Classification and Expression Profile of the U-Box E3 Ubiquitin Ligase Enzyme Gene Family in Maize (Zea mays L.)
Abstract
:1. Introduction
2. Results
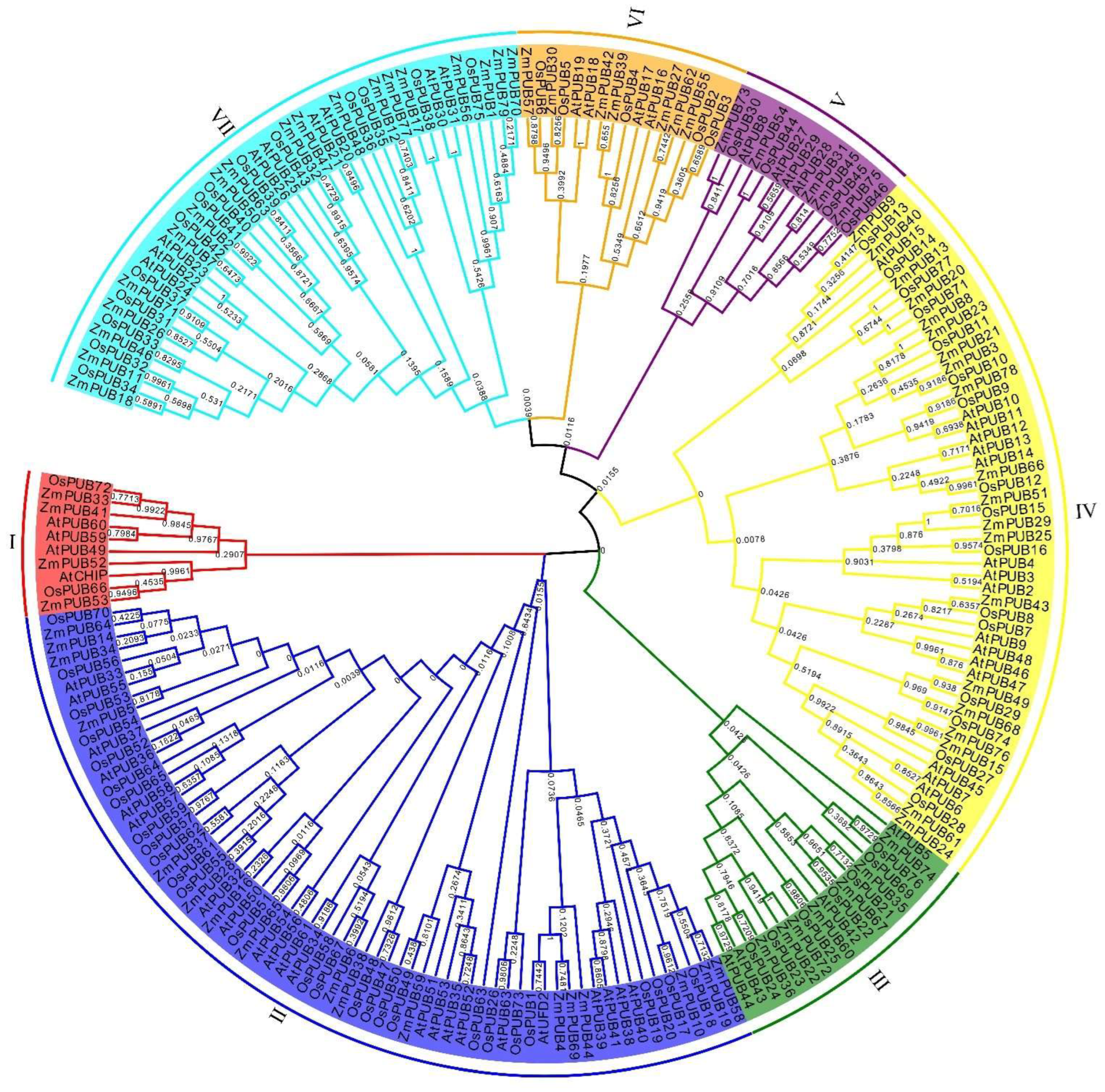
2.1. Identification and Phylogenetic Analysis of PUB Genes in Maize
2.2. Genetic Structures and Conserved Motifs Analyses of the PUB Gene Family
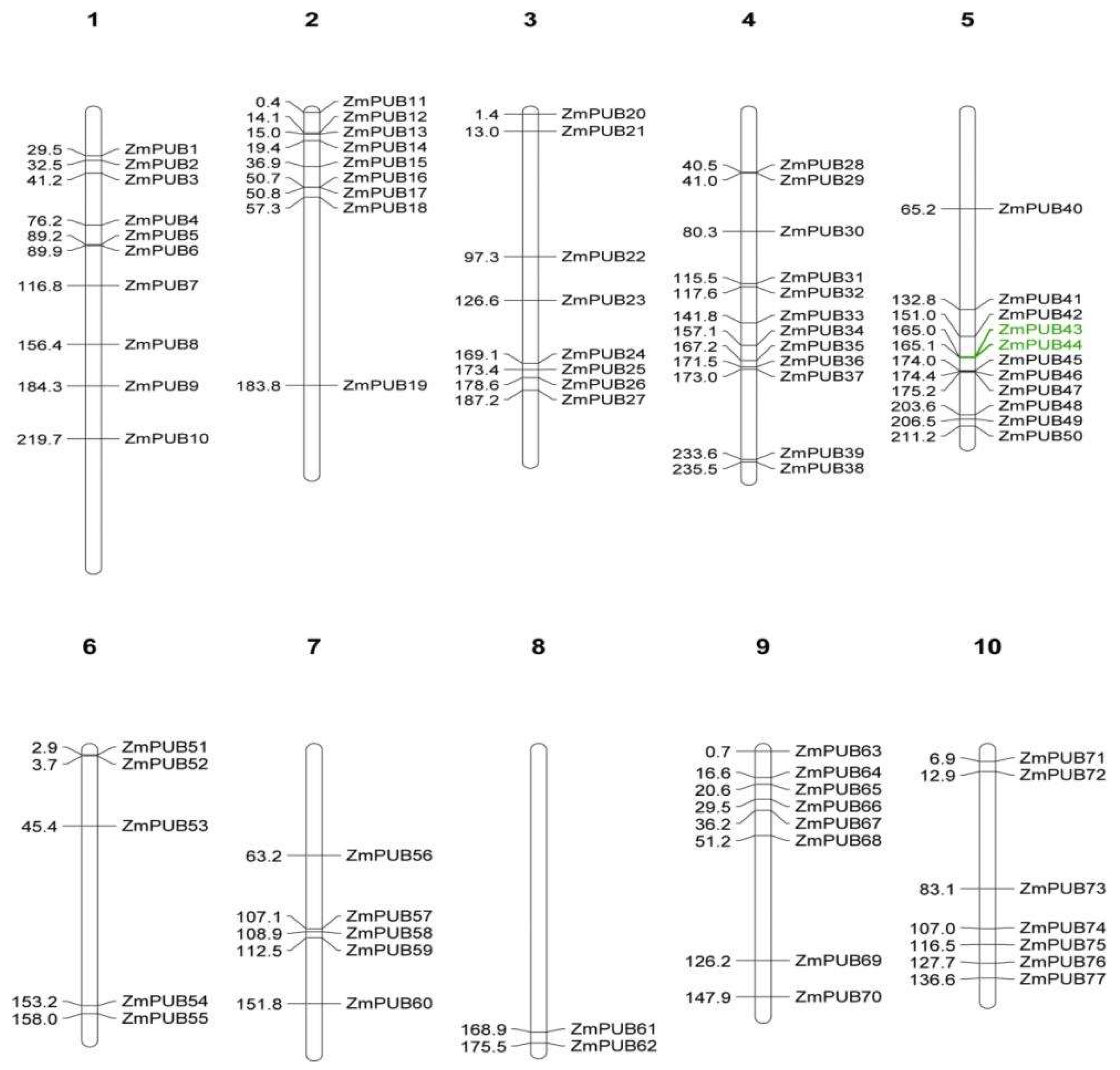
2.3. Chromosomal Locations, Duplications, and Synteny Analysis of the Maize PUB Gene Family
2.4. Cis-Acting Regulatory Element Analysis
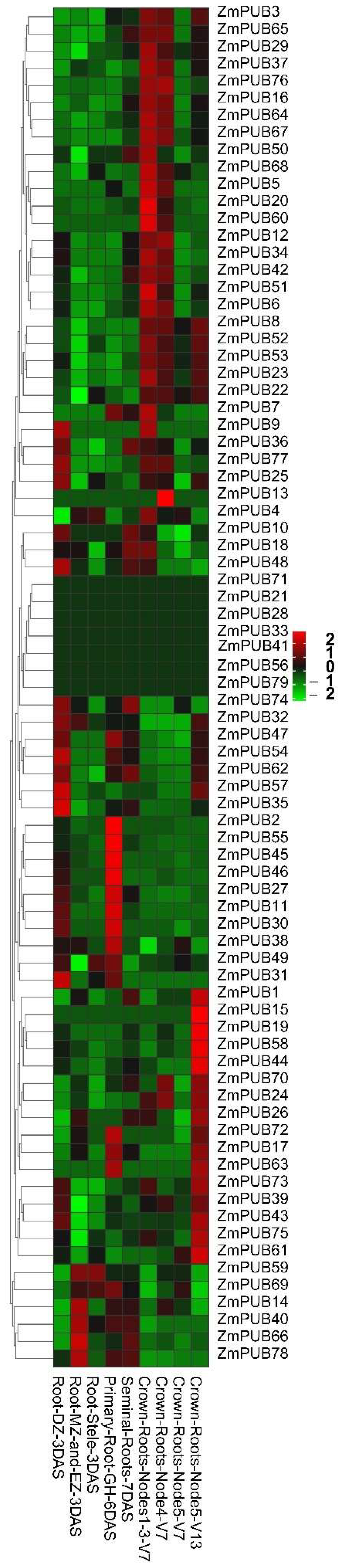
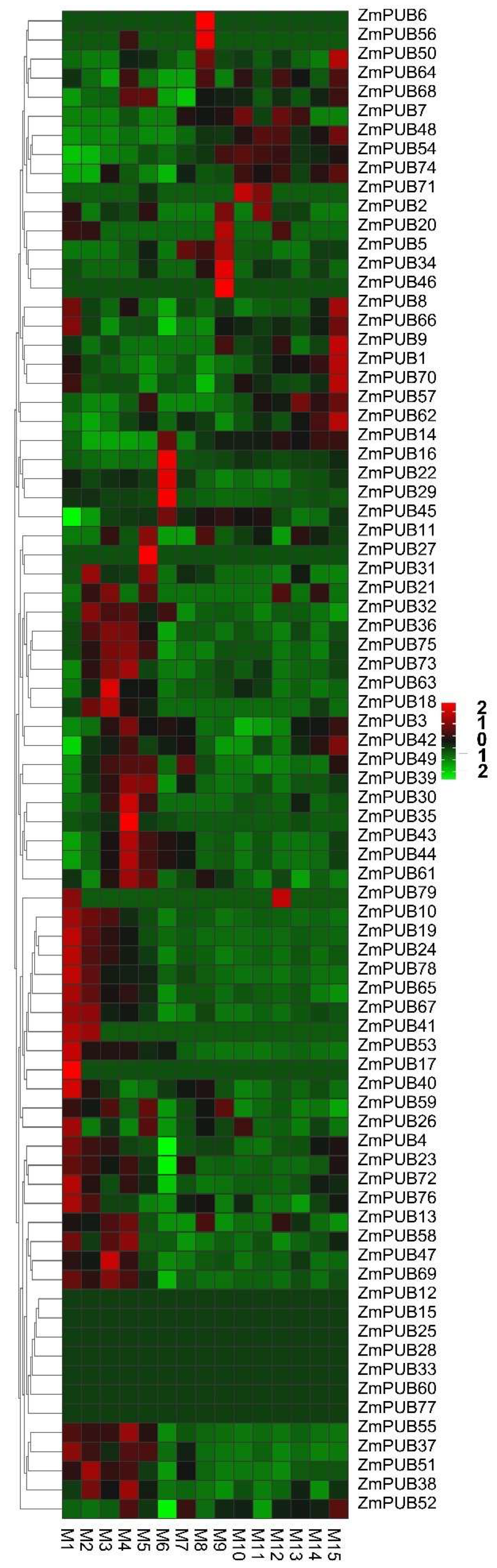
2.5. Expression Profiles of ZmPUB Genes during Root Development
2.6. Expression Profiles of ZmPUB Genes during Leaf Development
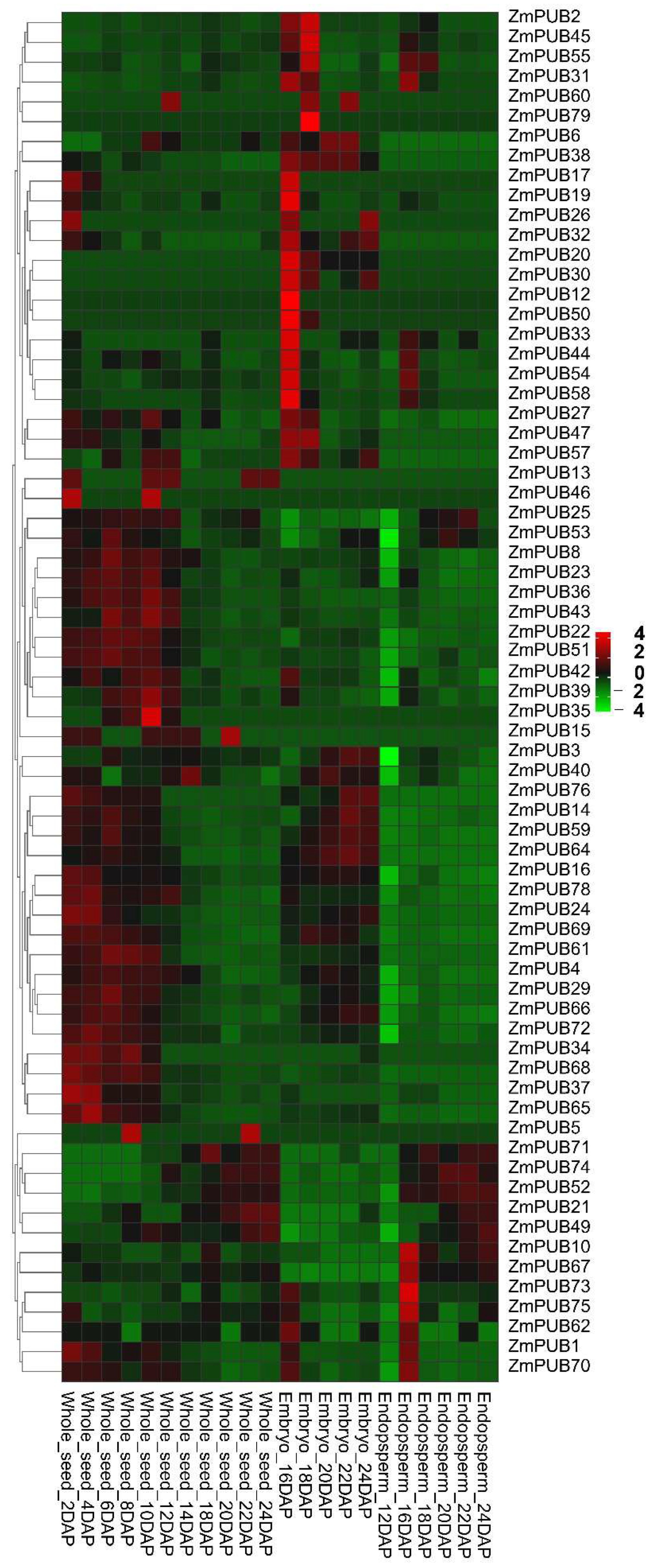
2.7. Expression Profiles of ZmPUB Genes during Seed Development
2.8. Expression Profiles of ZmPUB Genes during Abiotic Stress
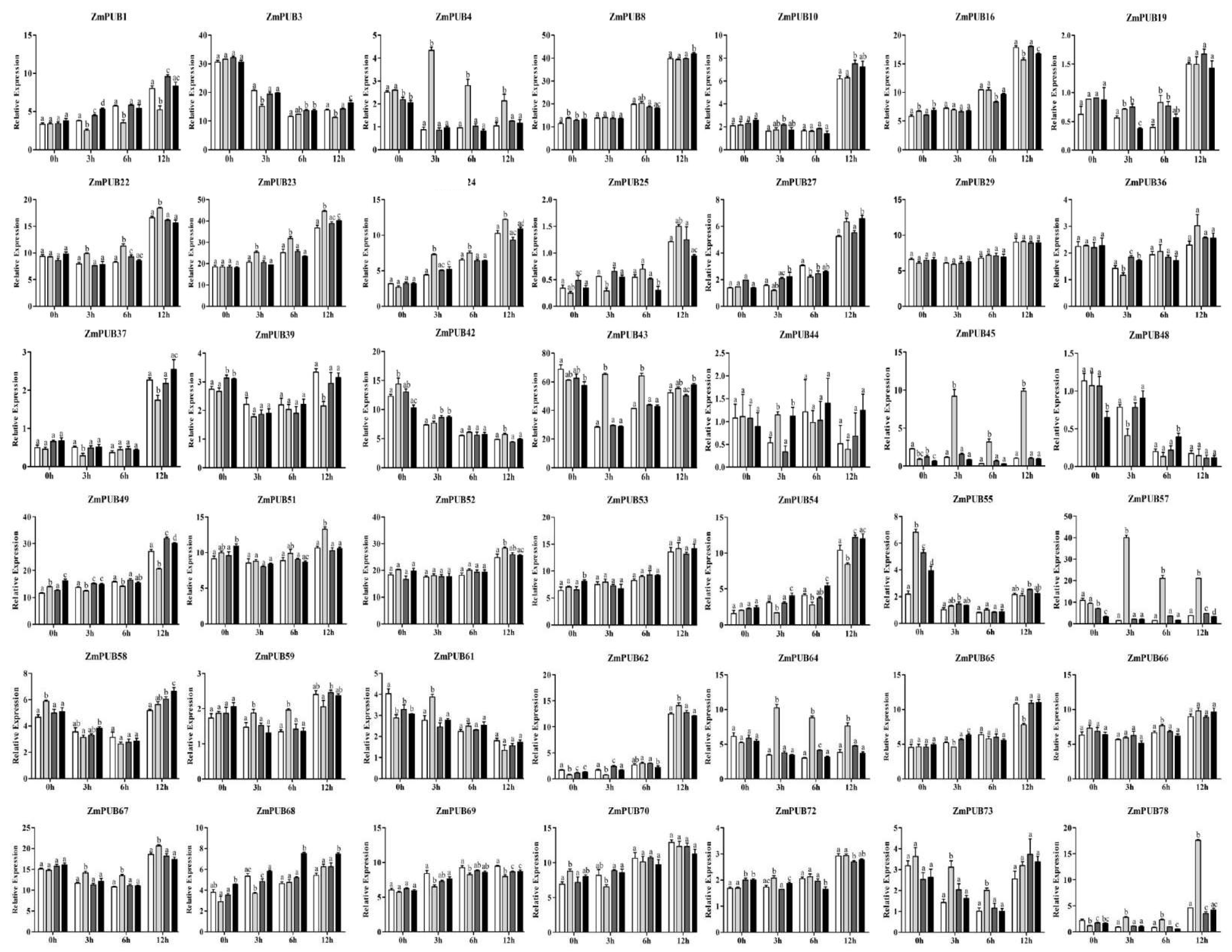
2.9. Expression Profiles of ZmPUB Genes during Hormonal Treatment
3. Discussion
4. Materials and Methods
4.1. Screening for PUB Genes in the Maize Genome
4.2. Multiple Alignments and Phylogenetic Analyses
4.3. Genetic Structure and Conserved Motif Analyses
4.4. Chromosomal Mapping, Gene Duplication (GD), and Synteny Analyses
4.5. Cis-Acting Regulatory Element Analysis
4.6. RNA-seq and Gene Expression Profile
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PUB | The U-box E3 |
| ZmPUB | The maize PUB |
| SD | Segmental duplication |
| TD | Tandem duplication |
| GD | Gene duplication |
| UPS | The ubiquitin proteasome system |
| E1 | The ubiquitin-activating enzyme |
| E2 | The ubiquitin-conjugating enzyme |
| E3 | The ubiquitin ligase enzyme |
| HECT | Homologous to E6-AP COOH-Terminus |
| RING | Really Interesting New Gene |
| CRLs | Cullin-RING Ligase |
| LRR1 | Leucine rich repeat protein 1 |
| KIN7 | Kinase 7 |
| ABA | Abscisic acid |
| BR | Brassinosteroid |
| ETH | Ethylene |
| AUX | Auxin |
| SA | Salicylic acid |
| GA | Gibberellin |
| MeJA | Methyl jasmonate |
| JA | Jasmonic acid |
| HMM | Hidden Markov model |
| RNA-seq | RNA-sequencing |
| MW | molecular weight |
| PI | isoelectric point |
| NJ | Neighbor-joining |
| UTR | Untranslated region |
