Pathophysiology of Nociception and Rare Genetic Disorders with Increased Pain Threshold or Pain Insensitivity
Abstract
1. Introduction
2. Materials and Methods
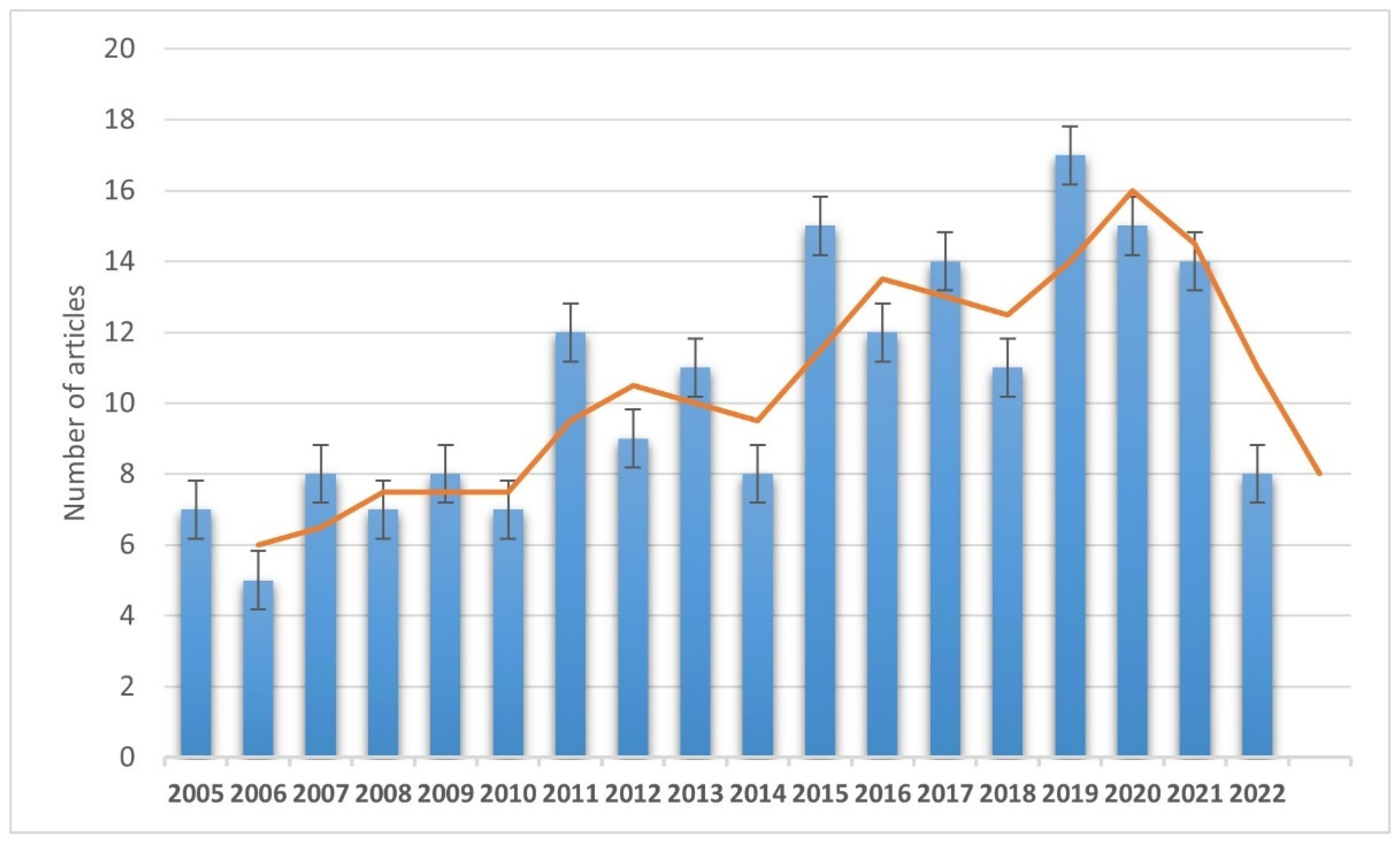
2.1. Search Strategy
2.2. Study Selection
3. Results
3.1. Features of Nociceptors
3.2. Genetic Disorders Featuring Increased Pain Threshold or Pain Insensitivity
3.2.1. Hereditary Sensory and Autonomic Neuropathies (HSANs)
HSAN-I Group
HSAN-II
HSAN-III
HSAN-IV
HSAN-V
HSAN Type VI
HSAN Type VII
HSAN Type VIII
HSAN Type IX
3.2.2. Numeric and Structural Chromosomal Abnormalities
Angelman Syndrome
Prader Willy Syndrome
Chromosome 15q Duplication Syndrome
Chromosome 4 Interstitial Deletion
4. Research and Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raja, S.N.; Carr, D.B.; Cohen, M.; Finnerup, N.B.; Flor, H.; Gibson, S.; Keefe, F.J.; Mogil, J.S.; Ringkamp, M.; Sluka, K.A.; et al. The revised International Association for the Study of Pain definition of pain: Concepts, challenges, and compromises. Pain 2020, 161, 1976–1982. [Google Scholar] [CrossRef] [PubMed]
- Zelaya, E.C.; Dahlhamer, M.J.; Lucas, W.J.; Connor, E. Chronic Pain and High-Impact Chronic Pain among U.S. Adults, 2019. The National Center for Health Statistics. Available online: https://www.cdc.gov/nchs/products/databriefs/db390.htm (accessed on 23 November 2021).
