Mapping Research on miRNAs in Cancer: A Global Data Analysis and Bibliometric Profiling Analysis
Abstract
:1. Introduction
2. Study Objective
3. Methods
3.1. Selection Criteria
- The keywords were chosen based on the number of queries and the bibliometric graph’s density variation.
- These keywords appear in the majority of miRNA in cancer research papers.
3.2. Inclusion Criteria
- miRNA expression papers focused on cancer prognosis or diagnosis or clinical cancer or clinical outcomes or treatments.
- The definition also comprises studies that are designed to evaluate the miRNAs that are downregulated or upregulated in cancer patients (specifically the magnitude of down/upregulation and their impact on patient prognosis).
- Manuscripts in all languages were included.
- All papers published in Scopus journals were included in the study to improve the precision of our search.
- All records, including errata, articles, book chapters, and conference papers, were included, making this a systematic review that included all sources related to miRNA in cancer and covered a broad range of research.
- Documents with clinical prognosis in cancer patients and patients’ survival or recurrence related to clinical diagnoses were considered.
3.3. Exclusion Criteria
3.4. Global Data and Bibliometric Profiling Analysis of Studies
- The form and language of published documents,
- Nation and institutional affiliation,
- Source/journal-title in which documents were published,
- Most active authors,
- Most cited papers,
- Collaboration trends and
3.5. Publication Productivity
3.6. Overview of the Research Output and Growth of miRNA-CANCER Research
3.7. Core Bibliometric Indicators
4. Results
4.1. General Data
4.2. Publications with Time
4.3. Countries
4.4. Most Frequent Terms
4.4.1. Authors
4.4.2. Frequently Cited Articles
4.4.3. Journals
4.4.4. Subject Area
5. Discussion
5.1. Key Findings
5.2. Journals That Published the Most Articles
5.3. Good Quality Publication Output
5.4. Strengths
5.5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Mostert, B.; Sieuwerts, A.M.; Martens, J.W.; Sleijfer, S. Diagnostic applications of cell-free and circulating tumor cell-associated miRNAs in cancer patients. Expert Rev. Mol. Diagn. 2011, 11, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Wiemer, E.A. The role of microRNAs in cancer: No small matter. Eur. J. Cancer 2007, 43, 1529–1544. [Google Scholar] [CrossRef]
- Shah, A.A.; Leidinger, P.; Blin, N.; Meese, E. miRNA: Small molecules as potential novel biomarkers in cancer. Curr. Med. Chem. 2010, 17, 4427–4432. [Google Scholar] [CrossRef]
