Virtual Screening of Alkaloid and Terpenoid Inhibitors of SMT Expressed in Naegleria sp.
Abstract
1. Introduction
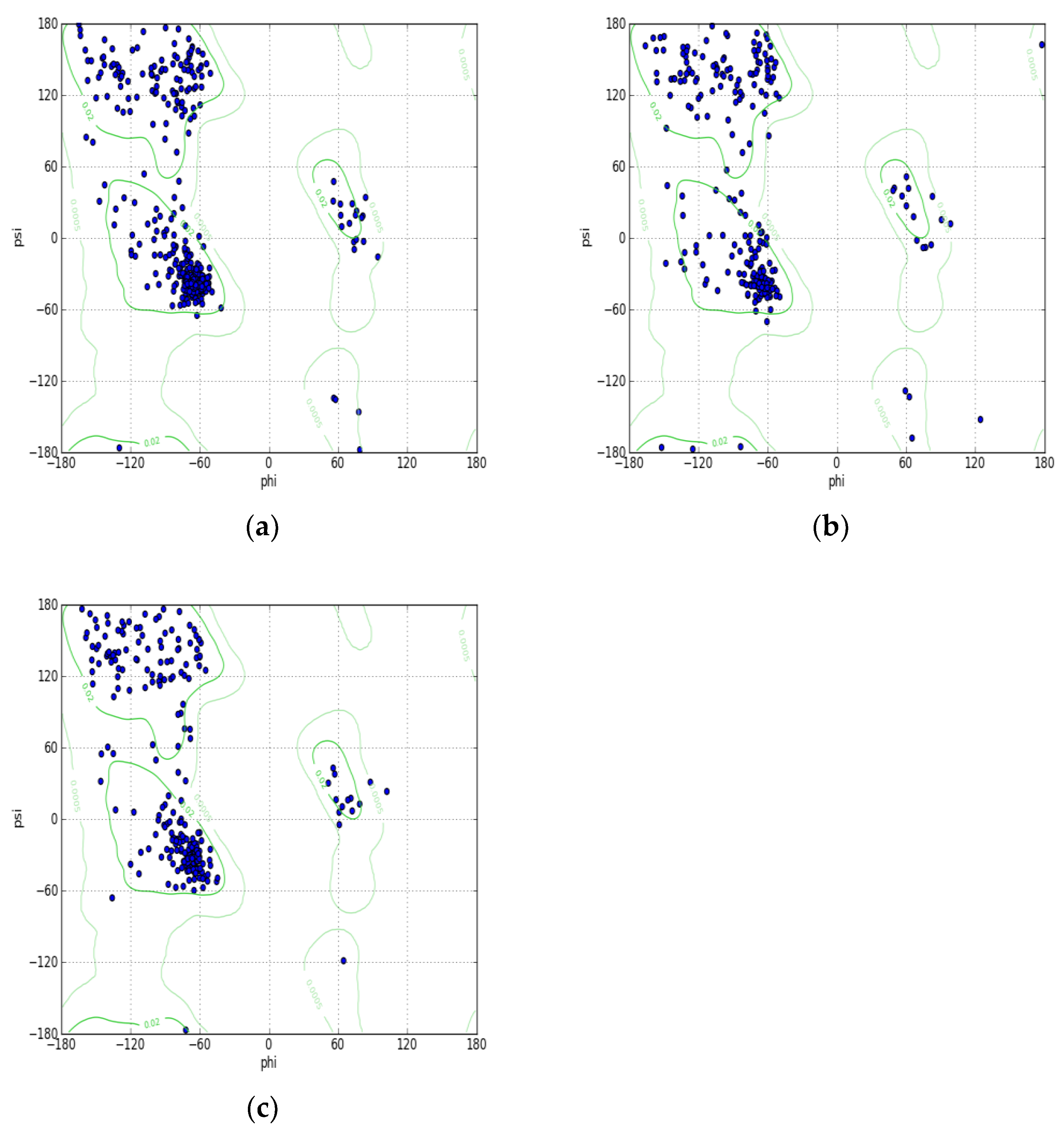
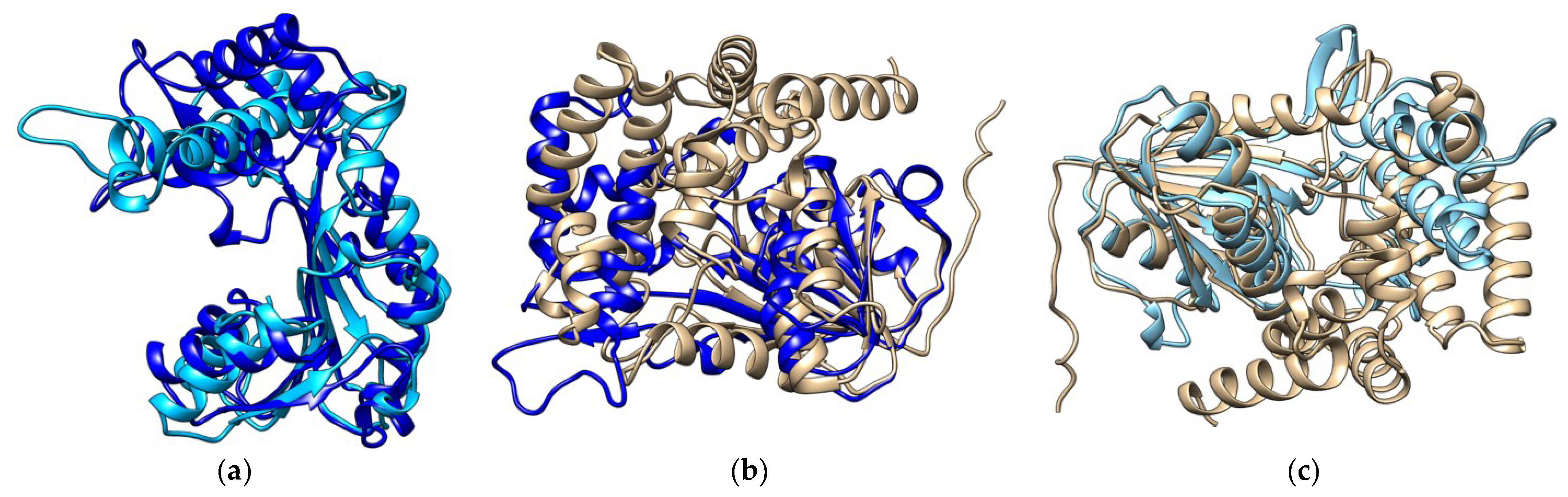
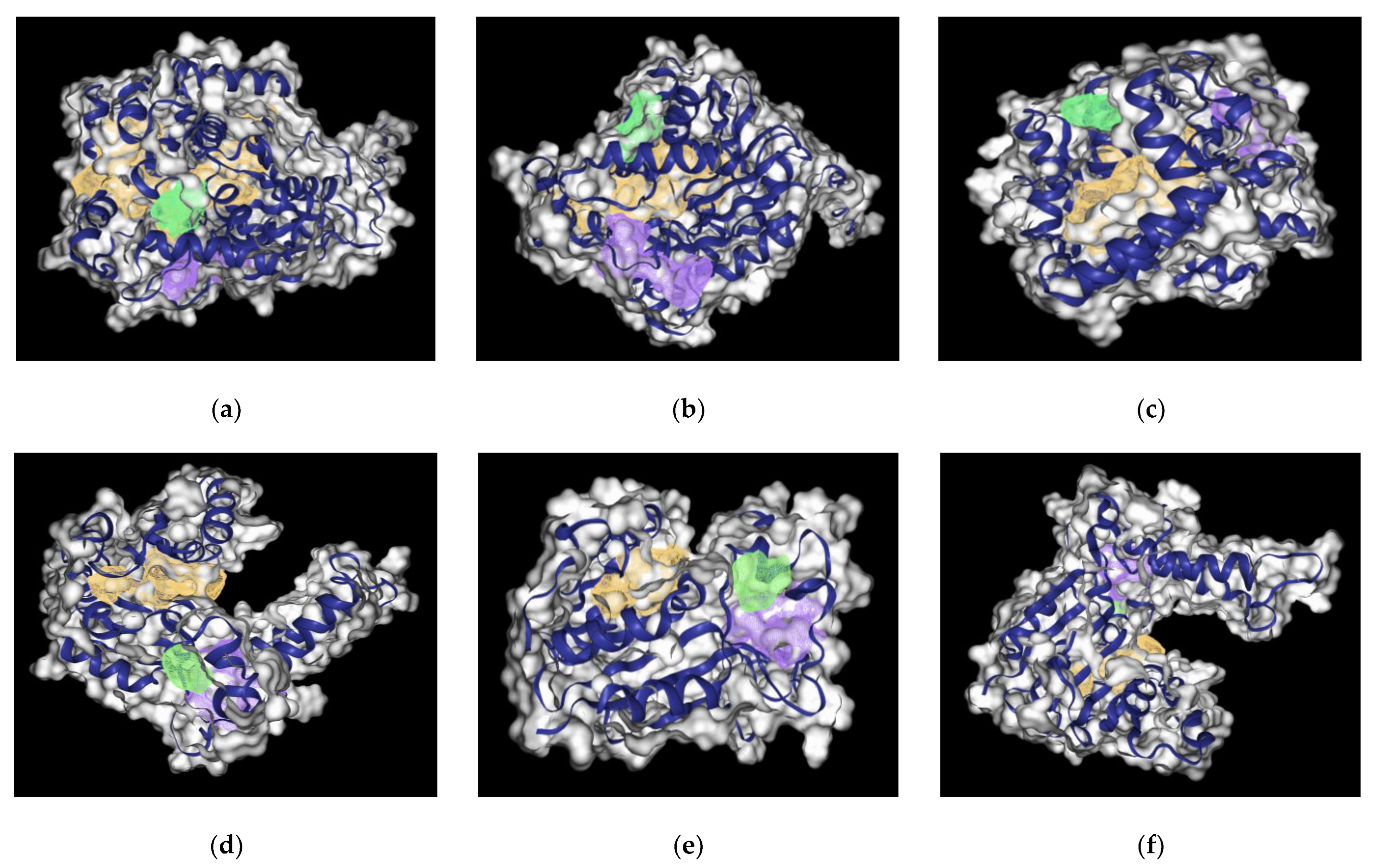
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Piñero, J.E.; Chávez-Munguía, B.; Omaña-Molina, M.; Lorenzo-Morales, J. Naegleria fowleri. Trends Parasitol. 2019, 35, 848–849. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, R.; Khan, N.A. Is ritual cleansing a missing link between fatal infection and brain-eating amoebae? Clin. Infect. Dis. 2012, 54, 1817–1818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Capewell, L.G.; Harris, A.M.; Yoder, J.S.; Cope, J.R.; Eddy, B.A.; Roy, S.L.; Visvesvara, G.S.; Fox, L.M.; Beach, M.J. Diagnosis, Clinical Course, and Treatment of Primary Amoebic Meningoencephalitis in the United States, 1937–2013. J. Pediatr. Infect. Dis. Soc. 2014, 4, e68–e75. [Google Scholar] [CrossRef] [PubMed]
- Jahangeer, M.; Mahmood, Z.; Munir, N.; Waraich, U.; Tahir, I.M.; Akram, M.; Shah, S.M.A.; Zulfqar, A.; Zainab, R. Naegleria fowleri: Sources of infection, pathophysiology, diagnosis, and management; a review. Clin. Exp. Pharmacol. Physiol. 2019, 47, 199–212. [Google Scholar] [CrossRef]
