Transcriptome and Metabolome Analyses Provide Insights into the Flavonoid Accumulation in Peels of Citrus reticulata ‘Chachi’
Abstract
:1. Introduction
2. Results
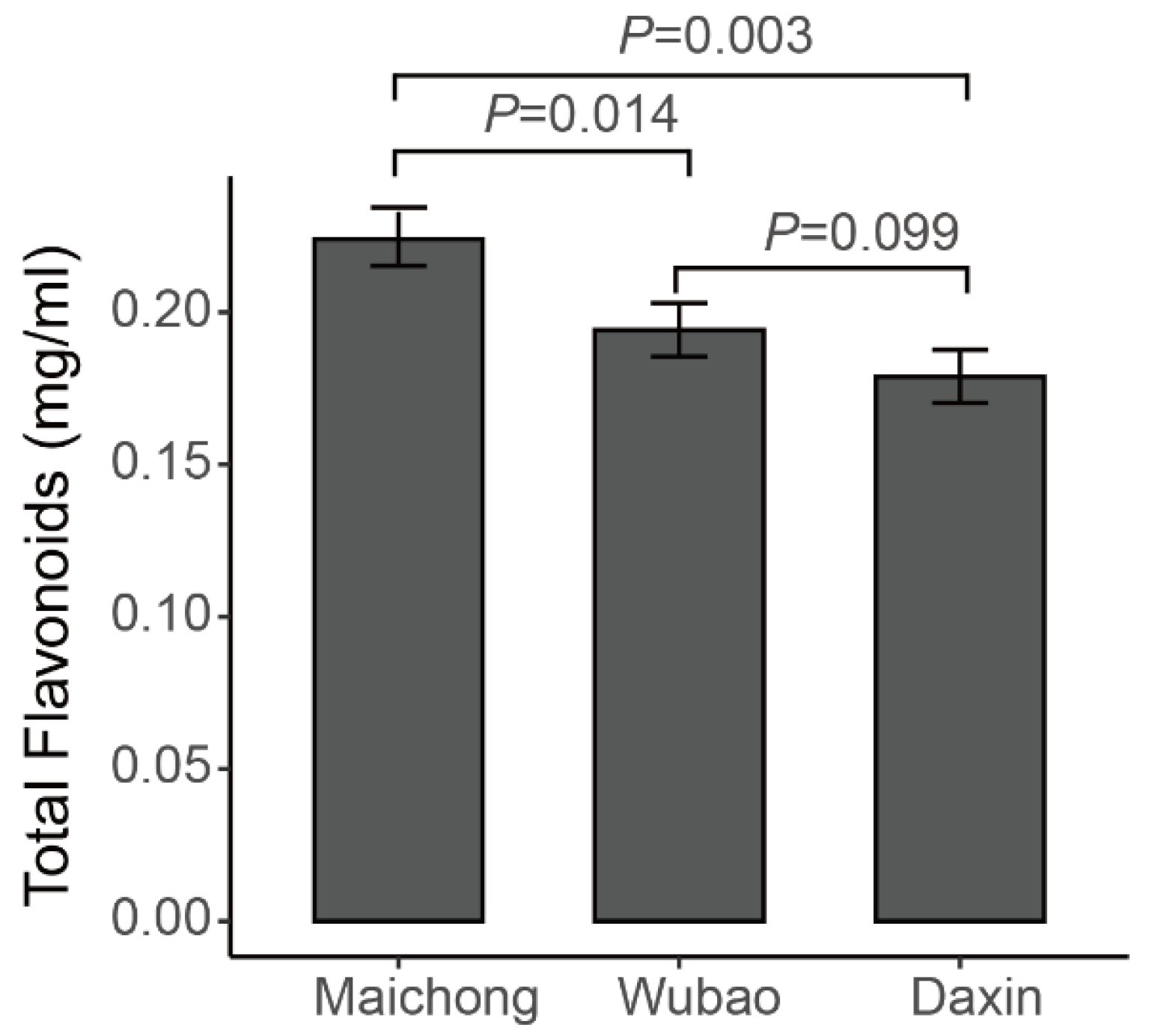
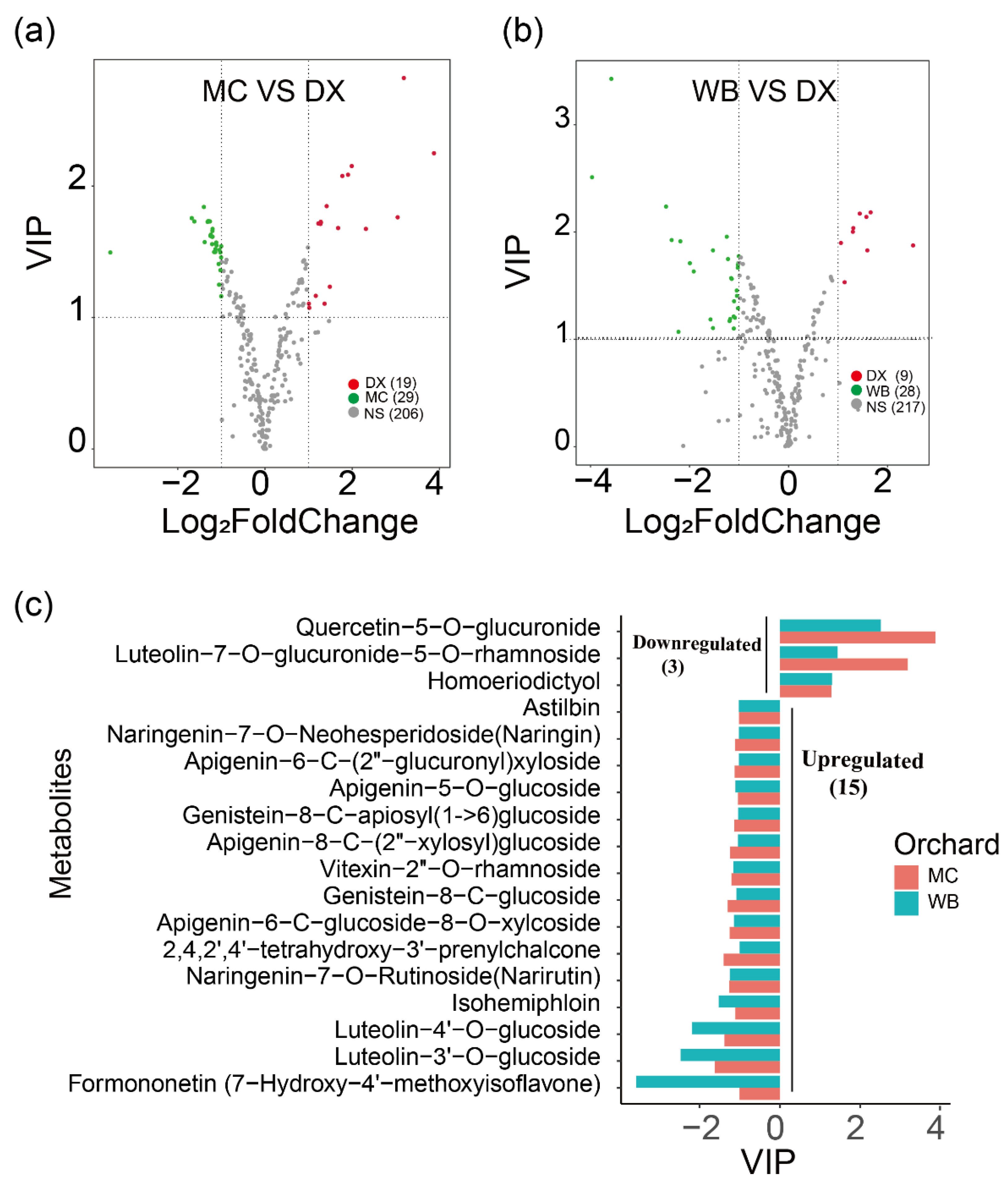
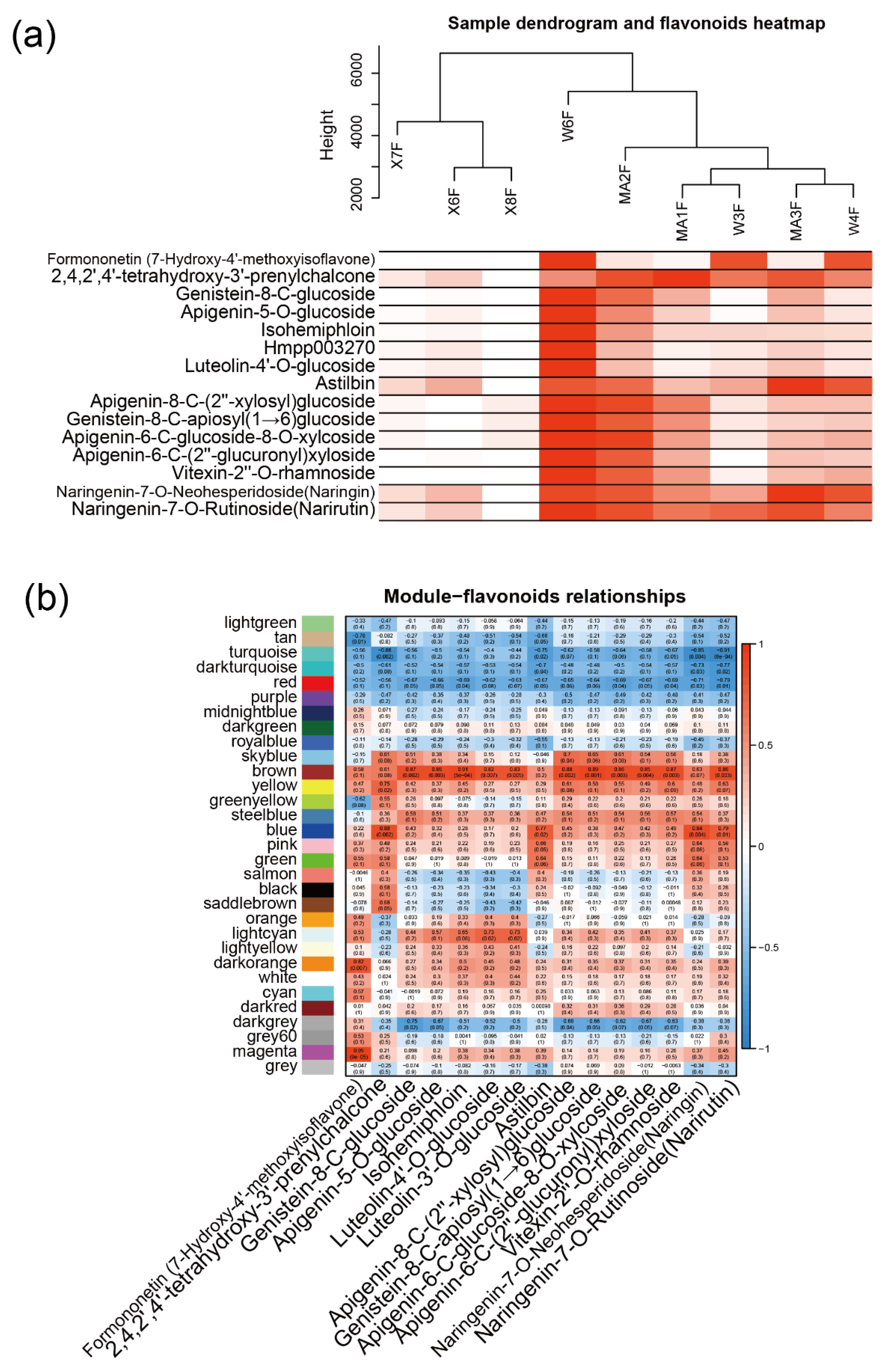
2.1. Qualitative and Quantitative Analysis of Flavonoid Metabolites
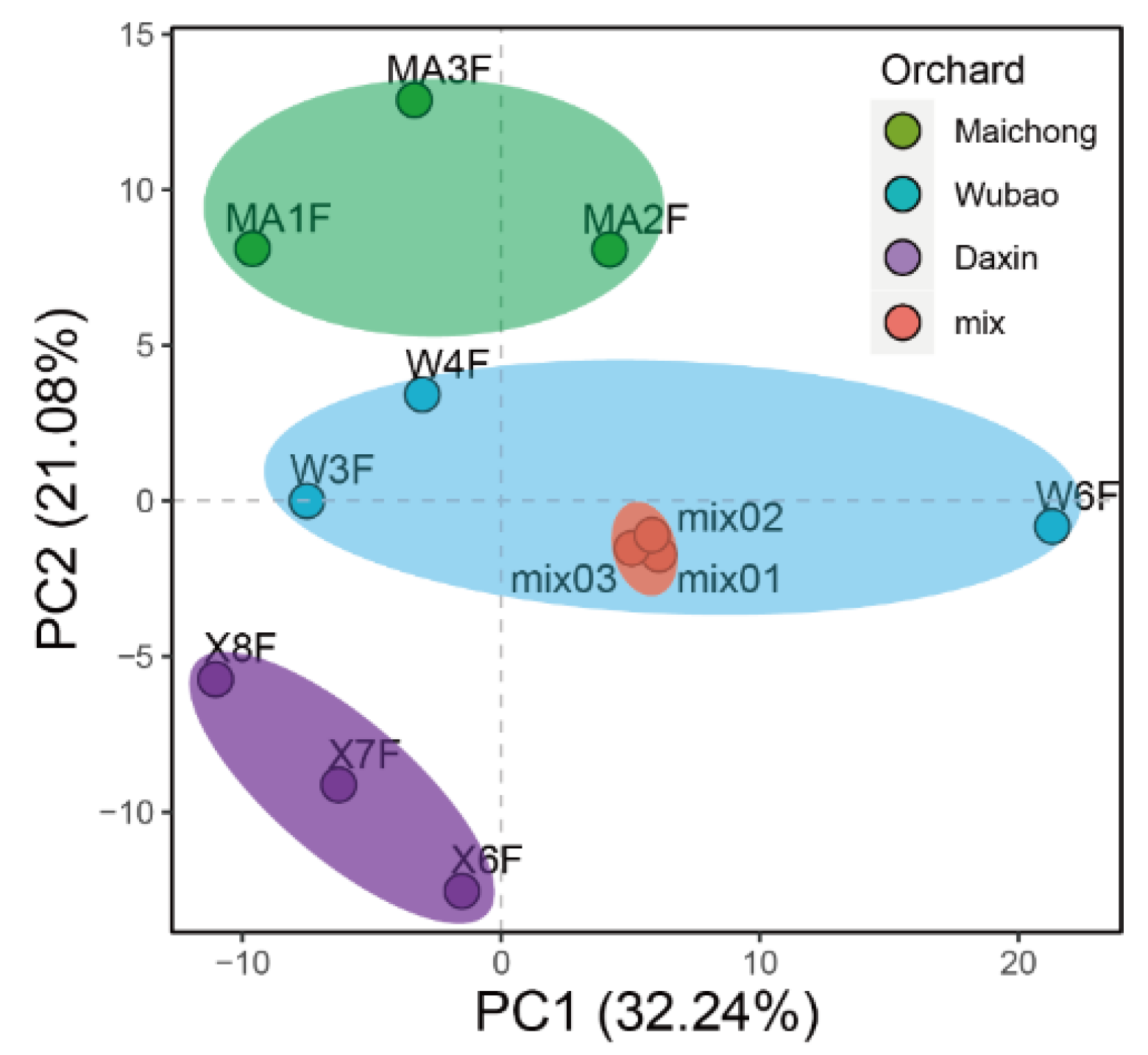
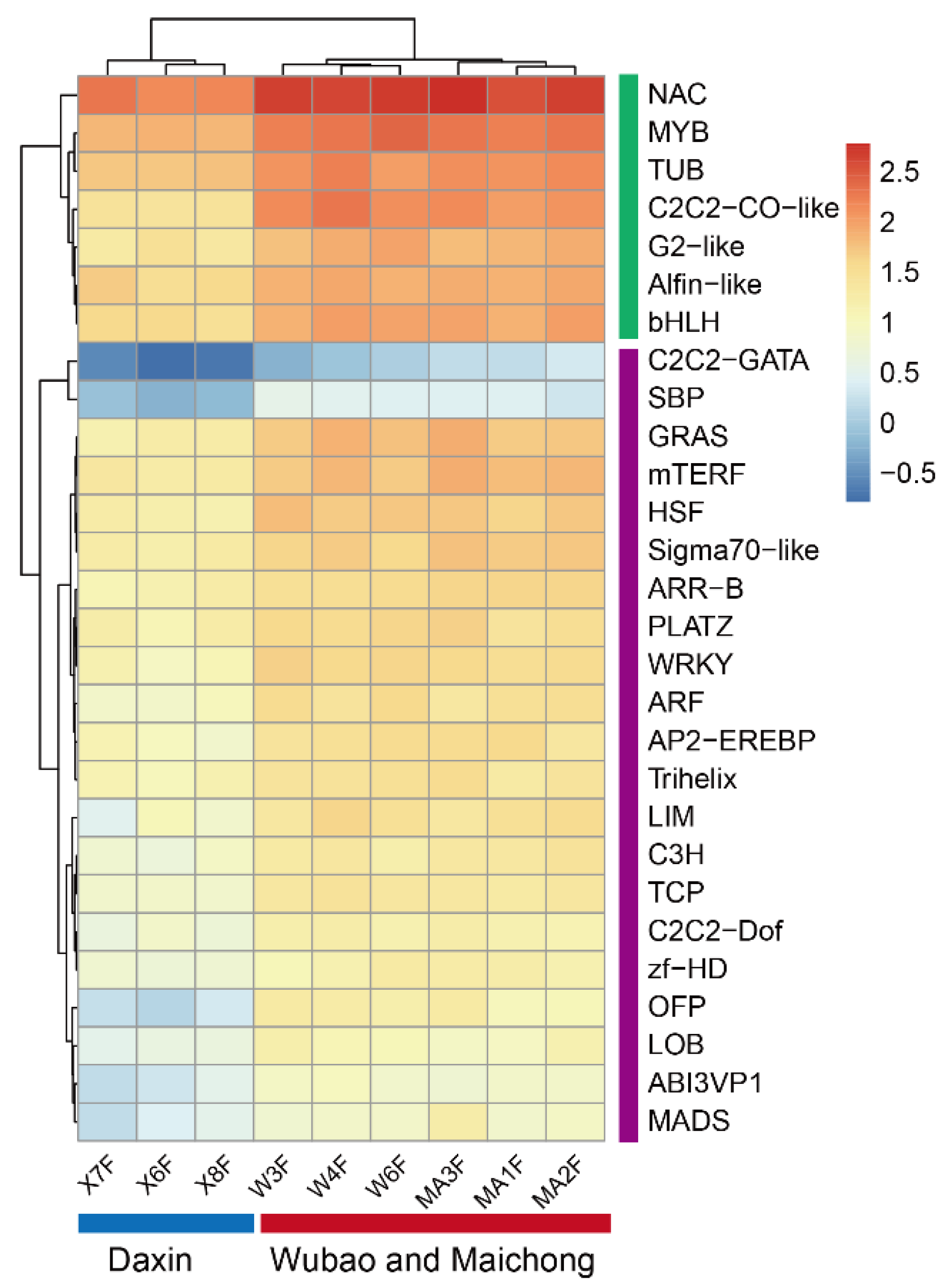
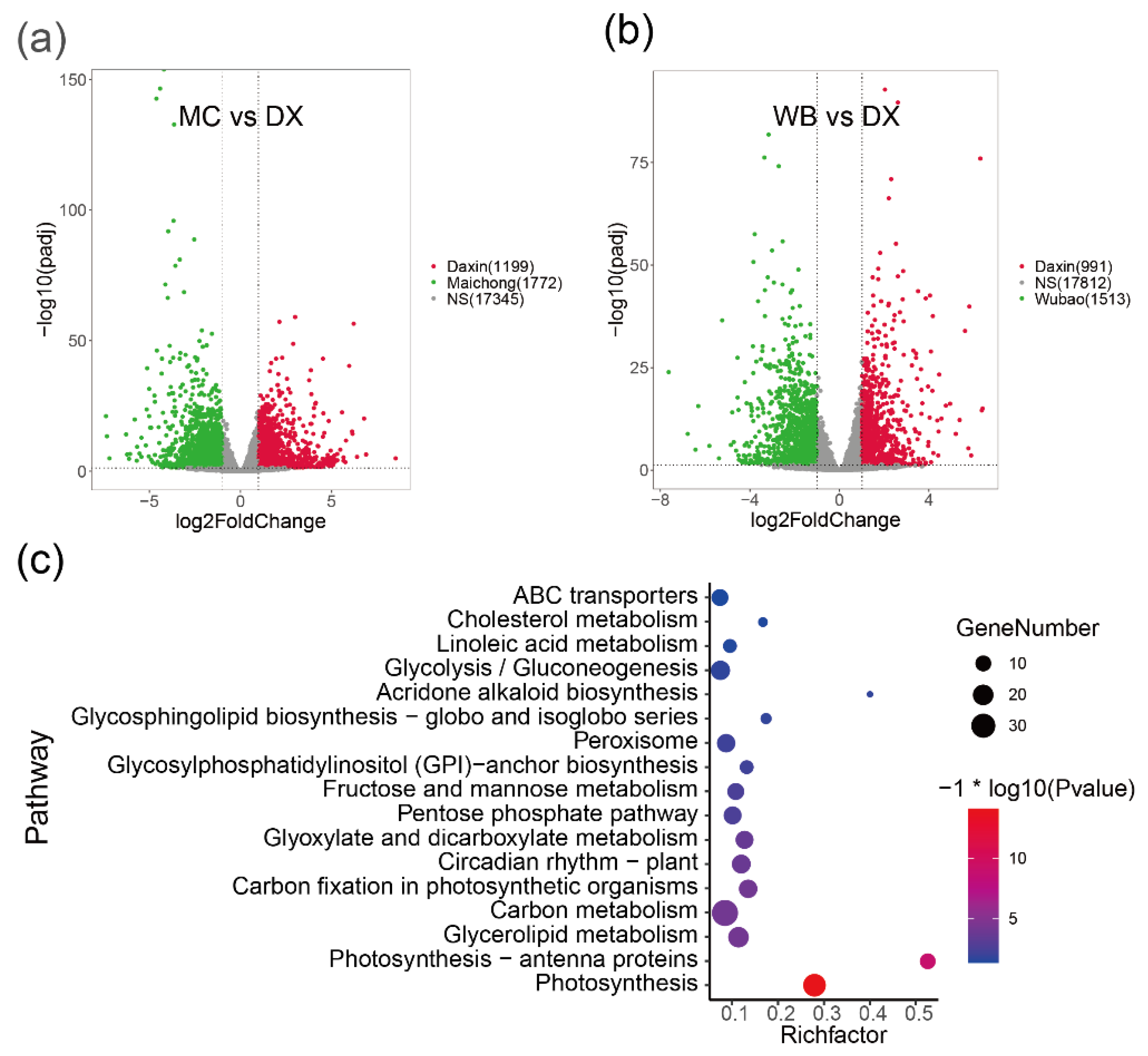
2.2. Differentially Expressed Genes between the Three Planting Regions
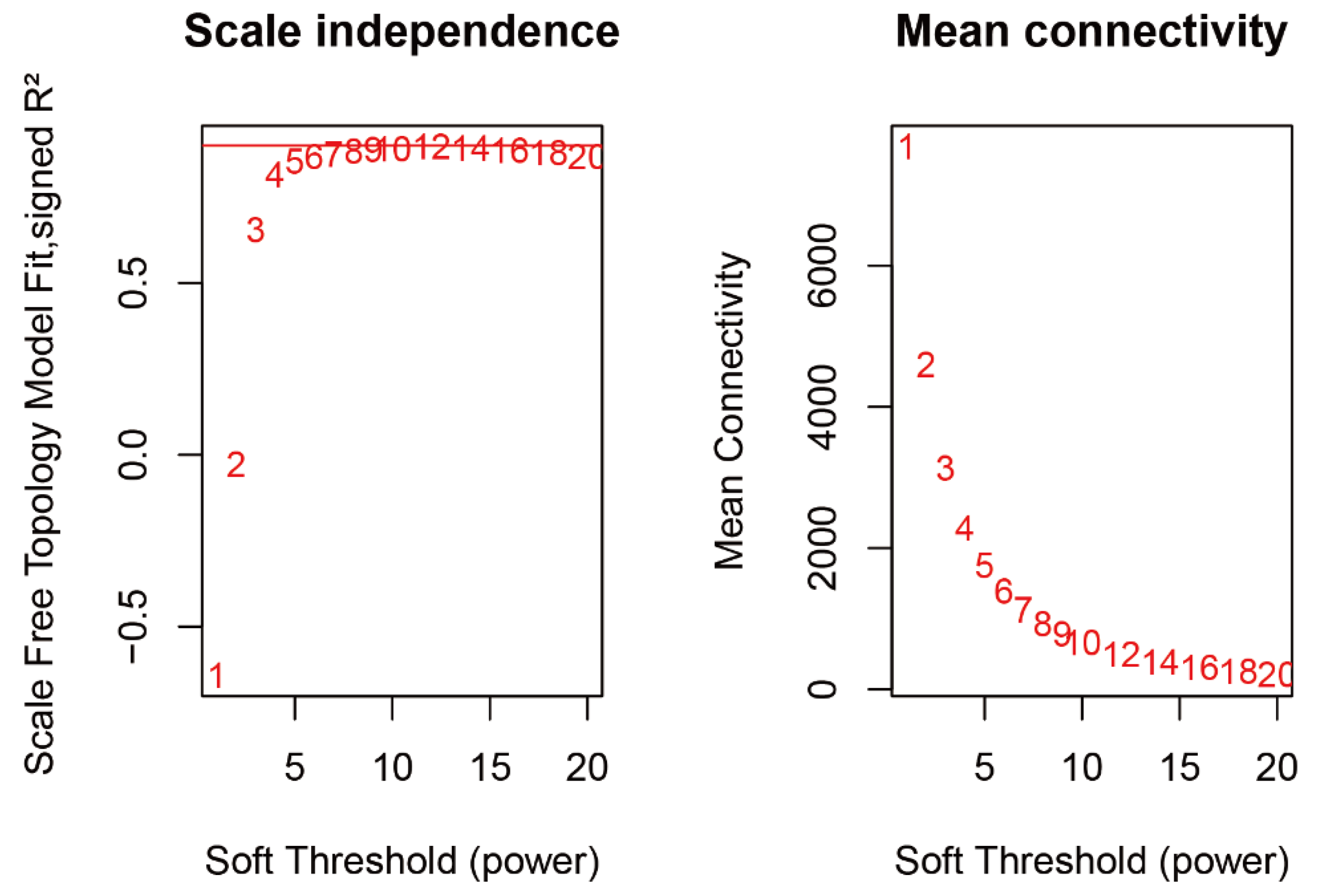
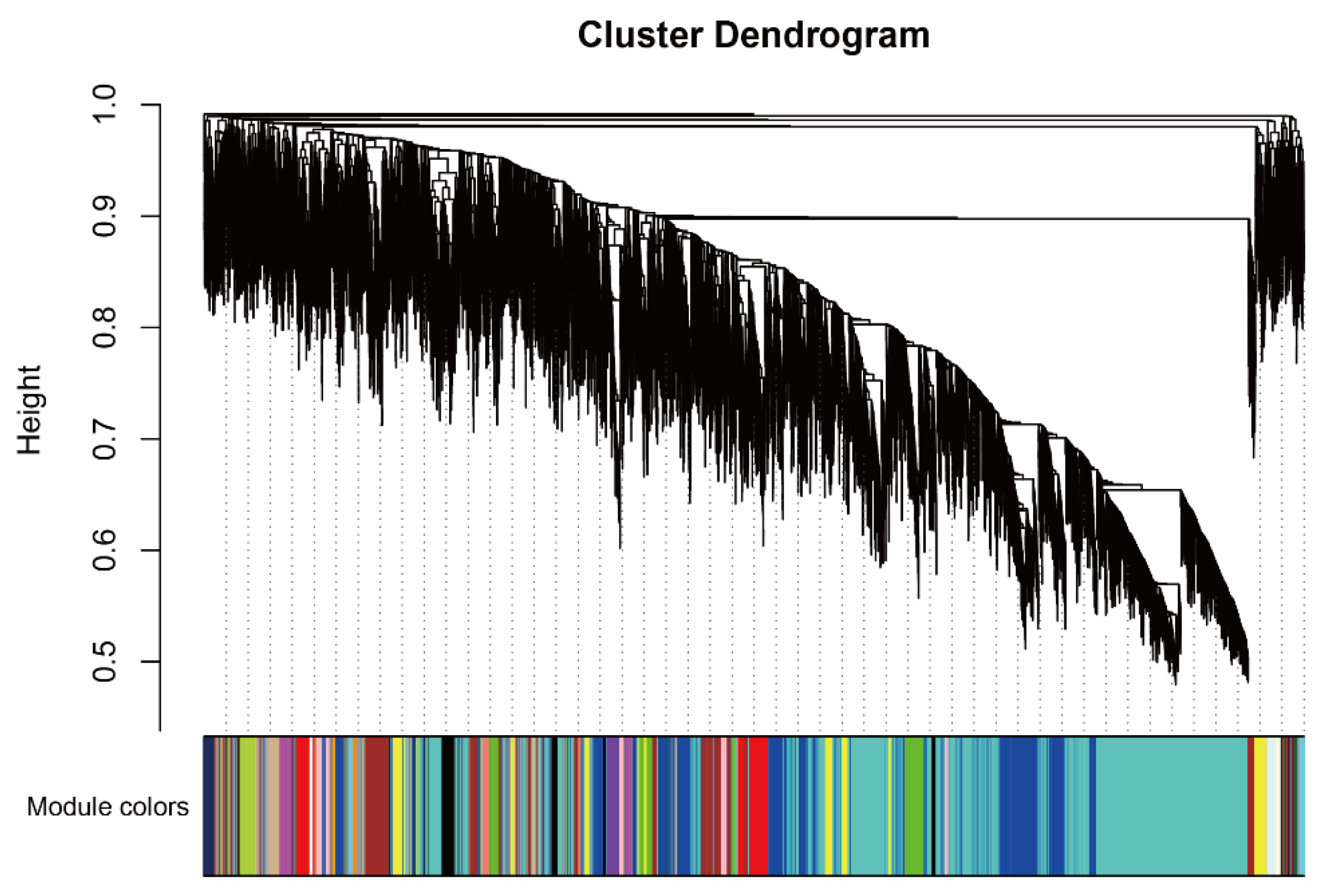
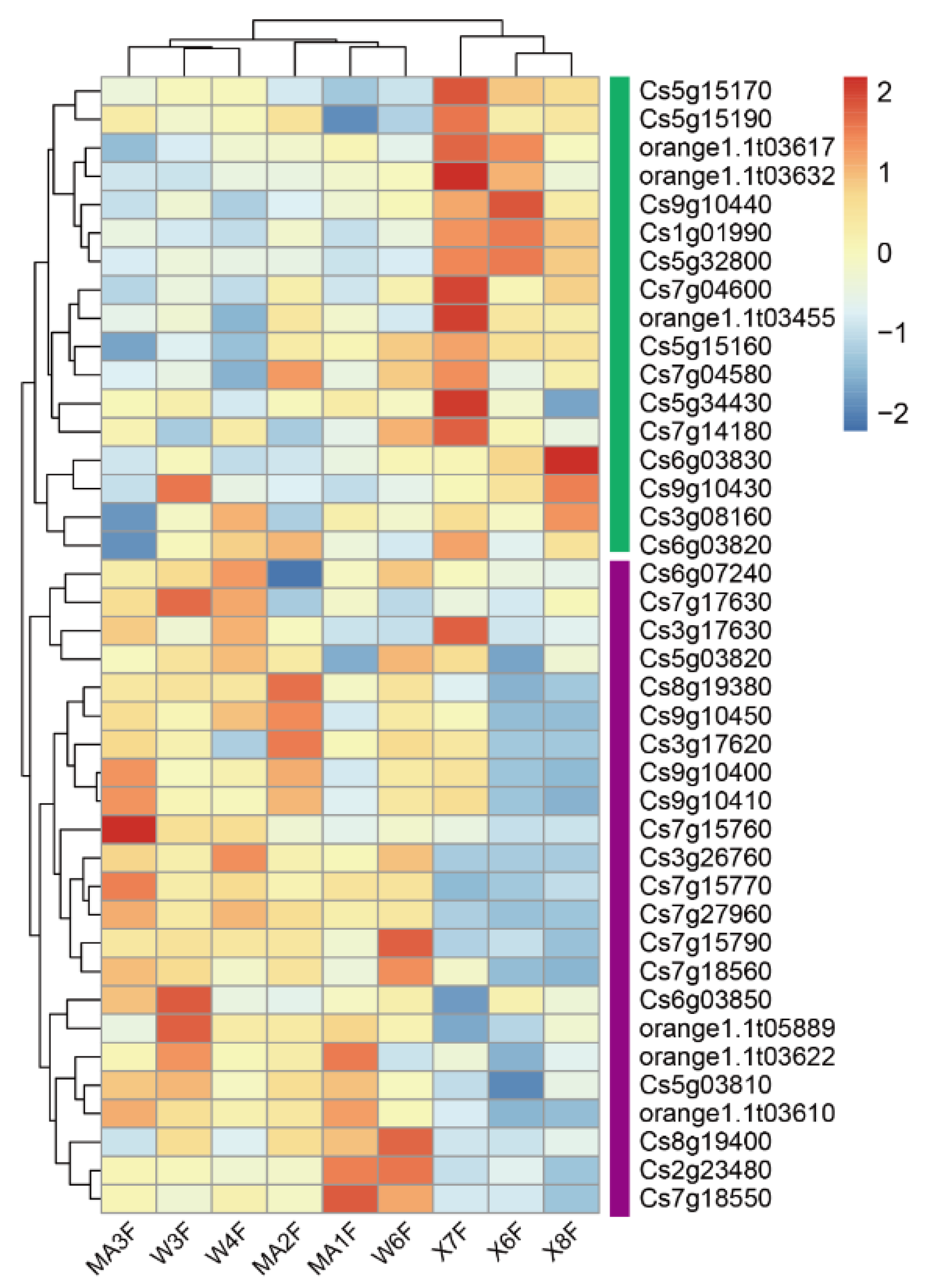
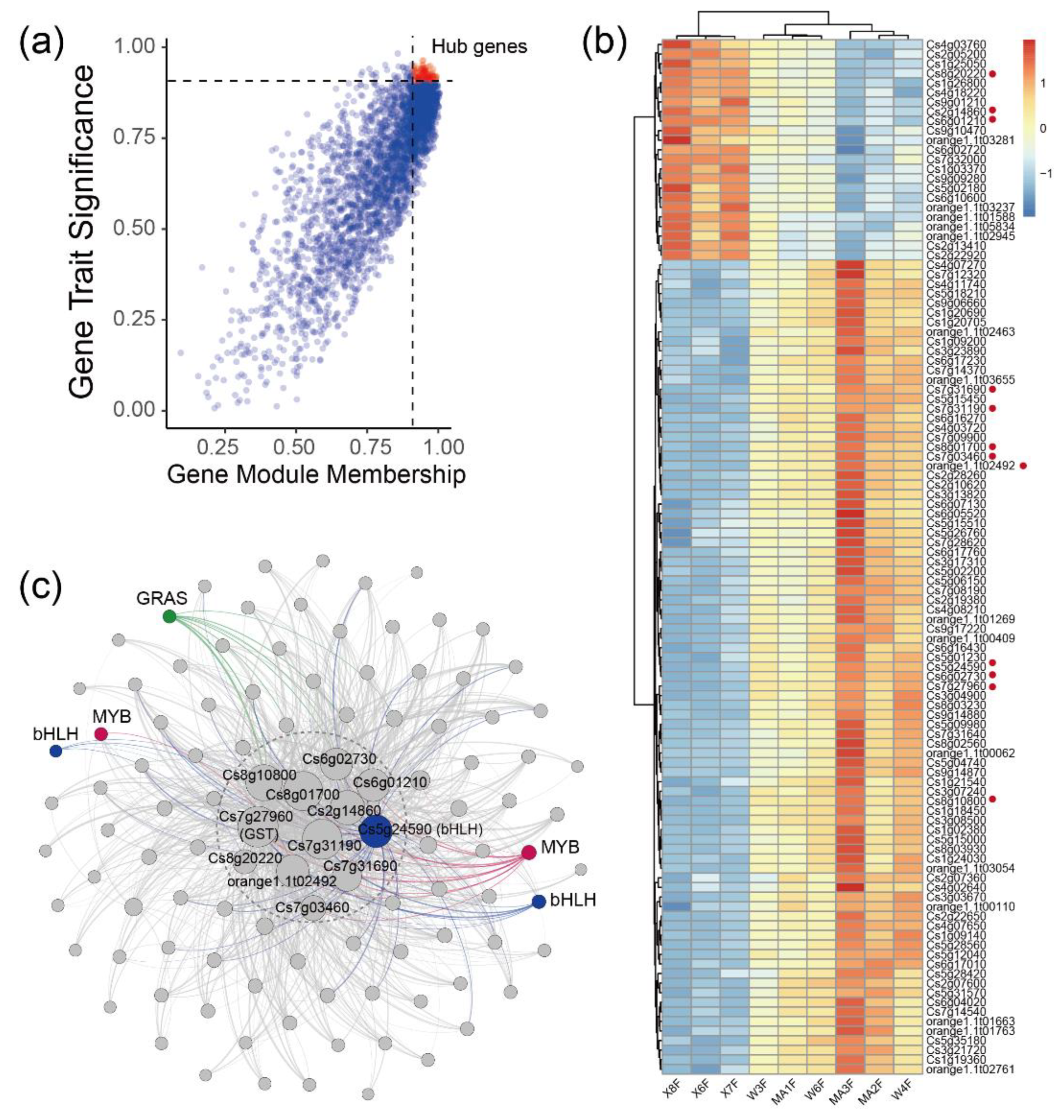
2.3. WGCNA Identifies Candidate Genes Associated with Flavonoid content
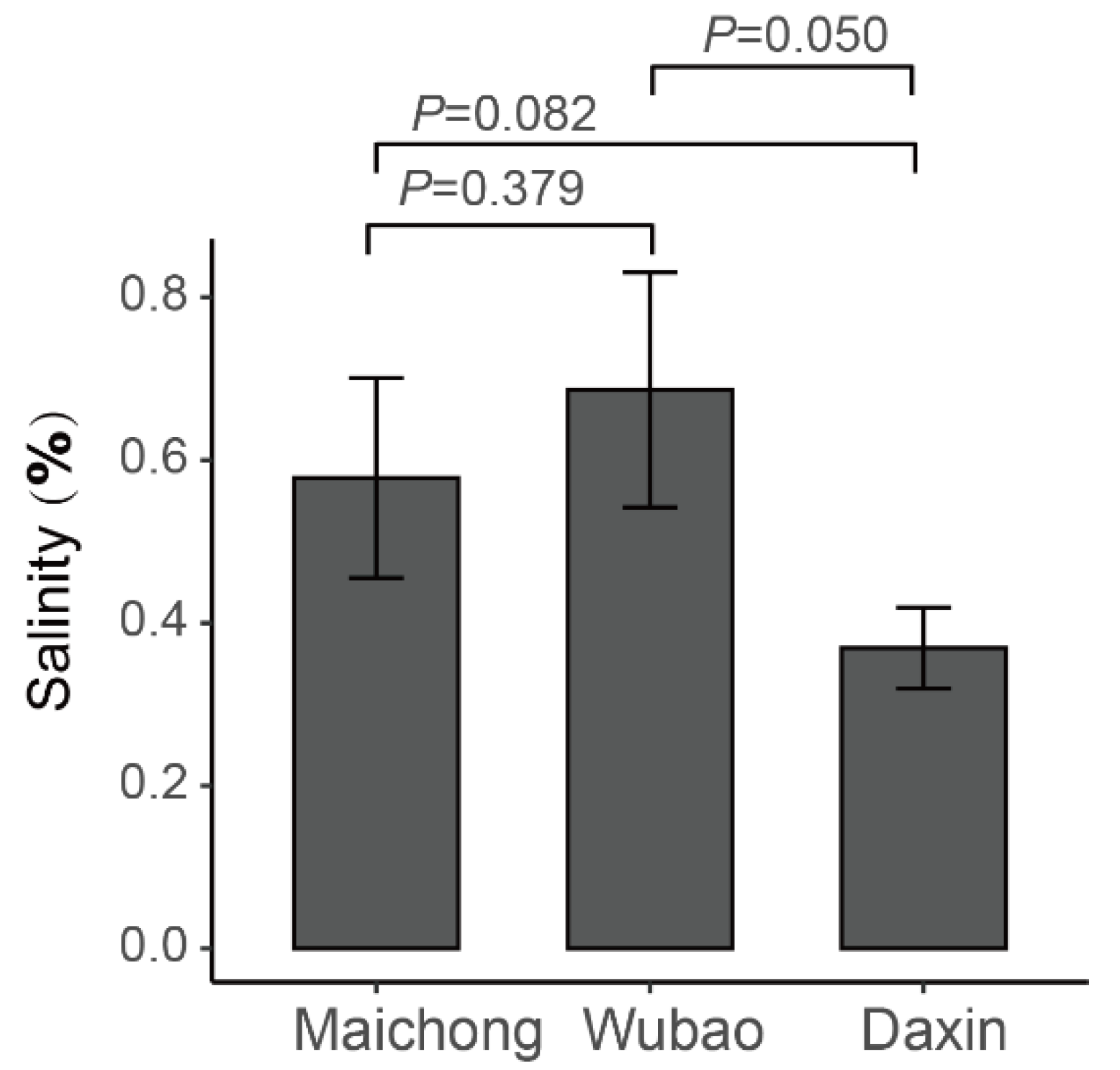
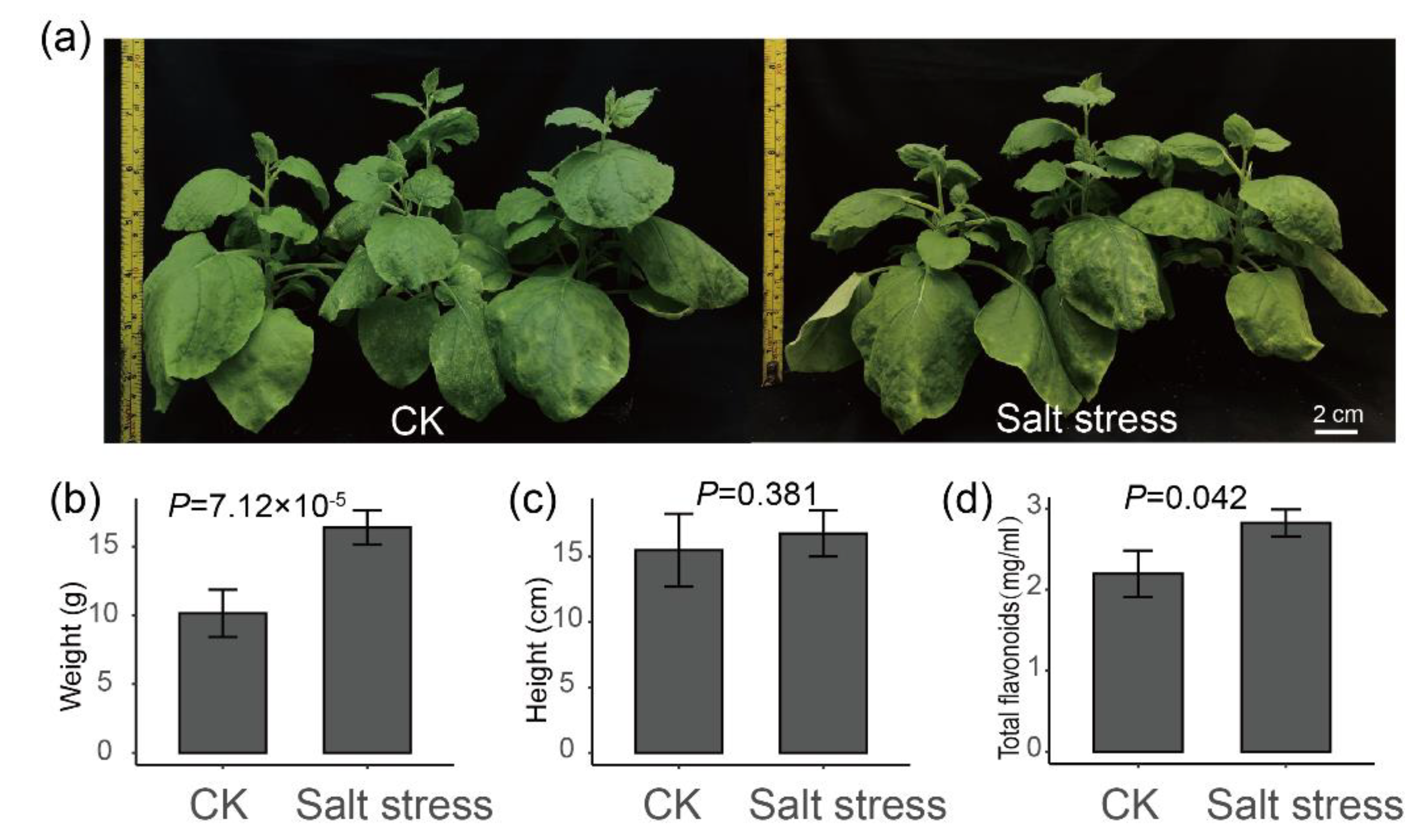
2.4. Salt Stress Plays an Important Role in the Accumulation of Flavonoids
2.5. Verification of Gene Expression Patterns by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation and Transcriptome Sequencing
4.3. Identification of Co-Expression Modules and Hub Genes
4.4. Metabolic Profiling
4.5. Tobacco Salt Stress Treatment
4.6. Validation of the RNA Sequence Data Using qRT-PCR
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Liu, E.H.; Zhao, P.; Duan, L.; Zheng, G.D.; Guo, L.; Yang, H.; Li, P. Simultaneous determination of six bioactive flavonoids in Citri Reticulatae Pericarpium by rapid resolution liquid chromatography coupled with triple quadrupole electrospray tandem mass spectrometry. Food Chem. 2013, 141, 3977–3983. [Google Scholar] [CrossRef] [PubMed]
