder: Neuropathology and animal models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef]
- Genovese, A.; Butler, M.G. Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD). Int. J. Mol. Sci. 2020, 21, 4726. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Wu, C.; Wang, J.; Sun, M. Autism spectrum disorder: Neurodevelopmental risk factors, biological mechanism, and precision therapy. Int. J. Mol. Sci. 2023, 24, 1819. [Google Scholar] [CrossRef]
- Craig, A.M.; Banker, G. Neuronal polarity. Annu. Rev. Neurosci. 1994, 17, 267–310. [Google Scholar] [CrossRef]
- da Silva, J.S.; Dotti, C.G. Breaking the neuronal sphere: Regulation of the actin cytoskeleton in neuritogenesis. Nat. Rev. Neurosci. 2002, 3, 694–704. [Google Scholar] [CrossRef]
- Arimura, N.; Kaibuchi, K. Neuronal polarity: From extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 2007, 8, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Poo, M.M. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 2013, 14, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Bray, D. Surface movements during the growth of single explanted neurons. Proc. Natl. Acad. Sci. USA 1970, 65, 905–910. [Google Scholar] [CrossRef]
- Rigby, M.J.; Gomez, T.M.; Puglielli, L. Glial cell-axonal growth cone interactions in neurodevelopment and regeneration. Front. Neurosci. 2020, 14, 203. [Google Scholar] [CrossRef] [PubMed]
- Worzfeld, T.; Offermanns, S. Semaphorins and plexins as therapeutic targets. Nat. Rev. Drug Discov. 2014, 13, 603–621. [Google Scholar] [CrossRef]
- Lin, L.; Lesnick, T.G.; Maraganore, D.M.; Isacson, O. Axon guidance and synaptic maintenance: Preclinical markers for neurodegenerative disease and therapeutics. Trends. Neurosci. 2009, 32, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Limoni, G.; Niquille, M. Semaphorins and Plexins in central nervous system patterning: The key to it all? Curr. Opin. Neurobiol. 2021, 66, 224–232. [Google Scholar] [CrossRef]
- Zang, Y.; Chaudhari, K.; Bashaw, G.J. New insights into the molecular mechanisms of axon guidance receptor regulation and signaling. Curr. Top. Dev. Biol. 2021, 142, 147–196. [Google Scholar]
- Matsuoka, R.L.; Chivatakarn, O.; Badea, T.C.; Samuels, I.S.; Cahill, H.; Katayama, K.; Kumar, S.R.; Suto, F.; Chédotal, A.; Peachey, N.S.; et al. Class 5 transmembrane semaphorins control selective mammalian retinal lamination and function. Neuron 2011, 71, 460–473. [Google Scholar] [CrossRef]
- Goldberg, J.L.; Vargas, M.E.; Wang, J.T.; Mandemakers, W.; Oster, S.F.; Sretavan, D.W.; Barres, B.A. An oligodendrocyte lineage-specific semaphorin, Sema5A, inhibits axon growth by retinal ganglion cells. J. Neurosci. 2004, 24, 4989–4999. [Google Scholar] [CrossRef]
- Melin, M.; Carlsson, B.; Anckarsater, H.; Rastam, M.; Betancur, C.; Isaksson, A.; Gillberg, C.; Dahl, N. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology 2006, 54, 64–69. [Google Scholar] [CrossRef]
- Zhang, B.; Willing, M.; Grange, D.K.; Shinawi, M.; Manwaring, L.; Vineyard, M.; Kulkarni, S.; Cottrell, C.E. Multigenerational autosomal dominant inheritance of 5p chromosomal deletions. Am. J. Med. Genet. A 2016, 170, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Morishita, R.; Nagata, K.I. Autism spectrum disorder-associated genes and the development of dentate granule cells. Med. Mol. Morphol. 2017, 50, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Mosca-Boidron, A.L.; Gueneau, L.; Huguet, G.; Goldenberg, A.; Henry, C.; Gigot, N.; Pallesi-Pocachard, E.; Falace, A.; Duplomb, L.; Thevenon, J.; et al. A de novo microdeletion of SEMA5A in a boy with autism spectrum disorder and intellectual disability. Eur. J. Hum Genet. 2016, 24, 838–843. [Google Scholar] [CrossRef]