- Livshits, G.; Ni Lochlainn, M.; Malkin, I.; Bowyer, R.; Verdi, S.; Steves, C.J.; Williams, F. Shared genetic influence on frailty and chronic widespread pain: A study from TwinsUK. Age Ageing 2018, 47, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Lischka, A.; Lassuthova, P.; Çakar, A.; Record, C.J.; Van Lent, J.; Baets, J.; Dohrn, M.F.; Senderek, J.; Lampert, A.; Bennett, D.L.; et al. Genetic pain loss disorders. Nat. Rev. Dis. Prim. 2022, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Middleton, S.J.; Barry, A.M.; Comini, M.; Li, Y.; Ray, P.R.; Shiers, S.; Themistocleous, A.C.; Uhelski, M.L.; Yang, X.; Dougherty, P.M.; et al. Studying human nociceptors: From fundamentals to clinic. Brain 2021, 144, 1312–1335. [Google Scholar] [CrossRef]
- McEntire, D.M.; Kirkpatrick, D.R.; Dueck, N.P.; Kerfeld, M.J.; Smith, T.A.; Nelson, T.J.; Reisbig, M.D.; Agrawal, D.K. Pain transduction: A pharmacologic perspective. Expert Rev. Clin. Pharmacol. 2016, 9, 1069–1080. [Google Scholar] [CrossRef]
- Magerl, W.; Krumova, E.K.; Baron, R.; Tölle, T.; Treede, R.-D.; Maier, C. Reference data for quantitative sensory testing (QST): Refined stratification for age and a novel method for statistical comparison of group data. Pain 2010, 151, 598–605. [Google Scholar] [CrossRef]
- Michoud, F.; Seehus, C.; Schönle, P.; Brun, N.; Taub, D.; Zhang, Z.; Jain, A.; Furfaro, I.; Akouissi, O.; Moon, R.; et al. Epineural optogenetic activation of nociceptors initiates and amplifies in-flammation. Nat. Biotechnol. 2021, 39, 179–185. [Google Scholar] [CrossRef]
- Nagi, S.S.; Marshall, A.G.; Makdani, A.; Jarocka, E.; Liljencrantz, J.; Ridderström, M.; Shaikh, S.; O’Neill, F.; Saade, D.; Donkervoort, S.; et al. An ultrafast system for signaling mechanical pain in human skin. Sci. Adv. 2019, 5, eaaw1297. [Google Scholar] [CrossRef]
- Schmidt, R.; Schmelz, M.; Forster, C.; Ringkamp, M.; Torebjork, E.; Handwerker, H. Novel classes of responsive and unresponsive C nociceptors in human skin. J. Neurosci. 1995, 15, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Cintra, R.R.; Lins, L.C.R.F.; Medeiros, K.A.A.L.; Souza, M.F.; Gois, A.M.; Bispo, J.M.; Melo, M.S.; Leal, P.C.; Meurer, Y.S.; Ribeiro, A.M.; et al. Nociception alterations precede motor symptoms in a progressive model of parkinsonism induced by reserpine in middle-aged rats. Brain Res. Bull. 2021, 171, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Green, P.G.; Levine, J.D. Neuropathic pain-like alterations in muscle nociceptor function associated with vibration-induced muscle pain. Pain 2010, 151, 460–466. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ronchetti, S.; Migliorati, G.; Delfino, D. Association of inflammatory mediators with pain perception. Biomed. Pharmacother. 2017, 96, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Themistocleous, A.C.; Ramirez, J.D.; Shillo, P.R.; Lees, J.G.; Selvarajah, D.; Orengo, C.; Tesfaye, S.; Rice, A.S.C.; Bennett, D.L.H. The Pain in Neuropathy Study (PiNS): A cross-sectional ob-servational study determining the somatosensory phenotype of painful and painless diabetic neuropathy. Pain 2016, 157, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, V.F.; Karlsson, P.; Drummond, P.D.; Schaldemose, E.L.; Terkelsen, A.J.; Jensen, D.T.S.; Knudsen, L.F. Bilaterally Reduced Intraepidermal Nerve Fiber Density in Unilateral CRPS-I. Pain Med. 2018, 19, 2021–2030. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.; Fletcher, D.; Martin, F.; Orlikowski, D.; Sharshar, T.; Chauvin, M.; Bouhassira, D.; Attal, N. Small fibre impairment predicts neuropathic pain in Guillain–Barré syndrome. Pain 2010, 151, 53–60. [Google Scholar] [CrossRef]