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, One Sections; China Medical Science Press: Beijing, China, 2020; pp. 197–200. [Google Scholar]
- Zheng, Y.; Zeng, X.; Peng, W.; Wu, Z.; Su, W. Study on the Discrimination between Citri Reticulatae Pericarpium Varieties Based on HS-SPME-GC-MS Combined with Multivariate Statistical Analyses. Molecules 2018, 23, 1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Zhang, J.; Zhang, Y.; Su, H.; Qiu, X.; Gong, L.; Huang, J.; Bai, J.; Huang, Z.; Xu, W. Chemical and genetic discrimination of commercial Guangchenpi (Citrus reticulata ’Chachi’) by using UPLC-QTOF-MS/MS based metabolomics and DNA barcoding approaches. Rsc Adv. 2019, 9, 23373–23381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.Y.; Zeng, X.; Peng, W.; Wu, Z.; Su, W.W. Characterisation and classification of Citri Reticulatae Pericarpium varieties based on UHPLC-Q-TOF-MS/MS combined with multivariate statistical analyses. Phytochem. Anal. 2019, 30, 278–291. [Google Scholar] [CrossRef]
- Yu, X.; Sun, S.; Guo, Y.; Liu, Y.; Yang, D.; Li, G.; Lü, S. Citri Reticulatae Pericarpium (Chenpi): Botany, ethnopharmacology, phytochemistry, and pharmacology of a frequently used traditional Chinese medicine. J. Ethnopharmacol. 2018, 220, 265–282. [Google Scholar] [CrossRef]
- Qian, Z.M.; Huang, Q.; Feng, X.L.; Li, D.Q. Comparison of the chemical-antioxidant profiles of different parts of Citrus reticulata Blanco (Rutaceae) based on OLE-HPLC-DAD-MS/MS-ABTS assay. J. Food Meas. Charact. 2021, 15, 3873–3883. [Google Scholar] [CrossRef]