- Heggie, T.W. Swimming with death: Naegleria fowleri infections in recreational waters. Travel Med. Infect. Dis. 2010, 8, 201–206. [Google Scholar] [CrossRef]
- Mahmood, K. Naegleria fowleri in Pakistan—An emerging catastrophe. J. Coll. Physicians Surg. Pakistan. 2015, 25, 159–160. [Google Scholar]
- Baig, A.M.; Khan, N.A. Novel chemotherapeutic strategies in the management of primary amoebic meningoencephalitis due to Naegleria fowleri. CNS Neurosci. Ther. 2014, 20, 289–290. [Google Scholar] [CrossRef]
- Yoder, J.S.; Eddy, B.A.; Visvesvara, G.S.; Capewell, L.; Beach, M.J. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962–2008. Epidemiol. Infect. 2010, 138, 968–975. [Google Scholar] [CrossRef]
- Shariq, A.; Afridi, F.I.; Farooqi, B.J.; Ahmed, S.; Hussain, A. Fatal primary meningoencephalitis caused by Naegleria fowleri. J. Coll. Physicians Surg. Pakistan. 2014, 24, 523–525. [Google Scholar]
- Moseman, E.A. Battling brain-eating amoeba: Enigmas surrounding immunity to Naegleria fowleri. PLoS Pathog. 2020, 16, e1008406. [Google Scholar] [CrossRef]