- Basbaum, A.I.; Bautista, D.M.; Scherrer, G.; Julius, D. Cellular and Molecular Mechanisms of Pain. Cell 2009, 139, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.M.; McAlexander, M.A.; Bíró, T.; Szallasi, A. Transient receptor potential channels as therapeutic targets. Nat. Rev. Drug Discov. 2011, 10, 601–620. [Google Scholar] [CrossRef]
- Sałat, K.; Filipek, B. Antinociceptive activity of transient receptor potential channel TRPV1, TRPA1, and TRPM8 antagonists in neurogenic and neuropathic pain models in mice. J. Zhejiang Univ. Sci. B 2015, 16, 167–178. [Google Scholar] [CrossRef]
- Cavanaugh, D.J.; Lee, H.; Lo, L.; Shields, S.D.; Zylka, M.J.; Basbaum, A.I.; Anderson, D.J. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc. Natl. Acad. Sci. USA 2009, 106, 9075–9080. [Google Scholar] [CrossRef]
- Shiers, S.; Klein, R.M.; Price, T.J. Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain 2020, 161, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Rostock, C.; Schrenk-Siemens, K.; Pohle, J.; Siemens, J. Human vs. mouse nociceptors—Similarities and differences. Neuroscience 2018, 387, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Bimonte, S.; Cascella, M.; Forte, C.A.; Esposito, G.; Cuomo, A. The Role of Anti-Nerve Growth Factor Monoclonal Antibodies in the Control of Chronic Cancer and Non-Cancer Pain. J. Pain Res. 2021, 14, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Sikandar, S.; Minett, M.S.; Millet, Q.; Santana-Varela, S.; Lau, J.; Wood, J.N.; Zhao, J. Brain-derived neurotrophic factor derived from sensory neurons plays a critical role in chronic pain. Brain 2018, 141, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Torck, A.; Quigley, L.; Wangzhou, A.; Neiman, M.; Rao, C.; Lam, T.; Kim, J.-Y.; Kim, T.H.; Zhang, M.Q.; et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: An RNA-seq–based resource for pain and sensory neuroscience research. Pain 2018, 159, 1325–1345. [Google Scholar] [CrossRef]
- Cascella, M.; Muzio, M.R. Potential application of the Kampo medicine goshajinkigan for prevention of chemotherapy-induced peripheral neuropathy. J. Integr. Med. 2017, 15, 77–87. [Google Scholar] [CrossRef]
- Cascella, M. Chemotherapy-induced peripheral neuropathy: Limitations in current prophylactic strategies and directions for future research. Curr. Med. Res. Opin. 2017, 33, 981–984. [Google Scholar] [CrossRef]
- Schwartzlow, C.; Kazamel, M. Hereditary Sensory and Autonomic Neuropathies: Adding More to the Classification. Curr. Neurol. Neurosci. Rep. 2019, 19, 52. [Google Scholar] [CrossRef]
- Ho, K.W.; Jerath, N.U. V144D mutation of SPTLC1 can present with both painful and painless phenotypes in hered-itary sensory and autonomic neuropathies type I. Case Rep. Genet. 2018, 2018, 1898151. [Google Scholar] [CrossRef]
- Houlden, H.; King, R.; Blake, J.; Groves, M.; Love, S.; Woodward, C.; Hammans, S.; Nicoll, J.; Lennox, G.; O’Donovan, D.G.; et al. Clinical, pathological and genetic characteri-zation of hereditary sensory and autonomic neuropathy type 1 (HSAN I). Brain 2006, 129, 411–425. [Google Scholar] [CrossRef]
- Rotthier, A.; Auer-Grumbach, M.; Janssens, K.; Baets, J.; Penno, A.; Almeida-Souza, L.; Van Hoof, K.; Jacobs, A.; De Vriendt, E.; Schlotter-Weigel, B.; et al. Mutations in the SPTLC2 Subunit of Serine Palmitoyltransferase Cause Hereditary Sensory and Autonomic Neuropathy Type I. Am. J. Hum. Genet. 2010, 87, 513–522. [Google Scholar] [CrossRef]