| GD | Gene duplication |
| SD | Segmental duplication |
| TD | Tandem duplication |
| Mya | Million years ago |
| FPKM | Fragment per kilobase of transcript per million mapped reads |
References
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell. Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Zhang, X.; Lin, H.K. Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene 2010, 29, 4493–4503. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The ubiquitin-proteasome proteolytic pathway. Cell 1994, 79, 13–21. [Google Scholar] [CrossRef]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Song, S.S.; Xie, D.X. The ubiquitin-proteosome pathway and plant development. Chin. Bull. Bot. 2006, 23, 564–577. [Google Scholar] [CrossRef]
- Vandemark, A.P.; Hill, C.P. Structural basis of ubiquitylation. Curr. Opin. Struct. Biol. 2002, 12, 822–830. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, s.; Ulrich, h.d.; Mayer, t.u.; Jentsch, s. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Hatakeyama, S.; Yada, M.; Matsumoto, M.; Ishida, N.; Nakayama, K.I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001, 276, 33111–33120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.R.; Chan, H.P.; Venu, R.C.; Gough, J.; Wang, G.L. Classification, expression pattern, and E3 ligase activity assay of rice u-box-containing proteins. Mol. Plant Breed. 2008, 1, 800–815. [Google Scholar] [CrossRef] [PubMed]
- Wiborg, J.; O’ Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef]
- Wang, C.; Duan, W.K.; Riquicho, A.R.; Jing, Z.G.; Liu, T.K.; Hou, X.L.; Li, Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genom. 2015, 290, 2241–2260. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Cong, Y.; Wang, T.; Zhong, X.; Yang, S.; Li, Y.; Gai, J. Genome-Wide Identifcation of Soybean U-Box E3 Ubiquitin Ligases and Roles of GmPUB8 in Negative Regulation of Drought Stress Response in Arabidopsis. Plant Cell Physiol. 2016, 57, 1189–1209. [Google Scholar] [CrossRef]
- Ryu, M.Y.; Cho, S.K.; Hong, Y.; Kim, J.; Yang, S.W. Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genom. 2019, 20, 326. [Google Scholar] [CrossRef]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 9581. [Google Scholar] [CrossRef]