- Ho, S.C.; Kuo, C.T. Hesperidin, nobiletin, and tangeretin are collectively responsible for the anti-neuroinflammatory capacity of tangerine peel (Citri reticulatae pericarpium). Food Chem Toxicol. 2014, 71, 176–182. [Google Scholar] [CrossRef]
- Liang, S.; Wen, Z.; Tang, T.; Liu, Y.; Dang, F.; Xie, T.; Wu, H. Study on flavonoid and bioactivity features of the pericarp of Citri Reticulatae ‘chachi’ during storage. Arab. J. Chem. 2022, 15, 1–13. [Google Scholar] [CrossRef]
- Liu, M.S.; Wang, J.; Deng, H.D.; Geng, L.L. Comparative study on main components and detection methods of Pericarpium Citri Reticulatae from different habitats. IOP Conf. Ser. Earth Environ. Sci. 2021, 705, 1–8. [Google Scholar] [CrossRef]
- Luo, M.; Luo, H.; Hu, P.; Yang, Y.; Wu, B.; Zheng, G. Evaluation of chemical components in Citri Reticulatae Pericarpium of different cultivars collected from different regions by GC-MS and HPLC. Food Sci. Nutr. 2018, 6, 400–416. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Bioch. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Shukla, S. Impact of various factors responsible for fluctuation in plant secondary metabolites. J. Appl. Res. Med. Aromat. Plants 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Li, Y.Q.; Kong, D.X.; Huang, R.S.; Liang, H.L.; Xu, C.G.; Wu, H. Variations in essential oil yields and compositions of Cinnamomum cassia leaves at different developmental stages. Ind. Crops Prod. 2013, 47, 92–101. [Google Scholar] [CrossRef]