- Zheng, W.; Yan, Z.; He, R.; Huang, Y.; Lin, A.; Huang, W.; Su, Y.; Li, S.; Zhang, V.W.; Xie, H. Identification of a novel DNMT1 mutation in a Chinese patient with hereditary sensory and autonomic neuropathy type IE. BMC Neurol. 2018, 18, 174. [Google Scholar] [CrossRef] [PubMed]
- Kornak, U.; Mademan, I.; Schinke, M.; Voigt, M.; Krawitz, P.; Hecht, J.; Barvencik, F.; Schinke, T.; Gießelmann, S.; Beil, F.T.; et al. Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain 2014, 137, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, F.B.; Simson, G.G.-V. Hereditary sensory and autonomic neuropathies: Types II, III, and IV. Orphanet J. Rare Dis. 2007, 2, 39. [Google Scholar] [CrossRef]
- Yuan, J.; Matsuura, E.; Higuchi, Y.; Hashiguchi, A.; Nakamura, T.; Nozuma, S.; Sakiyama, Y.; Yoshimura, A.; Izumo, S.; Takashima, H. Hereditary sensory and autonomic neuropathy type IID caused by an SCN9A mutation. Neurology 2013, 80, 1641–1649. [Google Scholar] [CrossRef]
- Eberhardt, M.; Nakajima, J.; Klinger, A.B.; Neacsu, C.; Hühne, K.; O’Reilly, A.O.; Kist, A.M.; Lampe, A.K.; Fischer, K.; Gibson, J.; et al. Inherited pain: Sodium channel Nav1.7 A1632T mutation causes erythromelalgia due to a shift of fast inactivation. J. Biol. Chem. 2014, 289, 1971–1980. [Google Scholar] [CrossRef]
- Carmi, S.; Hui, K.Y.; Kochav, E.; Liu, X.; Xue, J.; Grady, F.; Guha, S.; Upadhyay, K.; Ben-Avraham, D.; Mukherjee, S.; et al. Sequencing an Ashkenazi reference panel supports pop-ulationtargeted personal genomics and illuminates Jewish and European origins. Nat. Commun. 2014, 5, 4835. [Google Scholar] [CrossRef] [PubMed]
- Daneshjou, K.; Jafarieh, H.; Raaeskarami, S.-R. Congenital Insensitivity to Pain and Anhydrosis (CIPA) Syndrome: A Report of 4 Cases. Iran J. Pediatr. 2012, 22, 412–416. [Google Scholar] [PubMed]
- Indo, Y. Genetics of congenital insensitivity to pain with anhidrosis (CIPA) or hereditary sensory and autonomic neuropathy type IV. Clinical, biological and molecular aspects of mutations in TRKA(NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Clin. Auton. Res. 2002, 12, l20–l32. [Google Scholar]
- Echaniz-Laguna, A.; Altuzarra, C.; Verloes, A.; De La Banda, M.G.G.; Quijano-Roy, S.; Tudorache, R.A.; Jaxybayeva, A.; Myrzaliyeva, B.; Tazir, M.; Vallat, J.-M.; et al. NTRK1 gene-related congenital insensitivity to pain with anhidrosis: A nationwide multicenter retrospective study. Neurogenetics 2021, 22, 333–341. [Google Scholar] [CrossRef]
- Pérez-López, L.M.; Cabrera-González, M.; la Iglesia, D.G.-D.; Ricart, S.; Knörr-Giménez, G. Update Review and Clinical Presentation in Congenital Insensitivity to Pain and Anhidrosis. Case Rep. Pediatr. 2015, 2015, 589852. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, O.P.; Thornton, G.K.; Hertecant, J.; Houlden, H.; Nicholas, A.K.; Cox, J.J.; Rielly, M.; Al-Gazali, L.; Woods, C.G. A novel NGF mutation clarifies the molecular mechanism and ex-tends the phenotypic spectrum of the HSAN5 neuropathy. J. Med. Genet. 2011, 48, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Einarsdottir, E.; Carlsson, A.; Minde, J.; Toolanen, G.; Svensson, O.; Solders, G.; Holmgren, G.; Holmberg, D.; Holmberg, M. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum. Mol. Genet. 2004, 13, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Cinnamon, Y.; Jalas, C.; Shaag, A.; Maayan, C.; Axelrod, F.B.; Elpeleg, O. Hereditary sensory autonomic neu-ropathy caused by a mutation in dystonin. Ann. Neurol. 2012, 71, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, F.; Parisi, S.; Nolano, M.; Tao, F.; Paladino, S.; Pisciotta, C.; Tozza, S.; Nesti, C.; Rebelo, A.P.; Provitera, V.; et al. Novel mutations in dystonin provide clues to the pathomechanisms of HSAN-VI. Neurology 2017, 88, 2132–2140. [Google Scholar] [CrossRef] [PubMed]