- Li, Q.Y.; Zhang, Y.F.; Peng, J.; Wang, X.; Zhang, Z.Q.; Dai, X.S.; Jiang, D. Genome wide identification and expression analysis of the U-box gene family in Citrus. Sci. Agric. Sin. 2019, 52, 1942–1960. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, Y.; Lu, J. Overexpression of a stress-responsive U-box protein gene VaPUB afects the accumulation of resistance related proteins in Vitis vinifera ‘Tompson Seedless’. Plant Physiol. Biochem. 2016, 112, 53–63. [Google Scholar] [CrossRef]
- Liu, Y.C.; Zeng, L.Y.; Pan, J.; Qi, C.M.; Yuan, Z.H.; He, F.L.; Hu, K.J.; Zhang, X.W. Bioinformatics analysis of the plant U-box gene families (GrPUBs) in Gossypium raimondii. Mol. Plant Breed. 2018, 16, 4157–4171. [Google Scholar] [CrossRef]
- Zheng, X.W.; Shao, L.H.; Cong, L.I. Genome-wide screening and characterization of the U-box gene family in Medicago truncatula. Acta Pratacult. Sin. 2015, 24, 130–141. [Google Scholar] [CrossRef]
- Patterson, C. A new gun in town: The U-box is a ubiquitin ligase domain. Sci. STKE 2002, 116, pe4. [Google Scholar] [CrossRef] [PubMed]
- Farmer, L.M.; Book, A.J.; Lee, K.H.; Lin, Y.L.; Fu, H.; Vierstra, R.D. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 2010, 22, 124–142. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Qian, Q.; Xu, T.; Zhang, Y.; Dong, G.J.; Gao, T.; Xue, Q.; Xue, Y.B.; Li, J.M. The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G alpha subunit to regulate Brassinosteroid-mediated growth in rice. PLoS Genet. 2013, 9, e1003391. [Google Scholar] [CrossRef] [PubMed]
- Ilka, B.; Wojciech, U.; Aileen, P.; Christoph, D.; Shakhira, Z.; Mats, H. Semi-dwarf barley (Hordeum vulgare L.) brh2 and ari-l mutants are deficient in a U-box E3 ubiquitin ligase. Plant Growth Regul. 2018, 86, 223–234. [Google Scholar] [CrossRef]
- Kinoshita, A.; ten Hove, C.A.; Tabata, R.; Yamada, M.; Shimizu, N.; Ishida, T.; Yamaguchi, K.; Shigenobu, S.; Takebayashi, Y.; Iuchi, S.; et al. A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 2015, 142, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef]
- González-Lamothe, R.; Tsitsigiannis, D.I.; Ludwig, A.A.; Panicot, M.; Shirasu, K.; Jones, J.D. The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 2006, 18, 1067–1083. [Google Scholar] [CrossRef]