- Chakraborty, C.; Chin, K.-Y.; Das, S. miRNA-regulated cancer stem cells: Understanding the property and the role of miRNA in carcinogenesis. Tumor Biol. 2016, 37, 13039–13048. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.W.; Chen, L.; Man, Y.-G. miRNA biomarkers in breast cancer detection and management. J. Cancer 2011, 2, 116. [Google Scholar] [CrossRef] [PubMed]
- Harquail, J.; Benzina, S.; Robichaud, G.A. MicroRNAs and breast cancer malignancy: An overview of miRNA-regulated cancer processes leading to metastasis. Cancer Biomark. 2012, 11, 269–280. [Google Scholar] [CrossRef]
- Jay, C.; Nemunaitis, J.; Chen, P.; Fulgham, P.; Tong, A.W. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007, 26, 293–300. [Google Scholar] [CrossRef]
- Asaga, S.; Kuo, C.; Nguyen, T.; Terpenning, M.; Giuliano, A.E.; Hoon, D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011, 57, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sültmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Smyth, P.; Flavin, R.; Cahill, S.; Denning, K.; Aherne, S.; Guenther, S.M.; O’Leary, J.J.; Sheils, O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007, 7, 36. [Google Scholar] [CrossRef] [Green Version]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Liu, Z.; Liu, X.; Fu, Q.; Deng, T.; Lu, J.; Liu, Y.; Liang, Z.; Jiang, Q.; Cheng, C. miR-296-3p negatively regulated by nicotine stimulates cytoplasmic translocation of c-Myc via MK2 to suppress chemotherapy resistance. Mol. Ther. 2018, 26, 1066–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Guo, Q.; Lin, K.; Chen, H.; Chen, Y.; Xu, Y.; Lin, C.; Su, Y.; Chen, Y.; Chen, M. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci. 2020, 111, 1711. [Google Scholar] [CrossRef]
- Yang, J.; Wu, S.-P.; Wang, W.-J.; Jin, Z.-R.; Miao, X.-B.; Wu, Y.; Gou, D.-M.; Liu, Q.-Z.; Yao, K.-T. A novel miR-200c/c-myc negative regulatory feedback loop is essential to the EMT process, CSC biology and drug sensitivity in nasopharyngeal cancer. Exp. Cell Res. 2020, 111817. [Google Scholar] [CrossRef]
- Wan, F.-Z.; Chen, K.-H.; Sun, Y.-C.; Chen, X.-C.; Liang, R.-B.; Chen, L.; Zhu, X.-D. Exosomes overexpressing miR-34c inhibit malignant behavior and reverse the radioresistance of nasopharyngeal carcinoma. J. Transl. Med. 2020, 18, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xu, Y.; Wang, J.; Ying, H. Circulating miR-214-3p predicts nasopharyngeal carcinoma recurrence or metastasis. Clin. Chim. Acta 2020, 503, 54–60. [Google Scholar] [CrossRef]