- Rice, C.A.; Colon, B.L.; Chen, E.; Hull, M.V.; Kyle, D.E. Discovery of repurposing drug candidates for the treatment of diseases caused by pathogenic free-living amoebae. PLoS Negl. Trop. Dis. 2020, 14, e0008353. [Google Scholar] [CrossRef] [PubMed]
- Goswick, S.M.; Brenner, G.M. Activities of therapeutic agents against Naegleria fowleri in vitro and in a mouse model of primary amebic meningoencephalitis. J. Parasitol. 2003, 89, 837–842. [Google Scholar] [CrossRef]
- Alli, A.; Ortiz, J.F.; Cox, Á.M.; Armas, M.; Orellana, V.A. Miltefosine: A Miracle Drug for Meningoencephalitis Caused by Free-Living Amoebas. Cureus 2021, 13, e13698. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.; Tunac, J.B.; Galindo-Gómez, S.; Silva-Olivares, A.; Shibayama, M.; McKerrow, J.H. Corifungin, a new drug lead against Naegleria, identified from a high-throughput screen. Antimicrob. Agents Chemother. 2012, 56, 5450–5457. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jung, S.Y.; Lee, Y.J.; Song, K.J.; Kwon, D.; Kim, K.; Park, S.; Im, K.I.; Shin, H.J. Effect of therapeutic chemical agents in vitro and on experimental meningoencephalitis due to Naegleria fowleri. Antimicrob. Agents Chemother. 