- Li, S. Characterization of Soybean GmPUB1 Proteins That Interact with the Phytophthora sojae Effector Avr1b Protein. Master’s Thesis, Iowa State University, Ames, IA, USA, 2010. [Google Scholar] [CrossRef]
- Hwang, J.H.; Dong, H.S.; Kang, B.G.; Kwak, J.M.; Kim, W.T. Suppression of Arabidopsis AtPUB30 resulted in increased tolerance to salt stress during germination. Plant Cell Rep. 2014, 34, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Wang, Y.; Huang, L.; Du, F.; Zhao, X.; Li, Z.; Wang, W.; Fu, B. A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 2020, 102, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.X. Functional Analysis of Arabidopsis PUB19 U-box E3 Ubiquitin Ligase and Its Interacting Proteins. Master’s Thesis, Zhejiang University, Hangzhou, China, 2015. [Google Scholar]
- Seo, J.S.; Zhao, P.; Jung, C.; Chua, N.H. Plant U-box protein 10 negatively regulates abscisic acid response in Arabidopsis. Appl. Biol. Chem. 2019, 62, 39. [Google Scholar] [CrossRef]
- Wang, N.; Xing, Y.; Lou, Q.; Feng, P.; Liu, S.; Zhu, M.; Yin, W.; Fang, S.; Lin, Y.; Zhang, T.; et al. Dwarf and short grain 1, encoding a putative U-box protein regulates cell division and elongation in rice. J. Plant Physiol. 2016, 209, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Amador, V.; Monte, E.; García-Martínez, J.L.; Prat, S. Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 2001, 106, 343–354. [Google Scholar] [CrossRef]
- Monte, E.; Amador, V.; Russo, E.; Martínez-García, J.; Prat, S. Phor1: A U-box GA signaling component with a role in proteasome degradation? J. Plant Growth Regul. 2003, 22, 152–162. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An expanded maize gene expression atlas based on RNA sequencing and its Use to explore root development. Plant Genome 2016, 9, 1. [Google Scholar] [CrossRef]
- Wang, L.; Czedik-Eysenberg, A.; Mertz, R.A. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 2014, 32, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Childs, K.; Santoro, N.; Foster, C.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 2012, 159, 1730–1744. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 2013, 8, E61005. [Google Scholar] [CrossRef]