- Depienne, C.; Magnin, E.; Bouteiller, D.; Stevanin, G.; Saint-Martin, C.; Vidailhet, M.; Apartis, E.; Hirsch, E.; LeGuern, E.; Labauge, P.; et al. Familial cortical myoclonic tremor with epilepsy: The third locus (FCMTE3) maps to 5p. Neurology 2010, 74, 2000–2003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Z.; Lin, Z.; Zhang, R.; Lu, Y.; Su, W.; Li, F.; Xu, X.; Tu, M.; Lou, Y.; et al. De novo germline mutations in SEMA5A associated with infantile spasms. Front. Genet. 2019, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Gunn, R.K.; Huentelman, M.J.; Brown, R.E. Are Sema5a mutant mice a good model of autism? A behavioral analysis of sensory systems, emotionality and cognition. Behav. Brain Res. 2011, 225, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wang, S.H.; Song, J.; Mironova, Y.; Ming, G.L.; Kolodkin, A.L.; Giger, R.J. Semaphorin 5A inhibits synaptogenesis in early postnatal- and adult-born hippocampal dentate granule cells. Elife 2014, 3, e04390. [Google Scholar] [CrossRef]
- Hirose, M.; Ishizaki, T.; Watanabe, N.; Uehata, M.; Kranenburg, O.; Moolenaar, W.H.; Matsumura, F.; Maekawa, M.; Bito, H.; Narumiya, S. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 1998, 141, 1625–1636. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Yamauchi, J.; Sanbe, A.; Tanoue, A. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 2007, 313, 791–804. [Google Scholar] [CrossRef]
- Cano, E.; Mahadevan, L.C. Parallel signal processing among mammalian MAPKs. Trends. Biochem. Sci. 1995, 20, 117–122. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Lu, M.; Ravichandran, K.S. Dock180-ELMO cooperation in Rac activation. Methods Enzymol. 2006, 406, 388–402. [Google Scholar]
- Tam, C.; Kukimoto-Niino, M.; Miyata-Yabuki, Y.; Tsuda, K.; Mishima-Tsumagari, C.; Ihara, K.; Inoue, M.; Yonemochi, M.; Hanada, K.; Matsumoto, T.; et al. Targeting Ras-binding domain of ELMO1 by computational nanobody design. Commun. Biol. 2023, 6, 284. [Google Scholar] [CrossRef]
- Artigiani, S.; Conrotto, P.; Fazzari, P.; Gilestro, G.F.; Barberis, D.; Giordano, S.; Comoglio, P.M.; Tamagnone, L. Plexin-B3 is a functional receptor for semaphorin 5A. EMBO Rep. 2004, 5, 710–714. [Google Scholar] [CrossRef]
- Purohit, A.; Sadanandam, A.; Myneni, P.; Singh, R.K. Semaphorin 5A mediated cellular navigation: Connecting nervous system and cancer. Biochim. Biophys. Acta. 2014, 1846, 485–493. [Google Scholar] [CrossRef]
- Altuame, F.D.; Shamseldin, H.E.; Albatti, T.H.; Hashem, M.; Ewida, N.; Abdulwahab, F.; Alkuraya, F.S. PLXNA2 as a candidate gene in patients with intellectual disability. Am. J. Med. Genet. A 2021, 185, 3859–3865. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, M.; Fan, X.; Zhou, J.; Yang, J.; Wang, L.; Zhao, Z.; Dai, C.; Zhang, Z. SEMA5A-PLXNB3 axis promotes PDAC liver metastasis outgrowth through enhancing the Warburg effect. J. Immunol. Res. 2023, 2023, 3274467. [Google Scholar] [CrossRef]
- Su, S.C.; Hsin, C.H.; Lu, Y.T.; Chuang, C.Y.; Ho, Y.T.; Yeh, F.L.; Yang, S.F.; Lin, C.W. EF-24, a curcumin analog, inhibits cancer cell invasion in human nasopharyngeal carcinoma through transcriptional suppression of matrix metalloproteinase-9 gene expression. Cancers 2023, 15, 1552. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Kim, B.N.; Kim, J.W.; Shin, M.S.; Park, T.W.; Son, J.W.; Chung, U.S.; Park, M. Polymorphism in the promoter region of SEMA5A is associated with sociality traits in Korean subjects with autism apectrum disorders. Psychiatry Investig. 2017, 14, 876–878. [Google Scholar] [CrossRef] [PubMed]
- D’Aguanno, S.; Valentini, E.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Spagnuolo, M.; Buglioni, S.; Ercolani, C.; Falcone, I.; De Dominici, M.; et al. Semaphorin 5A drives melanoma progression: Role of Bcl-2, miR-204, and c-Myb. J. Exp. Clin. Cancer Res. 2018, 37, 278. [Google Scholar] [CrossRef]