- Luo, Y.; Zeng, W.; Huang, K.E.; Li, D.X.; Chen, W.; Yu, X.Q.; Ke, X.H. Discrimination of Citrus reticulata Blanco and Citrus reticulata ‘Chachi’ as well as the Citrus reticulata ‘Chachi’ within different storage years using ultra high performance liquid chromatography quadrupole/time-of-flight mass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2019, 171, 218–231. [Google Scholar]
- He, X.; Huang, S.; Wu, M.; Wu, M.; Zhang, Y.; Ma, Z.; Liu, L.; Cao, H. Simultaneous quantitative analysis of ten bioactive flavonoids in Citri Reticulatae Pericarpium Viride (Qing Pi) by ultrahigh-performance liquid chromatography and high-resolution mass spectrometry combined with chemometric methods. Phytochem. Anal. 2021, 32, 1152–1161. [Google Scholar] [CrossRef]
- Zheng, G.D.; Chao, Y.; Liu, M.; Yang, Y.; Zhang, D.; Wang, K.; Tao, Y.; Zhang, J.; Li, Y.; Wei, M. Evaluation of dynamic changes in the bioactive components in Citri Reticulatae Pericarpium (Citrus reticulata ‘Chachi’) under different harvesting and drying conditions. J. Sci. Food Agric. 2021, 101, 3280–3289. [Google Scholar] [CrossRef]
- Wang, G.; Chen, S.; Gu, G.; Qiu, J.; Chen, Y.; Jia, Z.; Tang, H. Comparison of the Content of Flavonoids, Total Phenols, and Carotenoids and Antioxidant Activity in Guang Citri reticulatae Pericarpium during the Aging Time. Pharmacogn. Mag. 2020, 16, 375–381. [Google Scholar]