2008, 52, 4010–4016. [Google Scholar] [CrossRef] [PubMed]
- Nes, W.D.; Zhou, W.; Ganapathy, K.; Liu, J.L.; Vatsyayan, R.; Chamala, S.; Hernandez, K.; Miranda, M. Sterol 24-C-methyltransferase: An enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch. Biochem. Biophys. 2009, 481, 210–218. [Google Scholar] [CrossRef]
- Bexkens, M.L.; Zimorski, V.; Sarink, M.J.; Wienk, H.; Brouwers, J.F.; de Jonckheere, J.F.; Martin, W.F.; Opperdoes, F.R.; van Hellemond, J.J.; Tielens, A.G.M. Lipids Are the Preferred Substrate of the Protist Naegleria gruberi, Relative of a Human Brain Pathogen. Cell Rep. 2018, 25, 537–543.e3. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Bar, E.E.; Chaudhry, A.; Lin, A.; Fan, X.; Schreck, K.; Matsui, W.; Piccirillo, S.; Vescovi, A.L.; DiMeco, F.; Olivi, A.; et al. Cyclopamine-Mediated Hedgehog Pathway Inhibition Depletes Stem-Like Cancer Cells in Glioblastoma. Stem Cells 2007, 25, 2524–2533. [Google Scholar] [CrossRef]