- Brenner, M.L.; Cheikh, N. The role of hormones in photosynthate partitioning and seed filling. Plant Horm. 1995, 649–670. [Google Scholar] [CrossRef]
- Jones, R.J.; Brenner, M.L. Distribution of abscisic acid in maize kernel during grain filling. Plant Physiol. 1987, 83, 905–909. [Google Scholar] [CrossRef]
- Wang, Y.F.; Cui, Z.H.; Ruan, Y.Y.; Ma, X.L.; Zhang, L.J. Changes in endogenous hormone of IAA, GA, ZR and ABA in kernels during grain-filling stage in different types of spring maize (Zea mays L.). Plant Physiol. Commun. 2006, 42, 225–228. [Google Scholar] [CrossRef]
- Waters, A.J.; Makarevitch, I.; Noshay, J.; Burghardt, L.T.; Hirsch, C.N.; Hirsch, C.D.; Springer, N.M. Natural variation for gene expression responses to abiotic stress in maize. Plant J. 2016, 89, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Kakumanu, A.; Ambavaram, M.; Klumas, C.; Krishnan, A.; Batlang, U.; Myers, E.; Grene, R.; Pereira, A. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 2012, 160, 846–867. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dong, C.; Sun, D.; Hu, Y.; Xie, J. Genome-Wide identification and analysis of U-Box E3 ubiquitin–protein ligase gene family in Banana. Int. J. Mol. Sci. 2018, 19, 3874. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Jo, B.-S.; Choi, S.S. Introns: The functional benefts of introns in genomes. Genom. Inf. 2015, 13, 112–118. [Google Scholar] [CrossRef]
- Yu, Y.; Li, X.Z.; Guo, D.L.; Zhang, H.L.; Li, G.R.; Li, X.Q.; Zhang, G.H. Genome-wide identifcation and analysis of the U-box family of E3 ligases in grapevine. Russ. J. Plant Physiol. 2016, 63, 835–848. [Google Scholar] [CrossRef]
- Chang, D.; Duda, T.F., Jr. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 2012, 29, 2019–2029. [Google Scholar] [CrossRef]
- Starr, T.K.; Jameson, S.C.; Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003, 21, 139–176. [Google Scholar] [CrossRef]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-box family genes in Medicago truncatula: Key elements in response to salt, cold, and drought stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef]