- Leipold, E.; Liebmann, L.; Korenke, G.C.; Heinrich, T.; Giesselmann, S.; Baets, J.; Ebbinghaus, M.; Goral, R.O.; Stödberg, T.; Hennings, J.C.; et al. A de novo gain-of-function mutation in SCN11A causes loss of pain perception. Nat. Genet. 2013, 45, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Priest, B.T.; Murphy, B.A.; Lindia, J.A.; Diaz, C.; Abbadie, C.; Ritter, A.M.; Liberator, P.; Iyer, L.M.; Kash, S.F.; Kohler, M.G.; et al. Contribution of the tetrodotoxin-resistant voltage-gated sodium channel nav1.9 to sensory transmission and nociceptive behavior. Proc. Natl. Acad. Sci. USA 2005, 102, 9382–9387. [Google Scholar] [CrossRef]
- Woods, C.G.; Babiker, M.O.E.; Horrocks, I.; Tolmie, J.; Kurth, I. The phenotype of congenital insensitivity to pain due to the NaV1.9 variant p.L811P. Eur. J. Hum. Genet. 2015, 23, 561–563. [Google Scholar] [CrossRef]
- Salvatierra, J.; Diaz-Bustamante, M.; Meixiong, J.; Tierney, E.; Dong, X.; Bosmans, F. A disease mutation reveals a role for NaV1.9 in acute itch. J. Clin. Investig. 2018, 128, 5434–5447. [Google Scholar] [CrossRef]
- Chen, Y.C.; Auer-Grumbach, M.; Matsukawa, S.; Zitzelsberger, M.; Themistocleous, A.C.; Strom, T.M.; Samara, C.; Moore, A.W.; Cho, L.T.; Young, G.T.; et al. Transcrip-tional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015, 47, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sharif, S.M.; Chen, Y.-C.; Valente, E.-M.; Ahmed, M.; Sheridan, E.; Bennett, C.; Woods, G. Clinical features for diagnosis and management of patients with PRDM12 congenital insensitivity to pain. J. Med. Genet. 2016, 53, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Oz-Levi, D.; Ben-Zeev, B.; Ruzzo, E.K.; Hitomi, Y.; Gelman, A.; Pelak, K.; Anikster, Y.; Reznik-Wolf, H.; Bar-Joseph, I.; Olender, T.; et al. Mutation in TECPR2 Reveals a Role for Autophagy in Hereditary Spastic Paraparesis. Am. J. Hum. Genet. 2012, 91, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Patwari, P.P.; Wolfe, L.F.; Sharma, G.D.; Berry-Kravis, E. TECPR2 mutation–associated respiratory dysregulation: More than central apnea. J. Clin. Sleep Med. 2020, 16, 977–982. [Google Scholar] [CrossRef]
- Neuser, S.; Brechmann, B.; Heimer, G.; Brösse, I.; Schubert, S.; O’Grady, L.; Zech, M.; Srivastava, S.; A Sweetser, D.; Dincer, Y.; et al. Clinical, neuroimaging and molecular spectrum of TECPR2-associated hereditary sensory and autonomic neuropathy with intellectual disability. Hum. Mutat. 2021, 42, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, L.G.; Lupski, J.R. Molecular Mechanisms for Constitutional Chromosomal Rearrangements in Humans. Annu. Rev. Genet. 2000, 34, 297–329. [Google Scholar] [CrossRef]
- Yang, L.; Shu, X.; Mao, S.; Wang, Y.; Du, X.; Zou, C. Genotype–Phenotype Correlations in Angelman Syndrome. Genes 2021, 12, 987. [Google Scholar] [CrossRef] [PubMed]
- Artigas-Pallarés, J.; Brun-Gasca, C.; Gabau-Vila, E.; Guitart-Feliubadaló, M.; Camprubí-Sánchez, C. Aspectos médicos e comportamentais da síndroma de Angelman. Rev. Neurol. 2005, 41, 649–656. [Google Scholar] [PubMed]
- Larson, A.M.; Shinnick, J.E.; Shaaya, E.A.; Thiele, E.A.; Thibert, R.L. Angelman syndrome in adulthood. Am. J. Med. Genet. Part A 2015, 167, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Cabo, R.; Tan, W.; Tayag, R.; Bird, L.M. Healthcare burden among individuals with Angelman syndrome: Findings from the Angelman Syndrome Natural History Study. Mol. Genet. Genom. Med. 2019, 7, e00734. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Bimonte, S.; Saettini, F.; Muzio, M.R. The challenge of pain assessment in children with cognitive disabilities: Features and clinical applicability of different observational tools. J. Paediatr. Child Health 2019, 55, 129–135. [Google Scholar] [CrossRef]