- Hamano, R.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Hara, J.; ho Moon, J.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 2011, 17, 3029–3038. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Li, Y.; Wang, X.; Yang, Y. Dysregulation of MiR-519d affects oral squamous cell carcinoma invasion and metastasis by targeting MMP3. J. Cancer 2019, 10, 2720. [Google Scholar] [CrossRef] [Green Version]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef] [Green Version]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [Green Version]
- Shin, V.Y.; Chu, K.-M. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 2014, 20, 10432. [Google Scholar] [CrossRef]
- Tie, Y.; Liu, B.; Fu, H.; Zheng, X. Circulating miRNA and cancer diagnosis. Sci. China Ser. C Life Sci. 2009, 52, 1117–1122. [Google Scholar] [CrossRef]
- Tong, A.; Nemunaitis, J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008, 15, 341–355. [Google Scholar] [CrossRef]
- Uzuner, E.; Ulu, G.T.; Gürler, S.B.; Baran, Y. The role of MiRNA in cancer: Pathogenesis, diagnosis, and treatment. In miRNomics; Springer: Cham, Switzerland, 2022; pp. 375–422. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Ferracin, M.; Veronese, A.; Negrini, M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 2010, 10, 297–308. [Google Scholar] [CrossRef]
- Pan, X.; Wang, Z.-X.; Wang, R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol. Ther. 2010, 10, 1224–1232. [Google Scholar] [CrossRef] [Green Version]
- Jayaraj, R.; Madhav, M.R.; Nayagam, S.G.; Kar, A.; Sathyakumar, S.; Mohammed, H.; Smiti, M.; Sabarimurugan, S.; Kumarasamy, C.; Priyadharshini, T. Clinical theragnostic relationship between drug-resistance specific miRNA expressions, chemotherapeutic resistance, and sensitivity in breast cancer: A systematic review and meta-analysis. Cells 2019, 8, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaraj, R.; Raymond, G.; Krishnan, S.; Tzou, K.S.; Baxi, S.; Ram, M.R.; Govind, S.K.; Chandramoorthy, H.C.; Abu-Khzam, F.N.; Shaw, P. Clinical theragnostic potential of diverse miRNA expressions in prostate cancer: A systematic review and meta-analysis. Cancers 2020, 12, 1199. [Google Scholar] [CrossRef]
- Kumarasamy, C.; Madhav, M.R.; Sabarimurugan, S.; Krishnan, S.; Baxi, S.; Gupta, A.; Gothandam, K.; Jayaraj, R. Prognostic value of miRNAs in head and neck cancers: A comprehensive systematic and meta-analysis. Cells 2019, 8, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madurantakam Royam, M.; Ramesh, R.; Shanker, R.; Sabarimurugan, S.; Kumarasamy, C.; Ramesh, N.; Gothandam, K.M.; Baxi, S.; Gupta, A.; Krishnan, S. Mirna predictors of pancreatic cancer chemotherapeutic response: A systematic review and meta-analysis. Cancers 2019, 11, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Royam, M.M.; Kumarasamy, C.; Baxi, S.; Gupta, A.; Ramesh, N.; Muthukaliannan, G.K.; Jayaraj, R. Current