- Pijuan, J.; Ortigoza-Escobar, J.D.; Ortiz, J.; Alcalá, A.; Calvo, M.J.; Cubells, M.; Hernando-Davalillo, C.; Palau, F.; Hoenicka, J. PLXNA2 and LRRC40 as candidate genes in autism spectrum disorder. Autism Res. 2021, 14, 1088–1100. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.; Liu, L.; Wang, Y.; Chen, H.; Fan, T.; Li, J.; Xia, K.; Sun, Z. Targeted sequencing and integrative analysis of 3195 Chinese patients with neurodevelopmental disorders prioritized 26 novel candidate genes. J. Genet. Genomics 2021, 48, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Steele, J.L.; Morrow, M.M.; Sarnat, H.B.; Alkhunaizi, E.; Brandt, T.; Chitayat, D.A.; DeFilippo, C.P.; Douglas, G.V.; Dubbs, H.A.; Elloumi, H.Z.; et al. Semaphorin-Plexin signaling: From axonal guidance to a new X-linked intellectual disability syndrome. Pediatr. Neurol. 2022, 126, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, C.; Veske, A.; Krejcova, S.; Rosenberger, G.; Finckh, U. Plexin B3 promotes neurite outgrowth, interacts homophilically, and interacts with Rin. BMC Neurosci. 2005, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lee, A.Y. Semaphorin 5A and plexin-B3 inhibit human glioma cell motility through RhoGDIa-mediated inactivation of Rac1 GTPase. J. Biol. Chem. 2010, 285, 32436–32445. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Law, J.W.; Lee, A.Y. Semaphorin 5A and plexin-B3 regulate human glioma cell motility and morphology through Rac1 and the actin cytoskeleton. Oncogene 2012, 31, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Hayashi, Y.; Wu, L.; Awaji, M.; Atri, P.; Varney, M.L.; Purohit, A.; Rachagani, S.; Batra, S.K.; Singh, R.K. Pathological and functional significance of Semaphorin-5A in pancreatic cancer progression and metastasis. Oncotarget 2017, 9, 5931–5943. [Google Scholar] [CrossRef]
- Xiao, J.B.; Li, X.L.; Liu, L.; Wang, G.; Hao, S.N.; Dong, H.J.; Wang, X.M.; Zhang, Y.F.; Liu, H.D. The association of semaphorin 5A with lymph node metastasis and adverse prognosis in cervical cancer. Cancer Cell Int. 2018, 18, 87. [Google Scholar] [CrossRef]
- Zuo, Q.; Yang, Y.; Lyu, Y.; Yang, C.; Chen, C.; Salman, S.; Huang, T.Y.; Wicks, E.E.; Jackson, W., 3rd; Datan, E.; et al. Plexin-B3 expression stimulates MET signaling, breast cancer stem cell specification, and lung metastasis. Cell Rep. 2023, 42, 112164. [Google Scholar] [CrossRef]
| Reagents or Materials | Companies or Sources | Cat. Nos. | Lot. Nos. | Concentrations Used |
|---|---|---|---|---|
| Key Antibodies | ||||
| Ant-growth-associated protein 43 (GAP43) | Santa Cruz Biotechnology (Santa Cruz, CA, USA) | sc-17790 | J0920 | Immunoblotting (IB), 1:5000 |
| Anti-Tau | Santa Cruz Biotechnology | sc-21796 | H2721 | IB, 1:500 |
| Anti-Tau | Santa Cruz Biotechnology | sc-21796 | G1222 | IB, 1:500 |
| Anti-actin (also called pan-β type actin) | MBL (Tokyo, Japan) | M177-3 | 007 | IB, 1:5000 |
| Anti-c-Jun N-terminal kinase (JNK, pan-JNK [p54 isoforms]) | Santa Cruz Biotechnology | sc-7345 | J2521 | IB, 1:250 |
| Anti-phospho-c-Jun N-terminal kinase (pJNK, pan-pJNK [phosphorylated p54 isoforms]) | Santa Cruz Biotechnology | sc-6254 | C1722 | IB, 1:500 |
| anti-Rac1 | Proteintech Japan (Tokyo, Japan) | 66122-1-Ig | 10011346 | IB, 1:500 |
| anti-His | MBL | D291-3 | 002 | IB, 1:500 |
| Anti-Lys-Asp-Glu-Leu (KDEL) | MBL | M181-3 | 004 | Immunofluorescence (IF), 1:200 |
| Anti-130 kDa Golgi membrane protein (GM130) | BD Biosciences (Franklin Lakes, NJ, USA) | 610823 | 8352796 | IF, 1:200 |
| Anti-cathepsin D | Abcam (Cambridge, UK) | ab75852 | GR260148-33 | IF, 1:200 |