- Fu, M.; An, K.; Xu, Y.; Chen, Y.; Wu, J.; Yu, Y.; Zou, B.; Xiao, G.; Ti, H. Effects of different temperature and humidity on bioactive flavonoids and antioxidant activity in Pericarpium Citri Reticulata (Citrus reticulata ‘Chachi’). LWT Food Sci. Technol. 2018, 93, 167–173. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, Y.; Liu, X.; Qi, J. Change in nutrients of Citri Reticulatae ‘Chachi’ in relation to browning during aging. J. Food Compos. Anal. 2022, 107, 1–7. [Google Scholar] [CrossRef]
- Zheng, G.; Chao, Y.; Luo, M.; Xie, B.; Zhang, D.; Hu, P.; Yang, X.; Yang, D.; Wei, M. Construction and Chemical Profile on “Activity Fingerprint” of Citri Reticulatae Pericarpium from Different Cultivars Based on HPLC-UV, LC/MS-IT-TOF, and Principal Component Analysis. Evid.Based Complement. Altern. Med. 2020, 2020, 4736152. [Google Scholar] [CrossRef] [Green Version]
- Zeng, S.L.; Li, S.Z.; Xiao, P.T.; Cai, Y.Y.; Chu, C.; Chen, B.Z.; Li, P.; Li, J.; Liu, E.H. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci. Adv. 2020, 6, eaax6208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.; Yeo, E.; Song, E.; Chang, Y.-H.; Han, B.-K.; Choi, H.-J.; Hwang, J. Bioconversion of Citrus unshiupeel extracts with cytolase suppresses adipogenic activity in 3T3-L1 cells. Nutr. Res. Pract. 2015, 9, 599–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Fang, S.; Yang, W.; Shang, X.; Fu, X. Light quality affects flavonoid production and related gene expression in Cyclocarya paliurus. J. Photochem. Photobiol. B 2018, 179, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Genzeng, R.; Jianghui, C.; Xiaodong, X.; Yukun, G.; Shilin, W.; Jinhua, C.; Hao, X. Flavonoid Biosynthesis Pathway Participating in Salt Resistance in a Landrace Sweet Sorghum Revealed by RNA-Sequencing Comparison With Grain Sorghum. J. Agric. Sci. 2019, 11, 63–76. [Google Scholar] [CrossRef]