- Sekulic, A.; Migden, M.R.; Oro, A.E.; Dirix, L.; Lewis, K.D.; Hainsworth, J.D.; Solomon, J.A.; Yoo, S.; Arron, S.T.; Friedlander, P.A.; et al. Efficacy and Safety of Vismodegib in Advanced Basal-Cell Carcinoma. N. Engl. J. Med. 2012, 366, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Iovine, V.; Mori, M.; Calcaterra, A.; Berardozzi, S.; Botta, B. One Hundred Faces of Cyclopamine. Curr. Pharm. Des. 2016, 22, 1658–1681. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Wang, P.; Wang, P.; Ma, C.; Kang, W. Antibacterial mechanism of chelerythrine isolated from root of Toddalia asiatica (Linn) Lam. BMC Complement. Altern. Med. 2018, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Fan, Y.; Liu, L.; Tao, W.; Shan, X.; Dong, Y.; Li, L.; Zhang, S.; Wang, H. Chelerythrine Attenuates the Inflammation of Lipopolysaccharide-Induced Acute Lung Inflammation Through NF-κB Signaling Pathway Mediated by Nrf2. Front. Pharmacol. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Valipour, M.; Zarghi, A.; Ebrahimzadeh, M.A.; Irannejad, H. Therapeutic potential of chelerythrine as a multi-purpose adjuvant for the treatment of COVID-19. Cell Cycle. 2021, 20, 2321–2336. [Google Scholar] [CrossRef]