- Young, B.M.; Cui, L.H.; Kyung, O.T.; Jung, Y.J.; Lee, A.; Park, K.Y.; Kang, B.G.; Kim, W.T. Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 16. [Google Scholar] [CrossRef]
- Peng, L.; Wan, X.; Huang, K.; Pei, L.; Xiong, J.; Li, X.; Wang, J. AtPUB48 E3 ligase plays a crucial role in the thermotolerance of Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 509, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, W.; Wu, Y.; Li, Q.; Zhang, G.; Shi, R.; Yang, J.; Wang, Y.; Wang, W. The involvement of wheat U-box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 2020, 62, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell Online 2008, 20, 1899–1914. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Ning, Y.; Shirsekar, G.; Cai, Y.; Wang, X.; Dai, L.; Wang, Z.; Liu, W.; Wang, G.L. The U-Box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol. 2012, 160, 28–37. [Google Scholar] [CrossRef]
- Liu, Y.B. Cytokinin and Auxin-Regulated Root Meristem Establishment during Arabidopsis Somatic Embryogenesis. Ph.D. Thesis, Shandong Agricultural University, Taian, China, 2011. [Google Scholar] [CrossRef]
- Baluška, F.; Parker, J.; Barlow, P. A role for gibberellic acid in orienting microtubules and regulating cell growth polarity in the maize root cortex. Planta 1993, 191, 149–157. [Google Scholar] [CrossRef]
- Liu, D.T.; Jing, Y.P.; Shi, H.X.; Zhong, T.T.; Wang, Z. Impact of IAA, GA3, and ABA on negative root phototropism and root growth of rice. Acta Agron. Sin. 2014, 40, 1658–1666. [Google Scholar] [CrossRef]
- Chai, M.M. Studies on Gibberellin’s Regulation on Auxin Biosynthesis and Transportation of Rice Root Tip and Cloning and Preliminary Analysis of Rice OsRLMG1. Master’s Thesis, Xinyang Normal University, Xinyang, China, 2020. [Google Scholar] [CrossRef]
- Xue, R.G.; Zhang, B. Increased endogenous methyl jasmonate altered leaf and root development in transgenic soybean plants. J. Genet. Genom. 2007, 4, 339–346. [Google Scholar] [CrossRef]
- Lin, M. Expression Profile Analysis of Leaf Senescence and Related Gene Function Studies in Cotton. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2014. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=1014326785.nh&DbName=CDFD2014 (accessed on 10 August 2022).