- Zoratti, L.; Karppinen, K.; Luengo Escobar, A.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Du, G.; Chen, J.; Zhou, J. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 2015, 5, 13477. [Google Scholar] [CrossRef] [Green Version]
- Cress, B.F.; Toparlak, Ö.D.; Guleria, S.; Lebovich, M.; Stieglitz, J.T.; Englaender, J.A.; Jones, J.A.; Linhardt, R.J.; Koffas, M.A. CRISPathBrick: Modular Combinatorial Assembly of Type II-A CRISPR Arrays for dCas9-Mediated Multiplex Transcriptional Repression in E. coli. ACS Synth. Biol. 2015, 4, 987–1000. [Google Scholar] [CrossRef]
- Frydman, A.; Weisshaus, O.; Bar-Peled, M.; Huhman, D.V.; Sumner, L.W.; Marin, F.R.; Lewinsohn, E.; Fluhr, R.; Gressel, J.; Eyal, Y. Citrus fruit bitter flavors: Isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 2004, 40, 88–100. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, J.; Xia, R. Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow). Sci. Hortic. 2010, 125, 110–116. [Google Scholar] [CrossRef]
- Chen, L.; Guo, H.; Lin, Y.; Cheng, H. Chalcone synthase EaCHS1 from Eupatorium adenophorum functions in salt stress tolerance in tobacco. Plant Cell Rep. 2015, 34, 885–894. [Google Scholar]