- Butler, M.G.; Miller, J.L.; Forster, J.L. Prader-Willi Syndrome—Clinical Genetics, Diagnosis and Treatment Approaches: An Update. Curr. Pediatr. Rev. 2019, 15, 207–244. [Google Scholar] [CrossRef]
- Angulo, M.A.; Butler, M.G.; Cataletto, M.E. Prader-Willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Investig. 2015, 38, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Priano, L.; Miscio, G.; Grugni, G.; Milano, E.; Baudo, S.; Sellitti, L.; Picconi, R.; Mauro, A. On the origin of sensory impair-ment and altered pain perception in Prader-Willi syndrome: A neurophysiological study. Eur. J. Pain 2009, 13, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Luchsinger, K.; Lau, H.; Hedlund, J.L.; Friedman, D.; Krushel, K.; Devinsky, O. Parental-reported pain insensitivity in Dup15q. Epilepsy Behav. 2016, 55, 124–127. [Google Scholar] [CrossRef]
- Cascella, M.; Muzio, M.R. Pain insensitivity in a child with a de novo interstitial deletion of the long arm of the chromosome 4: Case report. Rev. Chil. Pediatr. 2017, 88, 411–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shields, S.D.; Deng, L.; Reese, R.M.; Dourado, M.; Tao, J.; Foreman, O.; Chang, J.H.; Hackos, D.H. Insensitivity to Pain upon Adult-Onset Deletion of Nav1.7 or Its Blockade with Selective Inhibitors. J. Neurosci. 2018, 38, 10180–10201. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yu, G.; Su, R. Effects of ralfinamide in models of nerve injury and chemotherapy-induced neuropathic pain. Eur. J. Pharmacol. 2018, 823, 27–34. [Google Scholar] [CrossRef]
- Niu, H.-L.; Liu, Y.-N.; Xue, D.-Q.; Dong, L.-Y.; Liu, H.-J.; Wang, J.; Zheng, Y.-L.; Zou, A.-R.; Shao, L.-M.; Wang, K. Inhibition of Nav1.7 channel by a novel blocker QLS-81 for alleviation of neuropathic pain. Acta Pharmacol. Sin. 2021, 42, 1235–1247. [Google Scholar] [CrossRef]
- Chandra, S.; Wang, Z.; Tao, X.; Chen, O.; Luo, X.; Ji, R.R.; Bortsov, A.V. Computer-aided Discovery of a New Nav1.7 In-hibitor for Treatment of Pain and Itch. Anesthesiology 2020, 133, 611–627. [Google Scholar] [CrossRef]
- Waxman, S.G.; Merkies, I.S.J.; Gerrits, M.M.; Dib-Hajj, S.D.; Lauria, G.; Cox, J.J.; Wood, J.N.; Woods, C.G.; Drenth, J.P.H.; Faber, C.G. Sodium channel genes in pain-related disorders: Phenotype–genotype associations and recommendations for clinical use. Lancet Neurol. 2014, 13, 1152–1160. [Google Scholar] [CrossRef]
- Weiss, J.; Pyrski, M.; Jacobi, E.; Bufe, B.; Willnecker, V.; Schick, B.; Zizzari, P.; Gossage, S.J.; Greer, C.A.; Leinders-Zufall, T.; et al. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature 2011, 472, 186–190. [Google Scholar] [CrossRef]
- Pereira, V.; Millet, Q.; Aramburu, J.; Lopez-Rodriguez, C.; Gaveriaux-Ruff, C.; Wood, J.N. Analgesia linked to Nav1.7 loss of function requires μ- and δ-opioid receptors. Wellcome Open Res. 2018, 3, 101. [Google Scholar] [CrossRef] [PubMed]
- Sopacua, M.; Hoeijmakers, J.G.J.; Merkies, I.S.J.; Lauria, G.; Waxman, S.G.; Faber, C.G. Small-fiber neuropathy: Expanding the clinical pain universe. J. Peripher. Nerv. Syst. 2019, 24, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Hirose, M.; Kuroda, Y.; Murata, E. NGF/TrkA Signaling as a Therapeutic Target for Pain. Pain Pract. 2016, 16, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Long, P.; Wang, Y.; Ma, W. NTRK Fusions and TRK Inhibitors: Potential Targeted Therapies for Adult Glioblastoma. Front. Oncol. 2020, 10, 593578. [Google Scholar] [CrossRef] [PubMed]