- Tillhon, M.; Ortiz, L.M.G.; Lombardi, P.; Scovassi, A.I. Berberine: New perspectives for old remedies. Biochem. Pharmacol. 2012, 84, 1260–1267. [Google Scholar] [CrossRef]
- Kumar, A.; Ekavali; Chopra, K.; Mukherjee, M.; Pottabathini, R.; Dhull, D.K. Current knowledge and pharmacological profile of berberine: An update. Eur. J. Pharmacol. 2015, 761, 288–297. [Google Scholar] [CrossRef]
- Subedi, L.; Gaire, B.P. Tanshinone IIA: A phytochemical as a promising drug candidate for neurodegenerative diseases. Pharmacol. Res. 2021, 169, 105661. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, M.; Liu, J.N.; Zhao, X.; Zhang, Y.Q.; Fang, L. Tanshinone IIA: A Review of its Anticancer Effects. Front. Pharmacol. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Shang, Q.; Xu, H.; Huang, L. Tanshinone IIA: A promising natural cardioprotective agent. Evid. -Based Complement. Altern. Med. 2012, 2012, 716459. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, M.; Zhang, X.; Chen, L.; Wang, Y.; Xu, Y.; Xu, Y. Dissecting Chemical Composition and Cardioprotective Effects of Fuzhengkangfu Decoction against Doxorubicin-Induced Cardiotoxicity by LC-MS and Bioinformatics Approaches. ACS Omega 2020, 5, 14051–14060. [Google Scholar] [CrossRef] [PubMed]