- Lam, P.Y.; Wang, L.; Lui, A.C.W.; Liu, H.; Takeda-Kimura, Y.; Chen, M.X.; Zhu, F.Y.; Zhang, J.; Umezawa, T.; Tobimatsu, Y.; et al. Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol. 2022, 188, 1993–2011. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.S.; Xing, X.J.; Peng, R.H.; Zhu, B.; Gao, J.J.; Yao, Q.H. Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant 2016, 156, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant Flavonoids—Biosynthesis, Transport and Involvement in Stress Responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [Green Version]
- Loyall, L.; Uchida, K.; Braun, S.; Furuya, M.; Frohnmeyer, H. Glutathione and a UV Light–Induced Glutathione S-Transferase Are Involved in Signaling to Chalcone Synthase in Cell Cultures. Plant Cell 2000, 12, 1939–1950. [Google Scholar]
- Genzel, F.; Dicke, M.D.; Junker-Frohn, L.V.; Neuwohner, A.; Thiele, B.; Putz, A.; Usadel, B.; Wormit, A.; Wiese-Klinkenberg, A. Impact of Moderate Cold and Salt Stress on the Accumulation of Antioxidant Flavonoids in the Leaves of Two Capsicum Cultivars. J. Agric. Food Chem. 2021, 69, 6431–6443. [Google Scholar] [CrossRef]
- Singh, D.P.; Prabha, R.; Meena, K.K.; Sharma, L.; Sharma, A.K. Induced Accumulation of Polyphenolics and Flavonoids in Cyanobacteria under Salt Stress Protects Organisms through Enhanced Antioxidant Activity. Am. J. Plant Sci. 2014, 5, 726–735. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 1–18. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278–291. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]


| Gene | Former Primer | Reverse Primer |
|---|---|---|
| PAL | 5′-ACGTTGAAGGGAGCCATTT-3′ | 5′-ACGTTGAAGGGAGCCATTT-3′ |