- Chen, B.; Li, H.Y.; Liu, X.W.; Xia, B.; Sun, S.W.; Sun, Y.; Miao, H. Effects of different light intensities on morphogenesis and ultrastructure of gibasis pellucida leaf. Acta Pratacult. Sin. 2019, 28, 175–185. [Google Scholar] [CrossRef]
- Wu, Y.; Holding, D.R.; Messing, J. γ-Zeins are essential for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. USA 2010, 107, 12810–12815. [Google Scholar] [CrossRef]
- Sharma, M.; Pandey, G.K. OsPUB75, an Armadillo/U-box protein interacts with GSK3 kinase and functions as negative regulator of abiotic stress responses. Environ. Exp. Bot. 2018, 161, 388–398. [Google Scholar] [CrossRef]
- Acosta, M.A.; Ahumada, M.A.; Lassaga, S.L.; Casco, V.H. PUB16 gene expression under abiotic stress and their putative role as an ARM repeat protein in Arabidopsis thaliana self-pollination pathway. Adv. Biosci. Biotechnol. 2012, 3, 609–619. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, X.; Song, Q.; Sun, Y. Identification of key genes and modules in response to cadmium stress in different rice varieties and stem nodes by weighted gene co-expression network analysis. Sci. Rep. 2020, 10, 9525. [Google Scholar] [CrossRef]
- Bergler, J.; Hoth, S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 2011, 13, 725–730. [Google Scholar] [CrossRef]
- Gui, L.Y.; Liu, S.S.; Chen, Z.M. Plant resistance to insects induced by application of exogenous jasmonic acid and methyl jasmonate. Acta Entomol. Sin. 2004, 4, 507–514. [Google Scholar] [CrossRef]
- Pan, L.; Chen, X.; Xu, W.; Fan, S.; Wan, T.; Zhang, J.; Cai, Y.L. Methyl jasmonate induces postharvest disease resistance to decay caused by Alternaria alternata in sweet cherry fruit. Sci. Hortic. 2022, 292, 110624. [Google Scholar] [CrossRef]
- Yan, J.; Li, H.; Li, Y. Abscisic acid synthesis and root water uptake contribute to exogenous methyl jasmonate-induced improved tomato drought resistance. Plant Biotechnol. Rep. 2022, 16, 183–193. [Google Scholar] [CrossRef]
- Manisha, S.; Amarjeet, S.; Alka, S.; Amita, P.; Vinay, B.; Sanjay, K.; Akhilesh, T.K.; Girdhar, P.K. Comprehensive expression analysis of rice armadillo gene family during abiotic stress and development. DNA Res. 2014, 3, 267–283. [Google Scholar] [CrossRef]
- Kim, M.S.; Kang, K.K.; Cho, Y.G. Molecular and functional analysis of U-box E3 ubiquitin ligase gene family in rice (Oryza sativa). Int. J. Mol. Sci. 2021, 22, 12088. [Google Scholar] [CrossRef]
- Wilkinson, S.; Davies, W.J. ABA-based chemical signaling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002, 25, 195–210. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, T.; Rehman, A.U. Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J. Integr. Plant Biol. 2021, 63, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, W1, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M.A.J. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Yu, J.; Wang, J.; Wei, L.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C.; et al. The genomes of Oryza sativa: A History of Duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; de Peer, Y.V.; Rouzé, P.; Rombauts, S. Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; Leon, N.D.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. BioRxiv 2018, 274100. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]