evidence on miRNAs as potential theranostic markers for detecting chemoresistance in colorectal cancer: A systematic review and meta-analysis of preclinical and clinical studies. Mol. Diagn. Ther. 2019, 23, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Sabarimurugan, S.; Kumarasamy, C.; Baxi, S.; Devi, A.; Jayaraj, R. Systematic review and meta-analysis of prognostic microRNA biomarkers for survival outcome in nasopharyngeal carcinoma. PLoS ONE 2019, 14, e0209760. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Royam, M.M.; Das, A.; Das, S.; Gothandam, K.; Jayaraj, R. Systematic review and meta-analysis of the prognostic significance of miRNAs in melanoma patients. Mol. Diagn. Ther. 2018, 22, 653–669. [Google Scholar] [CrossRef]
- Shaw, P.; Senthilnathan, R.; Krishnan, S.; Suresh, D.; Shetty, S.; Muthukaliannan, G.K.; Mani, R.R.; Sivanandy, P.; Chandramoorthy, H.C.K.; Gupta, M.M. A Clinical Update on the Prognostic Effect of microRNA Biomarkers for Survival Outcome in Nasopharyngeal Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2021, 13, 4369. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Sabarimurugan, S.; Baxi, S. Diagnostic and prognostic value of microRNAs for cancers-strategies and approaches to improve the clinical utility. J. Cancer 2019, 10, 1252–1253. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Madhav, M.R.; Kumarasamy, C.; Gupta, A.; Baxi, S.; Krishnan, S.; Jayaraj, R. Prognostic value of MicroRNAs in stage II colorectal cancer patients: A systematic review and meta-analysis. Mol. Diagn. Ther. 2020, 24, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Sabarimurugan, S.; Kumarasamy, C.; Madhav, M.R.; Samiappan, S.; Jayaraj, R. The Significance of miRNAs as a Prognostic Biomarker for Survival Outcome in T Cell–Acute Lymphoblastic Leukemia Patients: A Systematic Review and Meta-Analysis. Cancer Manag. Res. 2020, 12, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, M.-H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free circulating miRNA biomarkers in cancer. J. Cancer 2012, 3, 432. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Gislefoss, R.; Haugen, A.; Langseth, H.; Staehler, P.; Lenhof, H.-P.; Meese, E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011, 8, 506–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Du, L.; Wang, L.; Li, J.; Liu, Y.; Zheng, G.; Qu, A.; Zhang, X.; Pan, H.; Yang, Y. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int. J. Cancer 2015, 136, 854–862. [Google Scholar] [CrossRef]