| 4CL | 5′-TTCCGATCAAAGTTGCCCGA-3′ | 5′-CGGCAGCGAATTTACGTGAG-3′ |
| CYP73A | 5′-AGGGAGTGGAGTTCGGATCA-3′ | 5′-CGTGAAGAAAGGGACGGTCA-3′ |
| CHS | 5′-GGCCTCAACCCATCTGTCAA-3′ | 5′-AGCATACGACTAGAACGCGG-3′ |
| CHI | 5′-ATGCAAGTCCCAGTGCATCA-3′ | 5′-TTGGGCAACGGATTGCAAAG-3′ |
| C12RT1 | 5′-GAGAGAGGTGGCCGACATTA-3′ | 5′-TCTTCGCACACTTGGATGCC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, J.; Peng, T.; Bai, M.; Bai, H.; Li, H.; Pan, H.; He, H.; Liu, H.; Wu, H. Transcriptome and Metabolome Analyses Provide Insights into the Flavonoid Accumulation in Peels of Citrus reticulata ‘Chachi’. Molecules 2022, 27, 6476. https://doi.org/10.3390/molecules27196476
Su J, Peng T, Bai M, Bai H, Li H, Pan H, He H, Liu H, Wu H. Transcriptome and Metabolome Analyses Provide Insights into the Flavonoid Accumulation in Peels of Citrus reticulata ‘Chachi’. Molecules. 2022; 27(19):6476. https://doi.org/10.3390/molecules27196476
Chicago/Turabian StyleSu, Jianmu, Tianhua Peng, Mei Bai, Haiyi Bai, Huisi Li, Huimin Pan, Hanjun He, Huan Liu, and Hong Wu. 2022. "Transcriptome and Metabolome Analyses Provide Insights into the Flavonoid Accumulation in Peels of Citrus reticulata ‘Chachi’" Molecules 27, no. 19: 6476. https://doi.org/10.3390/molecules27196476
APA StyleSu, J., Peng, T., Bai, M., Bai, H., Li, H., Pan, H., He, H., Liu, H., & Wu, H. (2022). Transcriptome and Metabolome Analyses Provide Insights into the Flavonoid Accumulation in Peels of Citrus reticulata ‘Chachi’. Molecules, 27(19), 6476. https://doi.org/10.3390/molecules27196476