| Ribosomal protein S6 (RPS6) | Santa Cruz Biotechnology | sc-74459 | D2921 | IB, 1:500 |
| Anti-Plexin-A2 | Santa Cruz Biotechnology | sc-39393 | F0719 | Neutralization, 1 microgram per 1 ml of medium |
| Anti-Plexin-A3 | Santa Cruz Biotechnology | sc-37466 | C1923 | Neutralization, 1 microgram per 1 ml of medium |
| Anti-Plexin-B3 | Atlas Antibodies (Bromma, Sweden) | HPA048046 | 23445 | Neutralization, 1 microgram per 1 ml of medium |
| IgG (Mouse control IgG) | Cosmo Bio (Tokyo, Japan) | 65124-1-lg | U0000635 | Neutralization, 1 microgram per 1 ml of medium |
| Anti-IgG (H+L chain) (Rabbit) pAb-HRP | MBL | 458 | 353 | IB, 1:5000 |
| Anti-IgG (H+L chain) (Mouse) pAb-HRP | MBL | 330 | 365 | IB, 1:5000 |
| Alexa Fluor TM 488 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific (Waltham, MA, USA) | A11001 | 774-9040 | IF, 1:500 |
| Alexa Fluor TM 594 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | A11005 | 226-8383 | IF, 1:500 |
| Alexa Fluor TM 488 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | A11008 | 075-1094 | IF, 1:500 |
| Alexa Fluor TM 594 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | A11012 | 201-8240 | IF, 1:500 |
| Chemicals | ||||
| Curcumin | Santa Cruz Biotechnology | sc-200509 | E3118 | Final concentration, 10 microM |
| Dimethyl sulfoxide (DMSO) | FUJIFILM Wako Pure Chemical Corporation (Tokyo, Japan) | 047-29353 | CDN0170 | Final concentration, less than 0.1% |
| Recombinant DNAs | ||||
| pCMV-SV40ori | A control constract deleted from Cat. No. 72035 (Addgene, Cambridge, MA, USA) | n.d. | n.d. | 1.25 microgram of DNA per 6 cm dish |
| pCMV-SV40ori-mouse Sema5A-His | Addgene | 72035 | n.d. | 1.25 microgram of DNA per 6 cm dish |
| pCMV-SV40ori-mouse Sema5A(R676C)-His | Generated in this study | n.d. | n.d. | 1.25 microgram of DNA per 6 cm dish |
| pCMV-SV40ori-mouse Sema5A(R951C)-His | Generated in this study | n.d. | n.d. | 1.25 microgram of DNA per 6 cm dish |
| pcDNA3.1-N-eGFP-RhoG-binding domain (RBD, amino acids 1-81) of human Elmo1 (synthesized in this study) | GenScript | J196YHL120-6 | RG245690 | 1.25 microgram of DNA per 6 cm dish |
| pET42a-conserved Cdc42 and Rac interactive binding (CRIB) domain of human Pak1 | Miyamoto, Y., Torii, T., Yamamori, N., Ogata, T., Tanoue, A., Yamauchi, J. Sci. Signal. 2013 6:ra15 | n.d. | n.d. | 2.5 microgram of DNA per 1 liter culture bottle for protein production |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okabe, M.; Miyamoto, Y.; Ikoma, Y.; Takahashi, M.; Shirai, R.; Kukimoto-Niino, M.; Shirouzu, M.; Yamauchi, J. RhoG-Binding Domain of Elmo1 Ameliorates Excessive Process Elongation Induced by Autism Spectrum Disorder-Associated Sema5A. Pathophysiology 2023, 30, 548-566. https://doi.org/10.3390/pathophysiology30040040
Okabe M, Miyamoto Y, Ikoma Y, Takahashi M, Shirai R, Kukimoto-Niino M, Shirouzu M, Yamauchi J. RhoG-Binding Domain of Elmo1 Ameliorates Excessive Process Elongation Induced by Autism Spectrum Disorder-Associated Sema5A. Pathophysiology. 2023; 30(4):548-566. https://doi.org/10.3390/pathophysiology30040040
Chicago/Turabian StyleOkabe, Miyu, Yuki Miyamoto, Yuta Ikoma, Mikito Takahashi, Remina Shirai, Mutsuko Kukimoto-Niino, Mikako Shirouzu, and Junji Yamauchi. 2023. "RhoG-Binding Domain of Elmo1 Ameliorates Excessive Process Elongation Induced by Autism Spectrum Disorder-Associated Sema5A" Pathophysiology 30, no. 4: 548-566. https://doi.org/10.3390/pathophysiology30040040
APA StyleOkabe, M., Miyamoto, Y., Ikoma, Y., Takahashi, M., Shirai, R., Kukimoto-Niino, M., Shirouzu, M., & Yamauchi, J. (2023). RhoG-Binding Domain of Elmo1 Ameliorates Excessive Process Elongation Induced by Autism Spectrum Disorder-Associated Sema5A. Pathophysiology, 30(4), 548-566. https://doi.org/10.3390/pathophysiology30040040