- Ren, A.; Dong, Y.; Tsoi, H.; Yu, J. Detection of miRNA as non-invasive biomarkers of colorectal cancer. Int. J. Mol. Sci. 2015, 16, 2810–2823. [Google Scholar] [CrossRef] [Green Version]
- Powrózek, T.; Krawczyk, P.; Kowalski, D.M.; Kuźnar-Kamińska, B.; Winiarczyk, K.; Olszyna-Serementa, M.; Batura-Gabryel, H.; Milanowski, J. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumor Biol. 2016, 37, 2049–2055. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Jin, X.; Wang, Z.; Wu, Y.; Zhao, D.; Chen, G.; Li, D.; Wang, X.; Cao, H. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res. Treat. 2015, 154, 423–434. [Google Scholar] [CrossRef]
- Mengual, L.; Lozano, J.J.; Ingelmo-Torres, M.; Gazquez, C.; Ribal, M.J.; Alcaraz, A. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int. J. Cancer 2013, 133, 2631–2641. [Google Scholar]
- Debernardi, S.; Massat, N.J.; Radon, T.P.; Sangaralingam, A.; Banissi, A.; Ennis, D.P.; Dowe, T.; Chelala, C.; Pereira, S.P.; Kocher, H.M. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am. J. Cancer Res. 2015, 5, 3455. [Google Scholar]
- Jayaraj, R.; Kumaraswamy, C.; Raymond, G.; Ram, M.R.; Govind, S.K.; Chandramoorthy, H.C.; Shaw, P. Diagnostic implications of miRNAs in Liquid Biopsy for Oral Squamous Cell Carcinoma (OSCC): Clinical validity and interpretation. Oral Oncol. 2020, 109, 104634. [Google Scholar] [CrossRef] [PubMed]
- Cacchiarelli, D.; Legnini, I.; Martone, J.; Cazzella, V.; d’Amico, A.; Bertini, E.; Bozzoni, I. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol. Med. 2011, 3, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Calin, G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [CrossRef]
- Buhagiar, A.; Borg, J.; Ayers, D. Overview of current microRNA biomarker signatures as potential diagnostic tools for leukaemic conditions. Non-Coding RNA Res. 2020, 5, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- McGuire, A.; Brown, J.A.; Kerin, M.J. Metastatic breast cancer: The potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015, 34, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Zubor, P.; Kubatka, P.; Dankova, Z.; Gondova, A.; Kajo, K.; Hatok, J.; Samec, M.; Jagelkova, M.; Krivus, S.; Holubekova, V. miRNA in a multiomic context for diagnosis, treatment monitoring and personalized management of metastatic breast cancer. Future Oncol. 2018, 14, 1847–1867. [Google Scholar] [CrossRef]
- Wang, W.-T.; Chen, Y.-Q. Circulating miRNAs in cancer: From detection to therapy. J. Hematol. Oncol. 2014, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Blenkiron, C.; Miska, E.A. miRNAs in cancer: Approaches, aetiology, diagnostics and therapy. Hum. Mol. Genet. 2007, 16, R106–R113. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]