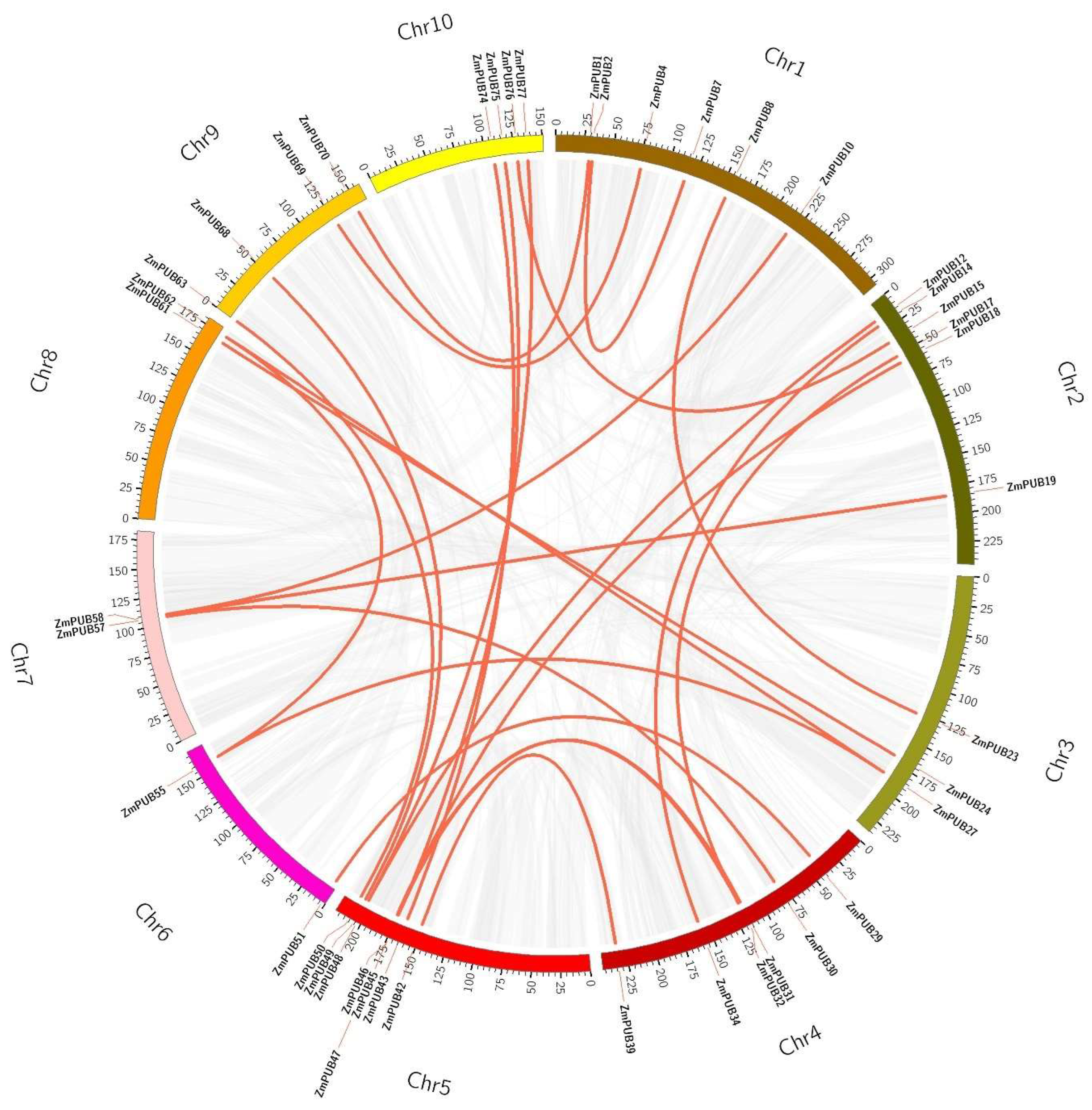



| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplicated Type | Purify Selection | Time (Mya) |
|---|---|---|---|---|---|---|
| ZmPUB43/ZmPUB44 | 0 | 0 | NA | Tandem | NA | NA |
| ZmPUB29/ZmPUB51 | 0.04 | 0.17 | 0.25 | Segmental | Purify selection | 13.07 |
| ZmPUB55/ZmPUB27 | 0.13 | 1.95 | 0.07 | Segmental | Purify selection | 150.27 |
| ZmPUB55/ZmPUB62 | 0.13 | 1.69 | 0.07 | Segmental | Purify selection | 129.99 |
| ZmPUB4/ZmPUB69 | 0.02 | 0.2 | 0.09 | Segmental | Purify selection | 15.01 |
| ZmPUB34/ZmPUB14 | 0.99 | 1.03 | 0.96 | Segmental | Purify selection | 79.21 |
| ZmPUB63/ZmPUB50 | 0.2 | 2.18 | 0.09 | Segmental | Purify selection | 167.62 |
| ZmPUB75/ZmPUB45 | 0.17 | 2.81 | 0.06 | Segmental | Purify selection | 215.77 |
| ZmPUB31/ZmPUB45 | 0.05 | 0.63 | 0.07 | Segmental | Purify selection | 48.32 |
| ZmPUB8/ZmPUB23 | 0.06 | 0.46 | 0.12 | Segmental | Purify selection | 35.08 |
| ZmPUB27/ZmPUB62 | 0.02 | 0.32 | 0.07 | Segmental | Purify selection | 24.28 |
| ZmPUB57/ZmPUB30 | 0.22 | 2.66 | 0.08 | Segmental | Purify selection | 204.83 |
| ZmPUB49/ZmPUB68 | 0.99 | 1.05 | 0.93 | Segmental | Purify selection | 81.07 |
| ZmPUB1/ZmPUB70 | 0.02 | 0.32 | 0.05 | Segmental | Purify selection | 24.67 |
| ZmPUB32/ZmPUB17 | 0.99 | 1.02 | 0.97 | Segmental | Purify selection | 78.39 |
| ZmPUB58/ZmPUB10 | 0 | 0 | NA | Segmental | NA | NA |
| ZmPUB58/ZmPUB19 | 1 | 1.01 | 0.99 | Segmental | Purify selection | 77.35 |
| ZmPUB46/ZmPUB18 | 0.23 | 2.41 | 0.1 | Segmental | Purify selection | 185.29 |
| ZmPUB77/ZmPUB48 | 0.18 | 2.37 | 0.08 | Segmental | Purify selection | 182.21 |
| ZmPUB32/ZmPUB47 | 0.03 | 0.37 | 0.09 | Segmental | Purify selection | 28.78 |
| ZmPUB2/ZmPUB7 | 0.33 | 2.04 | 0.16 | Segmental | Purify selection | 156.71 |
| ZmPUB61/ZmPUB24 | 0.05 | 0.17 | 0.27 | Segmental | Purify selection | 13.26 |
| ZmPUB43/ZmPUB74 | 0.99 | 1.04 | 0.95 | Segmental | Purify selection | 79.82 |
| ZmPUB42/ZmPUB39 | 0.03 | 0.18 | 0.15 | Segmental | Purify selection | 13.78 |
| ZmPUB12/ZmPUB48 | 0.18 | 2.03 | 0.09 | Segmental | Purify selection | 156.32 |
| ZmPUB76/ZmPUB15 | 0.96 | 1.09 | 0.88 | Segmental | Purify selection | 84.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhu, L.; Wu, Z.; Chen, J.; Wang, T.; Zhang, X.; Mei, G.; Wang, J.; Lv, G. Classification and Expression Profile of the U-Box E3 Ubiquitin Ligase Enzyme Gene Family in Maize (Zea mays L.). Plants 2022, 11, 2459. https://doi.org/10.3390/plants11192459
Li X, Zhu L, Wu Z, Chen J, Wang T, Zhang X, Mei G, Wang J, Lv G. Classification and Expression Profile of the U-Box E3 Ubiquitin Ligase Enzyme Gene Family in Maize (Zea mays L.). Plants. 2022; 11(19):2459. https://doi.org/10.3390/plants11192459
Chicago/Turabian StyleLi, Xiangnan, Longming Zhu, Zhenxing Wu, Jianjian Chen, Tingzhen Wang, Xiaoli Zhang, Gaofu Mei, Jian Wang, and Guihua Lv. 2022. "Classification and Expression Profile of the U-Box E3 Ubiquitin Ligase Enzyme Gene Family in Maize (Zea mays L.)" Plants 11, no. 19: 2459. https://doi.org/10.3390/plants11192459