- Tamim-Yecheskel, B.-C.; Fraiberg, M.; Kokabi, K.; Freud, S.; Shatz, O.; Marvaldi, L.; Subic, N.; Brenner, O.; Tsoory, M.; Eilam-Altstadter, R.; et al. A tecpr2 knockout mouse exhibits age-dependent neuroaxonal dystrophy associated with autophagosome accumulation. Autophagy 2021, 17, 3082–3095. [Google Scholar] [CrossRef]
- Bimonte, S.; Barbieri, A.; Cascella, M.; Rea, D.; Palma, G.; Del Vecchio, V.; Forte, C.A.; Del Prato, F.; Arra, C.; Cuomo, A. The effects of naloxone on human breast cancer progression: In vitro and in vivo studies on MDA.MB231 cells. Onco Targets Ther. 2018, 11, 185–191. [Google Scholar] [CrossRef]
- Desai, N.; Arora, N.; Gupta, A. Chemotherapy-Induced Peripheral Neuropathy. JAMA Intern. Med. 2022, 182, 766–767. [Google Scholar] [CrossRef]
- MacDonald, D.I.; Sikandar, S.; Weiss, J.; Pyrski, M.; Luiz, A.P.; Millet, Q.; Emery, E.C.; Mancini, F.; Iannetti, G.D.; Alles, S.R.; et al. A central mechanism of analgesia in mice and humans lacking the sodium channel NaV1.7. Neuron 2021, 109, 1497–1512.e6. [Google Scholar] [CrossRef]
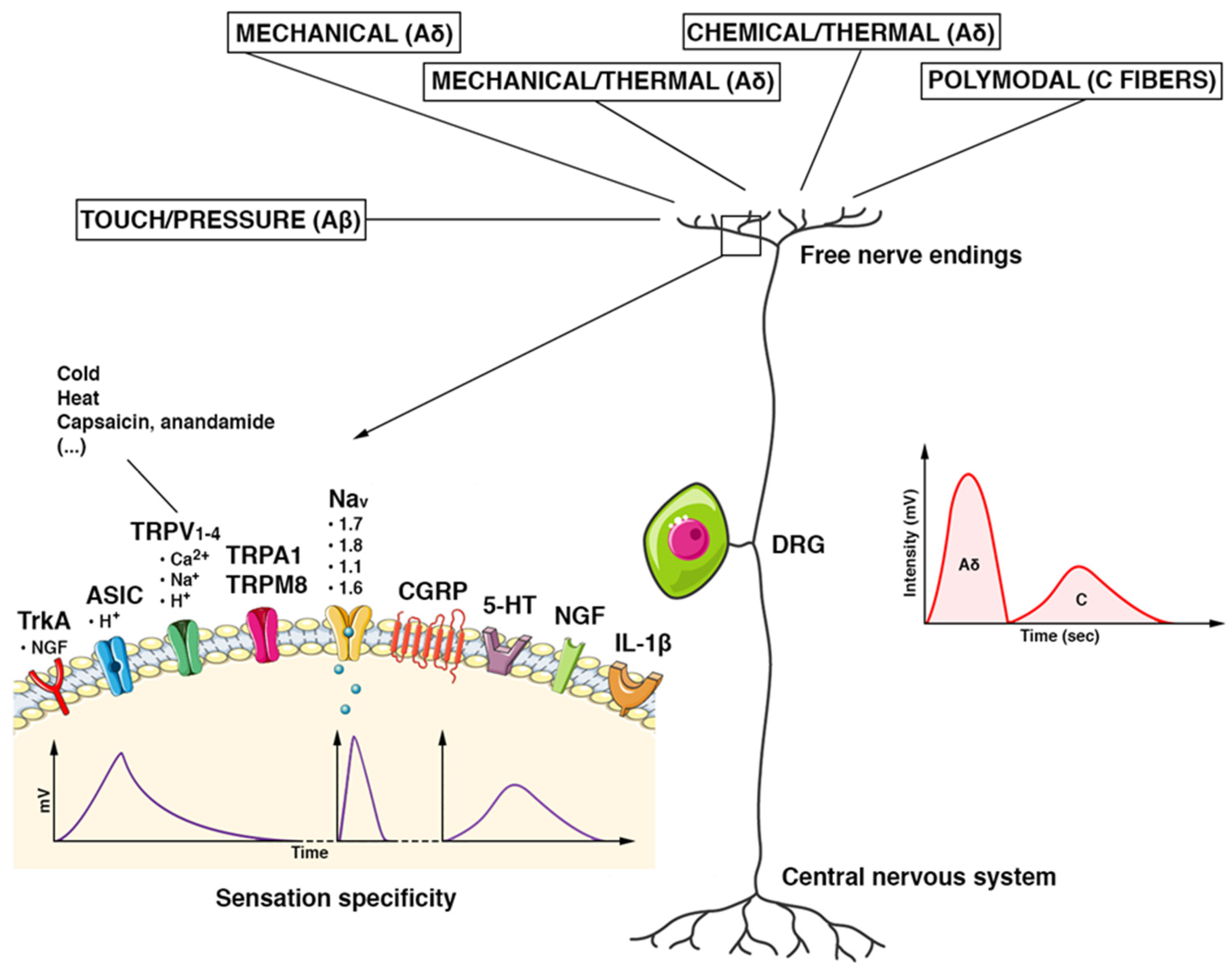
| Type | Structure | Fiber and Diameters | Axonal Conduction Velocities | Location | Stimuli | Act. Thresh. | Effects |
|---|---|---|---|---|---|---|---|
| Free nerve endings | Minimally specialized nerve endings | C | 0.5–2 m/s | Skin | Polymodal Temperature | High | Persisting pain sensations |
| Free nerve endings | Minimally specialized nerve endings | Aδ | 3–30 m/s | Skin | Pressure Temperature | High | Rapid, sharp pain |
| Free nerve endings | Minimally specialized nerve endings | Aβ | 6–12 m/s | Skin | Pressure | High | Rapid, sharp pain |
| Meissner’s corpuscles * | Encapsulated; dermal papillae of glabrous skin | Aβ 6–12 μm | — | Most common mechanoreceptors of “glabrous” skin | Touch, pressure (dynamic) | Low | Somatosensory acuity, (digital extremities and palmar skin) |
| Pacinian corpuscles | Encapsulated; onion-like covering | Aβ 6–12 μm | — | Skin, Subcutaneous tissues, viscera | Vibration (dynamic, 250 Hz) Deep pressure (quasi-static) | Low | Location of touch sensations |
| Merkel’s disks ° | Encapsulated; associated with peptide- releasing cells | Aβ | — | Skin, hair follicles | Low vibrations (5–15 Hz) | Low | Information on pressure, position, and deep static touch |
| Ruffini’s corpuscles | Enlarged dendritic endings. Elongated capsules | Aβ 6–12 μm | — | Skin (deep layers) | Mechanical deformation. Thermoreceptors | Low | Kinesthetic control |
| Muscle spindles | Highly specialized (intrafusal fibers) | Ia and II | — | Muscles | Muscle length | Low | Stretch reflex |
| Golgi tendon organs ^ | Highly specialized | Ib | — | Tendons | Muscle tension | Low | Golgi tendon reflex (sensory component) |
| Joint receptors | Minimally specialized | — | — | Joints | Joint excursion | Low | Limbs position and movement |
| Disease | Gene(s) | Encoding/Inheritance | Pain Features | Ref. |
|---|---|---|---|---|
| HSAN Type I | SPTLC1 (HSAN-IA); SPTLC2 (HSAN-IC) | Reduced activity of serine palmitoyltransferase; Autosomal dominant | Painless injuries to the tongue and limbs | [29,30,31,32,33] |
| HSAN Type II | WNK1 (HSAN-IIA) KIF1A FAM134B SCN9A (HSAN-IID) | TRPV4 (WNK1 gene); Nav1.7 (loss of function of SCN9A); Kinesines (KIF1A) Autosomal recessive | Congenital insensitivity to pain. Global pain insensitivity | [34,35,36] |