| Document Type | Number | Frequency % |
|---|---|---|
| Article | 14,572 | 78.08% |
| Review | 2909 | 15.59% |
| Book Chapter | 485 | 2.60% |
| Conference Paper | 266 | 1.43% |
| Short Survey | 178 | 0.95% |
| Article in Press | 89 | 0.48% |
| Note | 40 | 0.21% |
| Erratum | 31 | 0.17% |
| Letter | 26 | 0.14% |
| Editorial | 25 | 0.13% |
| Book | 17 | 0.09% |
| Retracted | 15 | 0.08% |
| Conference Review | 10 | 0.05% |
| Total | 18,663 | 100.00% |
| Language | Total No. of Articles | % of Articles | |
|---|---|---|---|
| 1st | English | 18,067 | 96.81% |
| 2nd | Chinese | 463 | 2.48% |
| 3rd | Japanese | 24 | 0.13% |
| 4th | Russian | 22 | 0.12% |
| 5th | German | 20 | 0.11% |
| 6th | Czech | 15 | 0.08% |
| 7th | French | 15 | 0.08% |
| 8th | Spanish | 13 | 0.07% |
| 9th | Polish | 9 | 0.05% |
| 10th | Persian | 7 | 0.04% |
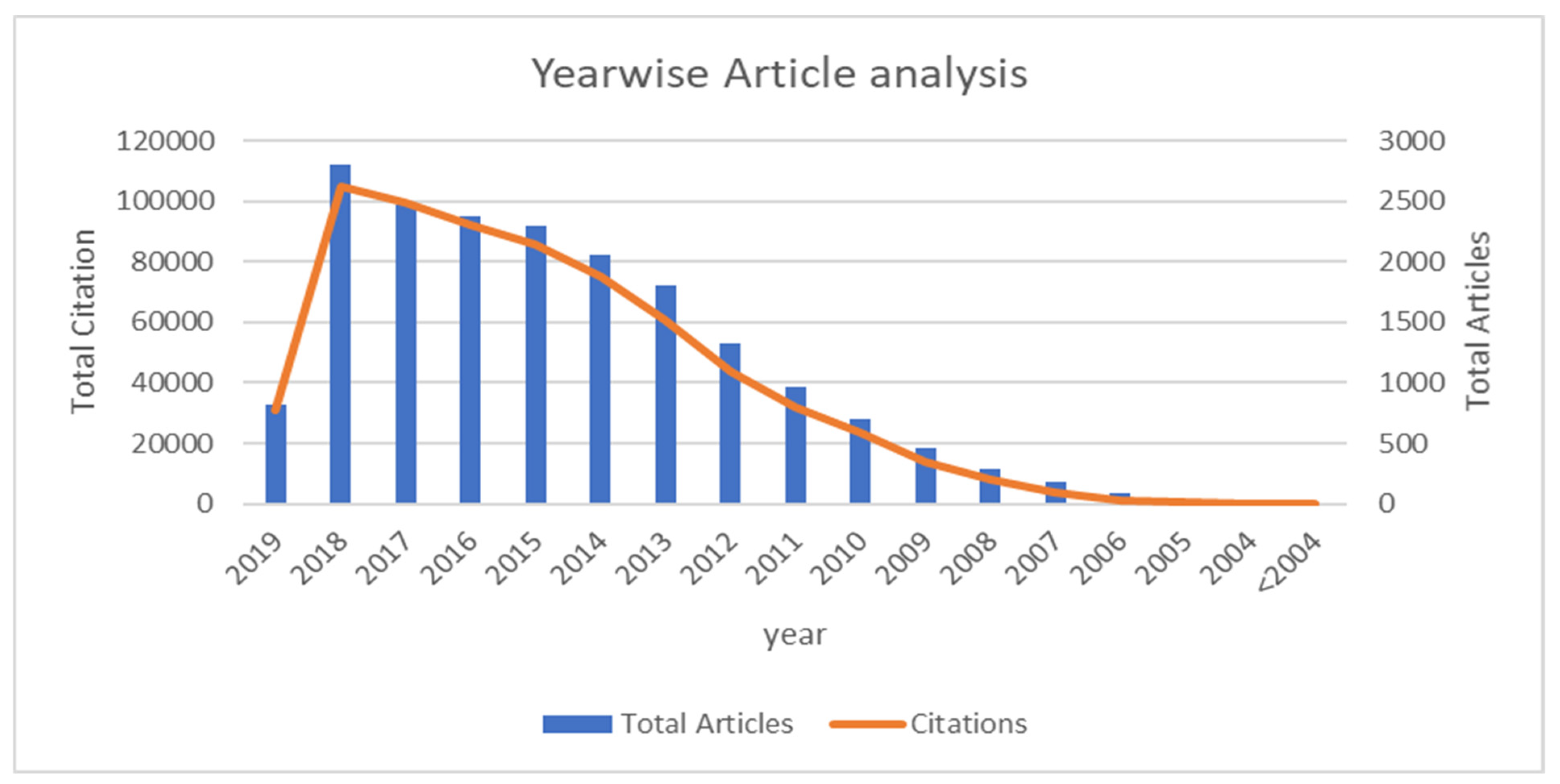
| Year | Open Access | Closed Access | Total Articles | Citations | Average Citation per Document |
|---|---|---|---|---|---|
| 2019 | 296 | 520 | 816 | 30,808 | 37.75490196 |
| 2018 | 1324 | 1486 | 2810 | 104,985 | 37.36120996 |
| 2017 | 1344 | 1132 | 2476 | 99,818 | 40.31421648 |
| 2016 | 1199 | 1175 | 2374 | 91,943 | 38.72914912 |
| 2015 | 1204 | 1091 | 2295 | 86,001 | 37.47320261 |
| 2014 | 1037 | 1026 | 2063 | 75,003 | 36.35627727 |
| 2013 | 882 | 920 | 1802 | 60,639 | 33.6509434 |
| 2012 | 672 | 651 | 1323 | 43,723 | 33.04837491 |
| 2011 | 438 | 533 | 971 | 32,016 | 32.97219361 |
| 2010 | 314 | 380 | 694 | 23,399 | 33.71613833 |
| 2009 | 216 | 238 | 454 | 13,884 | 30.5814978 |
| 2008 | 146 | 147 | 293 | 8028 | 27.39931741 |
| 2007 | 90 | 87 | 177 | 3607 | 20.37853107 |
| 2006 | 41 | 41 | 82 | 1249 | 15.23170732 |
| 2005 | 11 | 15 | 26 | 326 | 12.53846154 |
| 2004 | 2 | 2 | 4 | 51 | 12.75 |
| <2004 | 1 | 2 | 3 | 11 | 3.666666667 |
| Total (<2004–2019) | 9217 | 9446 | 18,663 | 675,491 | 483.9227894 |
| Average (<2004–2019) | 542.1764706 | 555.6470588 | 1097.823529 | 39,734.76 | 53.76919883 |
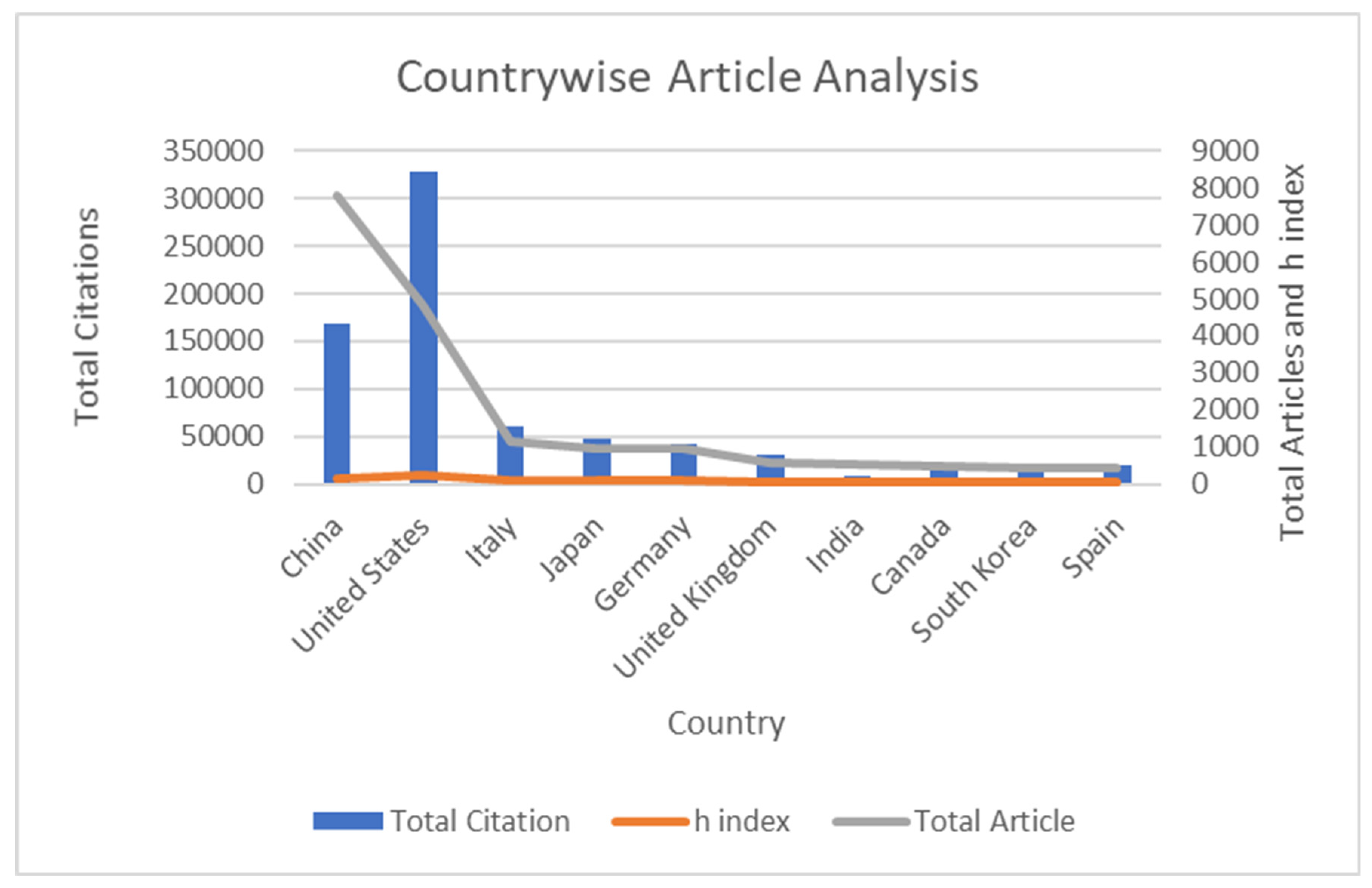
| Country Name | Total Citation | h Index | Total Article | % Article | % Citation |
|---|---|---|---|---|---|
| China | 168,314 | 145 | 7782 | 0.416974763 | 0.249172824 |
| United States | 327,538 | 244 | 4832 | 0.258908 | 0.48488877 |
| Italy | 61,473 | 108 | 1137 | 0.060922681 | 0.091004913 |
| Japan | 47,847 | 106 | 958 | 0.051331512 | 0.07083292 |
| Germany | 42,641 | 102 | 946 | 0.050688528 | 0.063125934 |
| United Kingdom | 30,325 | 76 | 567 | 0.030380968 | 0.04489327 |
| India | 8205 | 47 | 515 | 0.027594706 | 0.01214672 |
| Canada | 19,212 | 73 | 496 | 0.026576649 | 0.028441534 |
| South Korea | 14,240 | 59 | 459 | 0.024594117 | 0.021080962 |
| Spain | 19,244 | 66 | 422 | 0.022611584 | 0.028488907 |
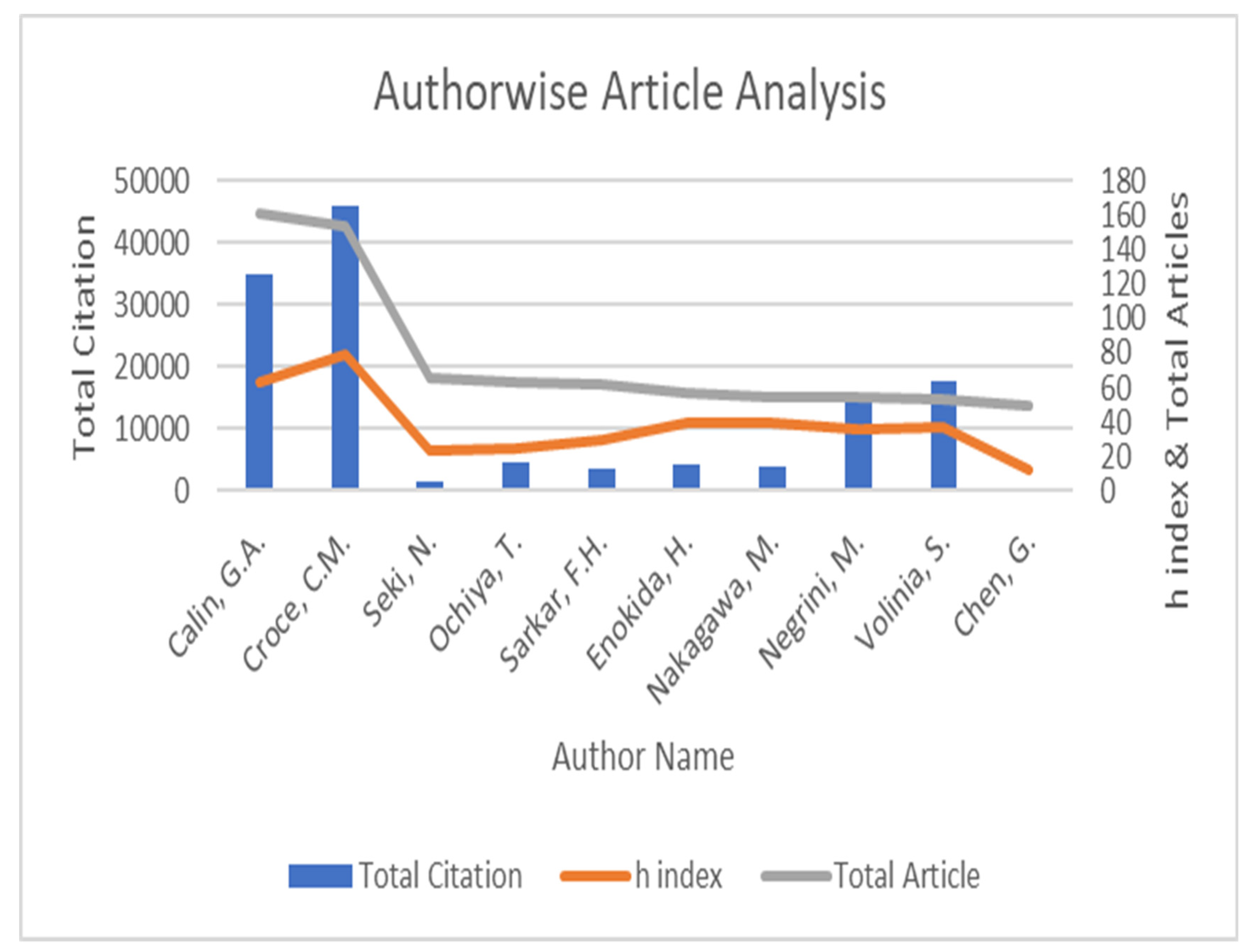
| Author Name | Total Citation | h Index | Total Article | % Article | % Citation | C/A |
|---|---|---|---|---|---|---|
| Calin, G.A. | 34,959 | 63 | 161 | 0.008627 | 0.05175347 | 217.136646 |
| Croce, C.M. | 46,117 | 79 | 154 | 0.008252 | 0.06827182 | 299.461039 |
| Seki, N. | 1597 | 24 | 65 | 0.003483 | 0.00236421 | 24.5692308 |
| Ochiya, T. | 4655 | 25 | 63 | 0.003376 | 0.00689128 | 73.8888889 |
| Sarkar, F.H. | 3723 | 30 | 62 | 0.003322 | 0.00551155 | 60.0483871 |
| Enokida, H. | 4110 | 40 | 57 | 0.003054 | 0.00608446 | 72.1052632 |
| Nakagawa, M. | 3953 | 39 | 55 | 0.002947 | 0.00585204 | 71.8727273 |
| Negrini, M. | 15,135 | 36 | 54 | 0.002893 | 0.02240592 | 280.277778 |
| Volinia, S. | 17,556 | 37 | 53 | 0.00284 | 0.02598998 | 331.245283 |
| Chen, G. | 497 | 12 | 49 | 0.002626 | 0.00073576 | 10.1428571 |
| Authors | Title | Year | Source Title | Document Type | Cited by |
|---|---|---|---|---|---|
| Lu, J. et al. [1] | MicroRNA expression profiles classify human cancers | 2005 | Nature | Article | 6712 |
| Calin, G.A. and Croce, C.M. [4] | MicroRNA signatures in human cancers | 2006 | Nature Reviews Cancer | Review | 5205 |
| Esquela-Kerscher, A. and Slack, F.J. [20] | Oncomirs—MicroRNAs with a role in cancer | 2006 | Nature Reviews Cancer | Review | 5058 |
| Mitchell, P.S. et al. [15] | Circulating microRNAs as stable blood-based markers for cancer detection | 2008 | Proceedings of the National Academy of Sciences of the United States of America | Article | 4606 |
| Volinia, S. et al. [71] | A microRNA expression signature of human solid tumours defines cancer gene targets | 2006 | Proceedings of the National Academy of Sciences of the United States of America | Article | 4193 |
| Iorio, M.V. et al. [72] | MicroRNA gene expression deregulation in human breast cancer | 2005 | Cancer Research | Article | 2928 |
| Johnson, S.M. et al. [73] | RAS is regulated by the let-7 microRNA family | 2005 | Cell | Article | 2729 |
| Landgraf, P. et al. [74] | A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing | 2007 | Cell | Article | 2417 |
| Yanaihara, N. et al. [75] | Unique microRNA molecular profiles in lung cancer diagnosis and prognosis | 2006 | Cancer Cell | Article | 2329 |
| Skog, J. et al. [76] | Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers | 2008 | Nature Cell Biology | Article | 2298 |
| Journal Name | Total Citation | h Index | Total Articles | % Citation | % Articles |
|---|---|---|---|---|---|