| HSAN Type III | IKBKAP | ELP1; Autosomal recessive | Pain insensitivity or reduced pain sensitivity | [28,37] |
| HSAN Type IV | TRKA | Autosomal recessive | Congenital insensitivity to pain with anhidrosis (CIPA); Global pain insensitivity | [38,39,40,41] |
| HSAN Type V | NGFβ | NGF; Autosomal recessive | Congenital insensibility to pain, lack of temperature sensing; reduced C-fibres | [42,43] |
| HSAN Type VI | DST | Dystonin; Autosomal recessive | Reduced pain sensitivity | [44,45] |
| HSAN Type VII | SCN11A | NaV1.9 channel. Gain of function with sustained depolarization Autosomal dominant | Decreased pain sensitivity | [46,47,48,49] |
| HSAN Type VIII | PRDM12 | PRDM12; Autosomal recessive | Non-global pain insensitivity | [50,51] |
| HSAN Type IX | TECPR2 | Autosomal recessive | Decreased pain sensitivity | [52,53,54] |
| Disease | Genetics | Encoding/Inheritance/Pain Features | Ref. |
|---|---|---|---|
| Angelman syndrome | Deletion or mutation in the maternal chromosome region containing the UBE3A gene | High resistance to pain in about two-thirds of patients (67%) | [56,57,58,59,60] |
| Prader Willy syndrome | Paternal 15q11.2–q13 deletion | A high pain threshold is a very common finding | [61,62,63] |
| Chromosome 15q duplication syndrome | Duplication of the PWACR region (15q11.2–q13.1) | High pain threshold in 87% | [64] |
| Chromosome 4 interstitial deletion | Cr4 del q31.3 to q32.2 | High pain threshold with preserved tactile sensitivity | [65] |
| Disease | Study type | Intervention(s) | Outcome | Study ID * |
|---|---|---|---|---|
| HSAN-I | RCT | L-serine supplementation (400 mg/kg/d, 2 years) | Disease severity ° | NCT01733407 |
| HSAN-III | RCT | Proprioceptive inputs (cortical representation) | Proprioception and Sensorimotor Control | NCT02876939 |
| HSAN-III | Observational Prospective | N/A | Database. Predictive biomarkers of disease progression and severity | NCT03920774 |
| Rare Disease | Observational Prospective | N/A | Genetics; Pain; Diagnosis (e.g., skin biopsy) | NCT02696746 |
| Rare Diseases ^ | Observational Prospective | N/A | Patient Registry & Natural History Study | NCT01793168 |
| Pain sensitivity alterations † | Observational Prospective | N/A | Pain score, Measures of quality of life, Neurophysiology, Imaging | NCT02696746 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cascella, M.; Muzio, M.R.; Monaco, F.; Nocerino, D.; Ottaiano, A.; Perri, F.; Innamorato, M.A. Pathophysiology of Nociception and Rare Genetic Disorders with Increased Pain Threshold or Pain Insensitivity. Pathophysiology 2022, 29, 435-452. https://doi.org/10.3390/pathophysiology29030035
Cascella M, Muzio MR, Monaco F, Nocerino D, Ottaiano A, Perri F, Innamorato MA. Pathophysiology of Nociception and Rare Genetic Disorders with Increased Pain Threshold or Pain Insensitivity. Pathophysiology. 2022; 29(3):435-452. https://doi.org/10.3390/pathophysiology29030035
Chicago/Turabian StyleCascella, Marco, Maria Rosaria Muzio, Federica Monaco, Davide Nocerino, Alessandro Ottaiano, Francesco Perri, and Massimo Antonio Innamorato. 2022. "Pathophysiology of Nociception and Rare Genetic Disorders with Increased Pain Threshold or Pain Insensitivity" Pathophysiology 29, no. 3: 435-452. https://doi.org/10.3390/pathophysiology29030035
APA StyleCascella, M., Muzio, M. R., Monaco, F., Nocerino, D., Ottaiano, A., Perri, F., & Innamorato, M. A. (2022). Pathophysiology of Nociception and Rare Genetic Disorders with Increased Pain Threshold or Pain Insensitivity. Pathophysiology, 29(3), 435-452. https://doi.org/10.3390/pathophysiology29030035