| Oncotarget | 15,560 | 54 | 829 | 2.30% | 4.44% |
| Plos One | 33,313 | 86 | 777 | 4.93% | 4.16% |
| Oncology Reports | 5462 | 37 | 326 | 0.81% | 1.75% |
| Oncology Letters | 2783 | 26 | 324 | 0.41% | 1.74% |
| Tumour Biology | 5916 | 17 | 303 | 0.88% | 1.62% |
| Molecular Medicine Reports | 2454 | 23 | 291 | 0.36% | 1.56% |
| Cancer Research | 41,876 | 106 | 280 | 6.20% | 1.50% |
| Scientific Reports | 3945 | 32 | 265 | 0.58% | 1.42% |
| Oncogene | 21,939 | 84 | 213 | 3.25% | 1.14% |
| International Journal Of Oncology | 5918 | 44 | 203 | 0.88% | 1.09% |
| Rank | Founding Sponsor | Number |
|---|---|---|
| 1 | National Natural Science Foundation of China | 1440 |
| 2 | National Natural Science Foundation of China (NSFC) | 1205 |
| 3 | National Institutes of Health (NIH) | 521 |
| 4 | National Institutes of Health | 433 |
| 5 | National Cancer Institute | 235 |
| 6 | National Cancer Institute (NCI) | 169 |
| 7 | National Research Foundation of Korea (NRF) | 160 |
| 8 | National Basic Research Program of China (973 Program) | 149 |
| 9 | Natural Science Foundation of Jiangsu Province | 146 |
| 10 | Associazione Italiana per la Ricerca sul Cancro | 122 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, P.; Lokhotiya, K.; Kumarasamy, C.; Sunil, K.; Suresh, D.; Shetty, S.; Muthukaliannan, G.K.; Baxi, S.; Mani, R.R.; Sivanandy, P.; et al. Mapping Research on miRNAs in Cancer: A Global Data Analysis and Bibliometric Profiling Analysis. Pathophysiology 2022, 29, 66-80. https://doi.org/10.3390/pathophysiology29010007
Shaw P, Lokhotiya K, Kumarasamy C, Sunil K, Suresh D, Shetty S, Muthukaliannan GK, Baxi S, Mani RR, Sivanandy P, et al. Mapping Research on miRNAs in Cancer: A Global Data Analysis and Bibliometric Profiling Analysis. Pathophysiology. 2022; 29(1):66-80. https://doi.org/10.3390/pathophysiology29010007
Chicago/Turabian StyleShaw, Peter, Kartik Lokhotiya, Chellan Kumarasamy, Krishnan Sunil, Deepa Suresh, Sameep Shetty, Gothandam Kodiveri Muthukaliannan, Siddhartha Baxi, Ravishankar Ram Mani, Palanisamy Sivanandy, and et al. 2022. "Mapping Research on miRNAs in Cancer: A Global Data Analysis and Bibliometric Profiling Analysis" Pathophysiology 29, no. 1: 66-80. https://doi.org/10.3390/pathophysiology29010007
APA StyleShaw, P., Lokhotiya, K., Kumarasamy, C., Sunil, K., Suresh, D., Shetty, S., Muthukaliannan, G. K., Baxi, S., Mani, R. R., Sivanandy, P., Chandramoorthy, H. C., Gupta, M. M., Samiappan, S., & Jayaraj, R. (2022). Mapping Research on miRNAs in Cancer: A Global Data Analysis and Bibliometric Profiling Analysis. Pathophysiology, 29(1), 66-80. https://doi.org/10.3390/pathophysiology29010007