- Gurudeeban, S.; Ramanathan, T.; Satyavani, K. Antimicrobial and Radical Scavenging Effects of Alkaloid Extracts from Rhizophora Mucronata. Pharm. Chem. J. 2015, 49, 34–37. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Tyagi, R.; Sanchita; Tripathi, S.; Pati, S.; Srivastava, S.K.; Darokar, M.P.; Sharma, A. Antibiotics potentiating potential of catharanthine against superbug Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 2018, 36, 4270–4284. [Google Scholar] [CrossRef] [PubMed]
- Stratton, C.F.; Newman, D.J.; Tan, D.S. Cheminformatic comparison of approved drugs from natural product versus synthetic origins. Bioorg. Med. Chem. Lett. 2015, 25, 4802–4807. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1–13. [Google Scholar] [CrossRef]
- Debnath, A.; Calvet, C.M.; Jennings, G.; Zhou, W.; Aksenov, A.; Luth, M.R.; Abagyan, R.; Nes, W.D.; McKerrow, J.H.; Podust, L.M. CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM). PLoS Negl. Trop. Dis. 2017, 11, e0006104. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Visualizer, D.S. Dassault Systèmes; BIOVIA: San Diego, CA, USA, 2020. [Google Scholar]
- Kuo, K.-W.; Hsu, S.-H.; Li, Y.-P.; Lin, W.-L.; Liu, L.-F.; Chang, L.-C.; Lin, C.-C.; Lin, C.-N.; Sheu, H.-M. Anticancer activity evaluation of the Solanum glycoalkaloid solamargine. Biochem. Pharmacol. 2000, 60, 1865–1873. [Google Scholar] [CrossRef]
- Saavedra, A.; Fernández-García, S.; Cases, S.; Puigdellívol, M.; Alcalá-Vida, R.; Martín-Flores, N.; Alberch, J.; Ginés, S.; Malagelada, C.; Pérez-Navarro, E. Chelerythrine promotes Ca2+-dependent calpain activation in neuronal cells in a PKC-independent manner. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The anti-cancer mechanisms of berberine: A review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef] [PubMed]




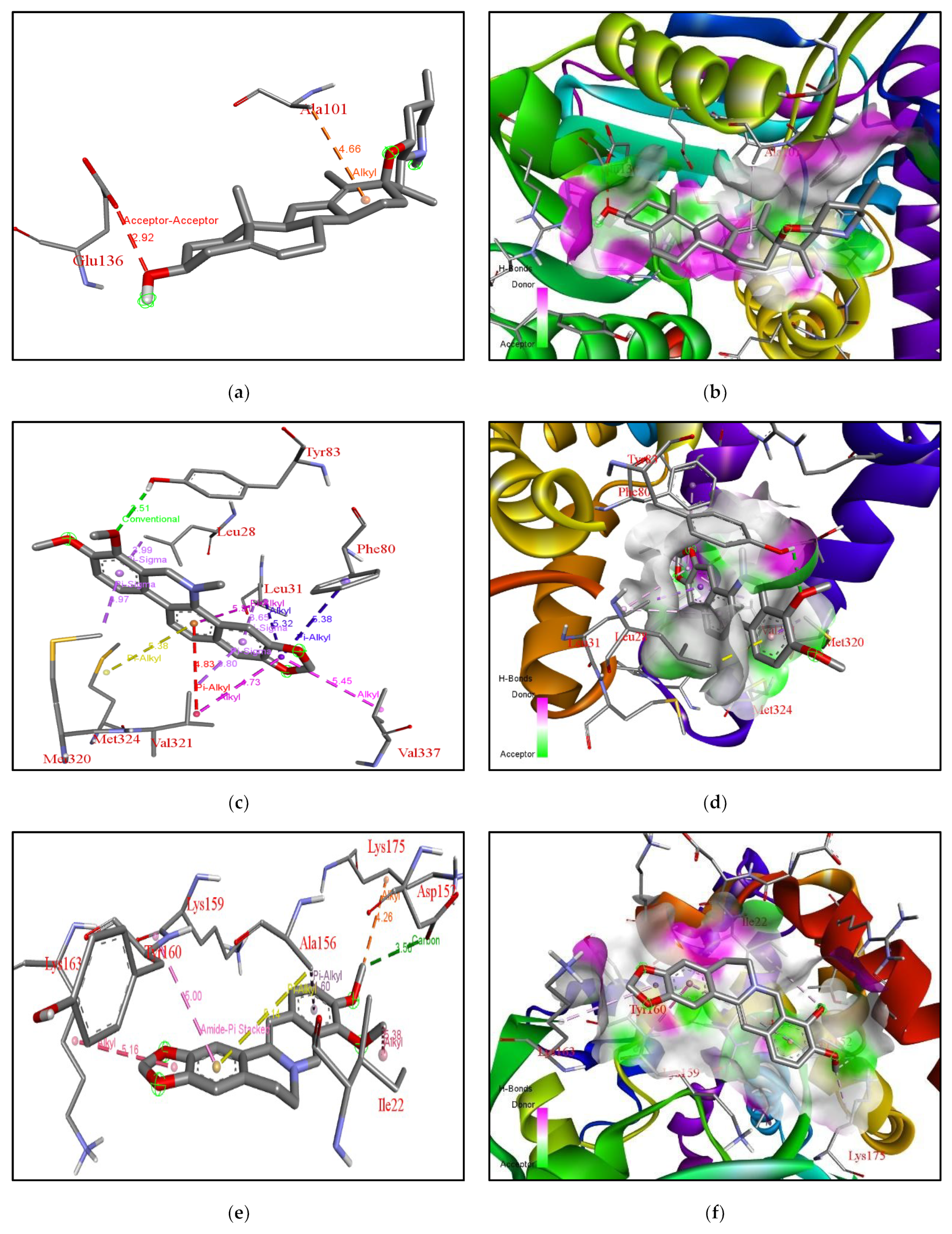

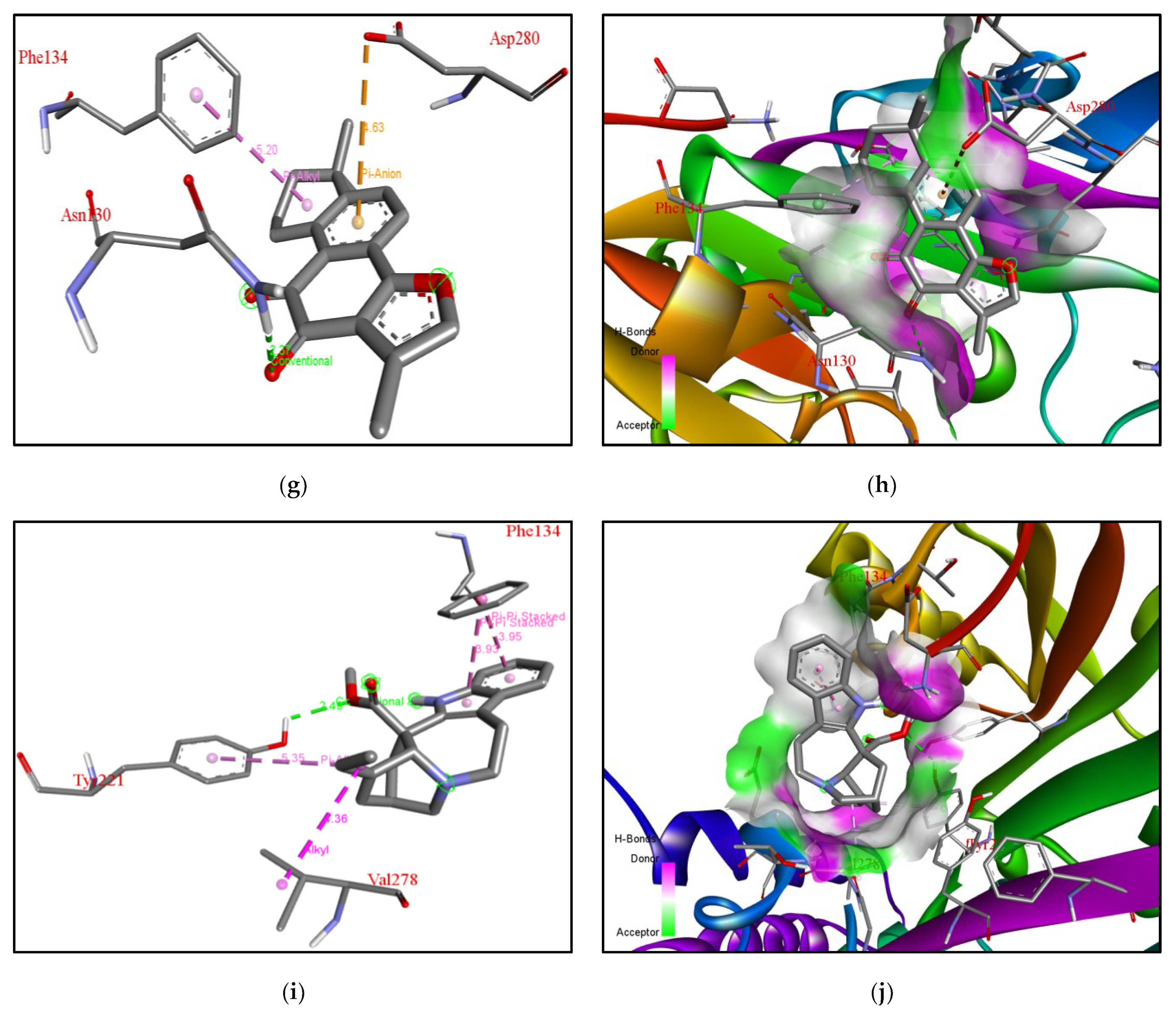
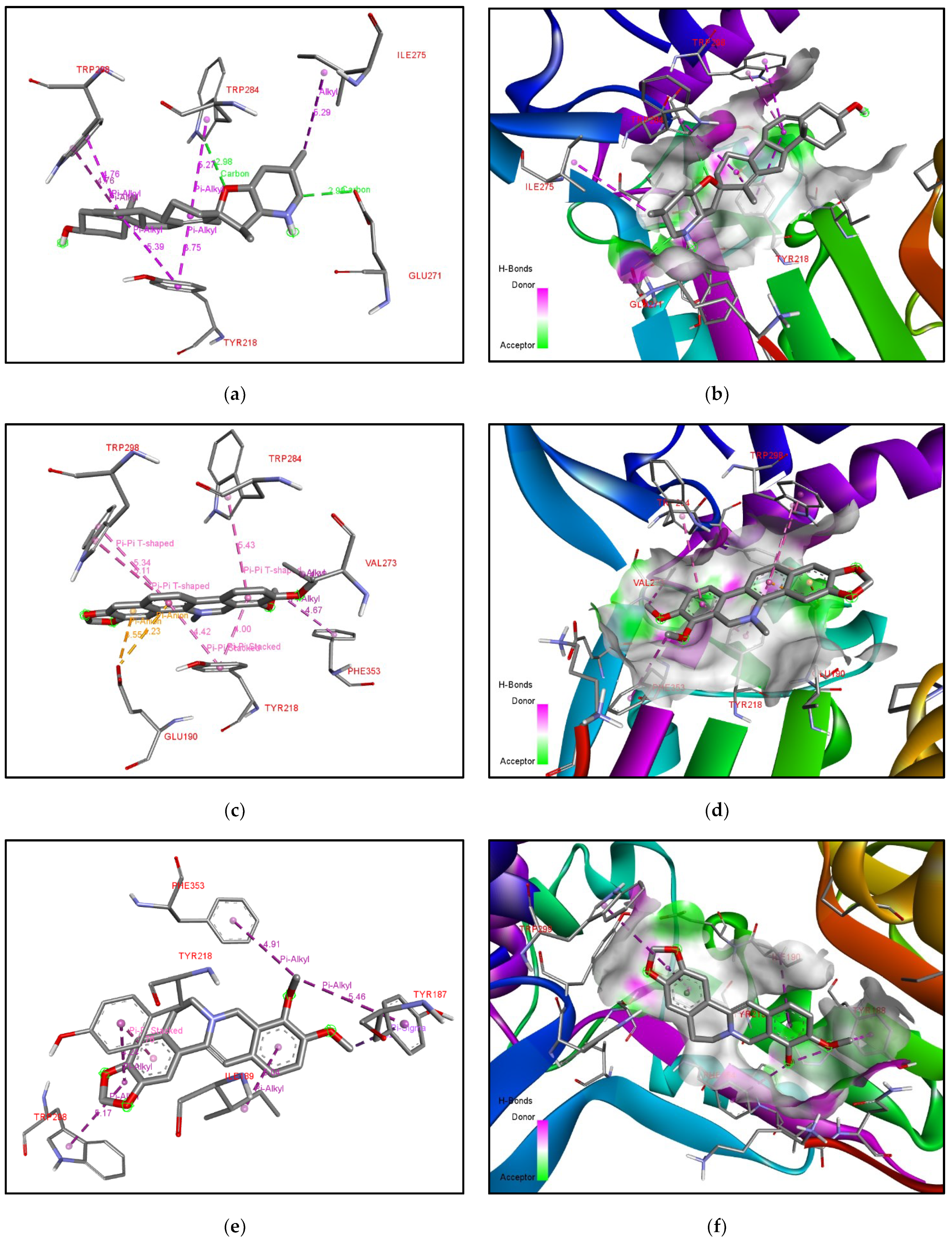
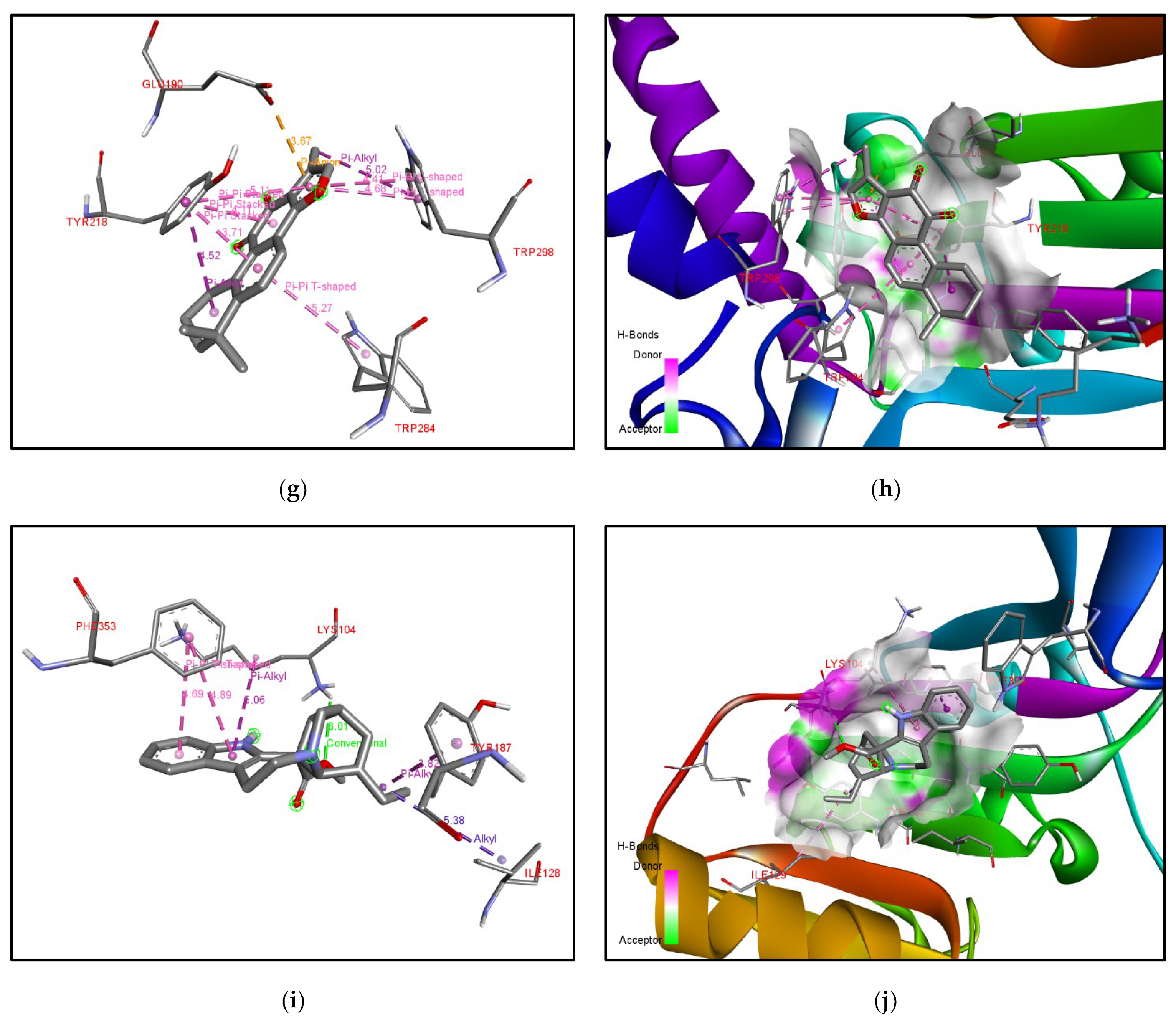
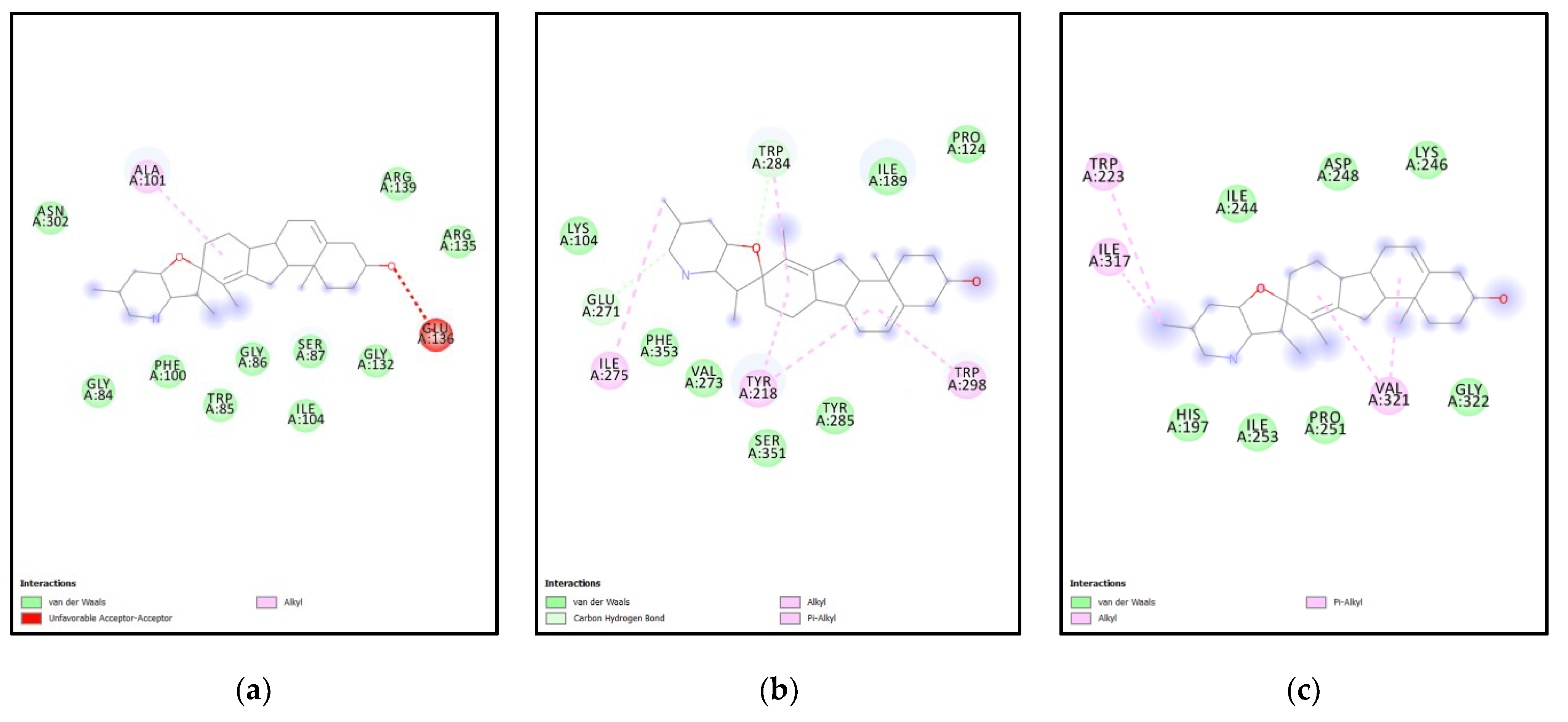
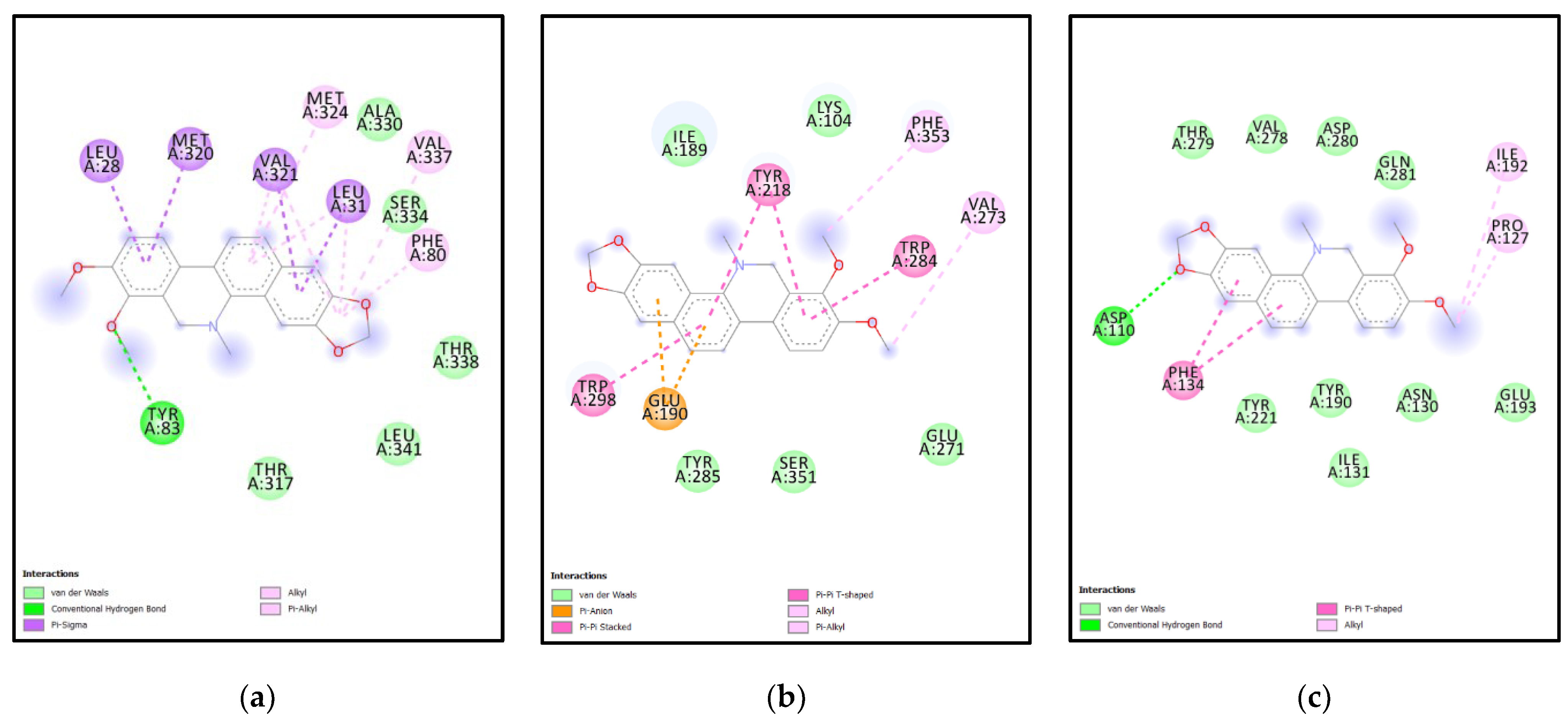
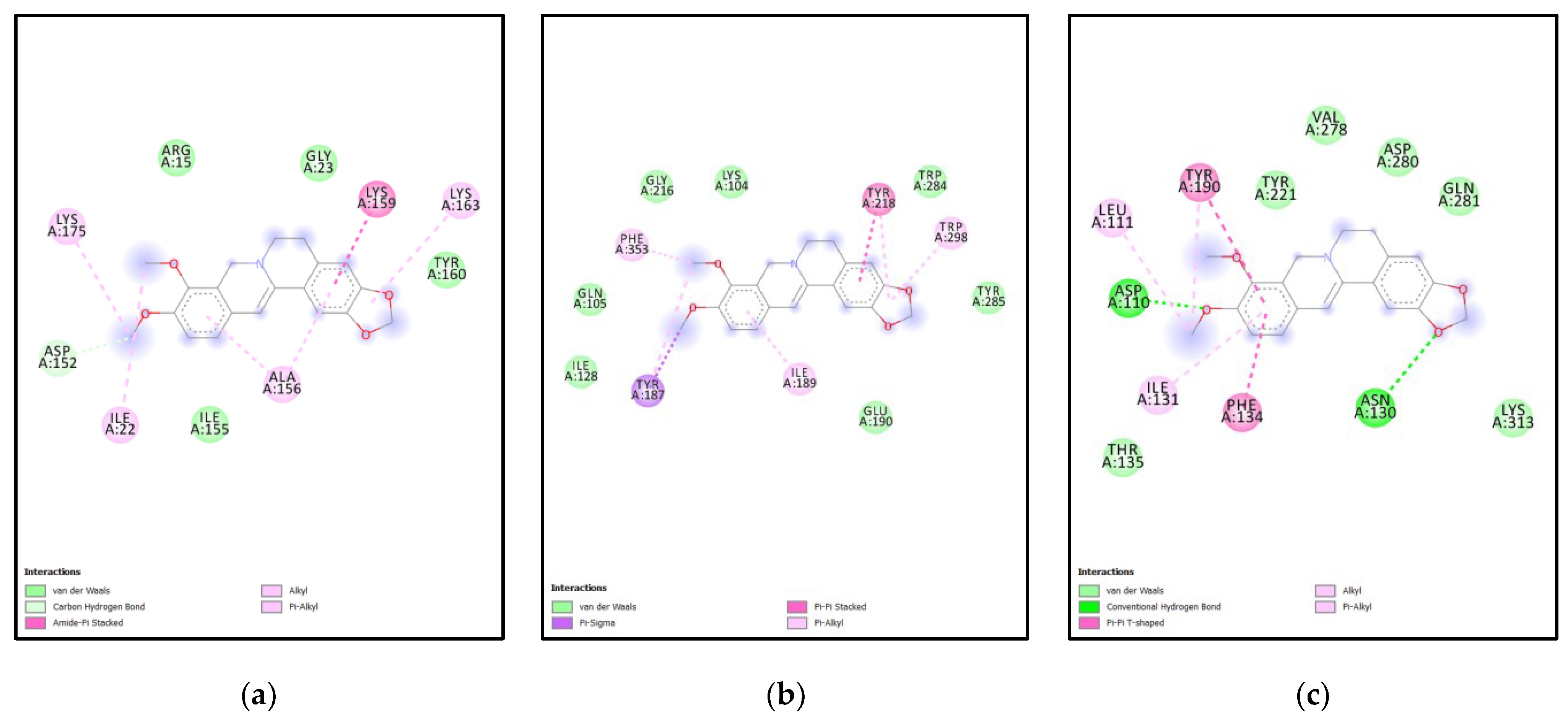
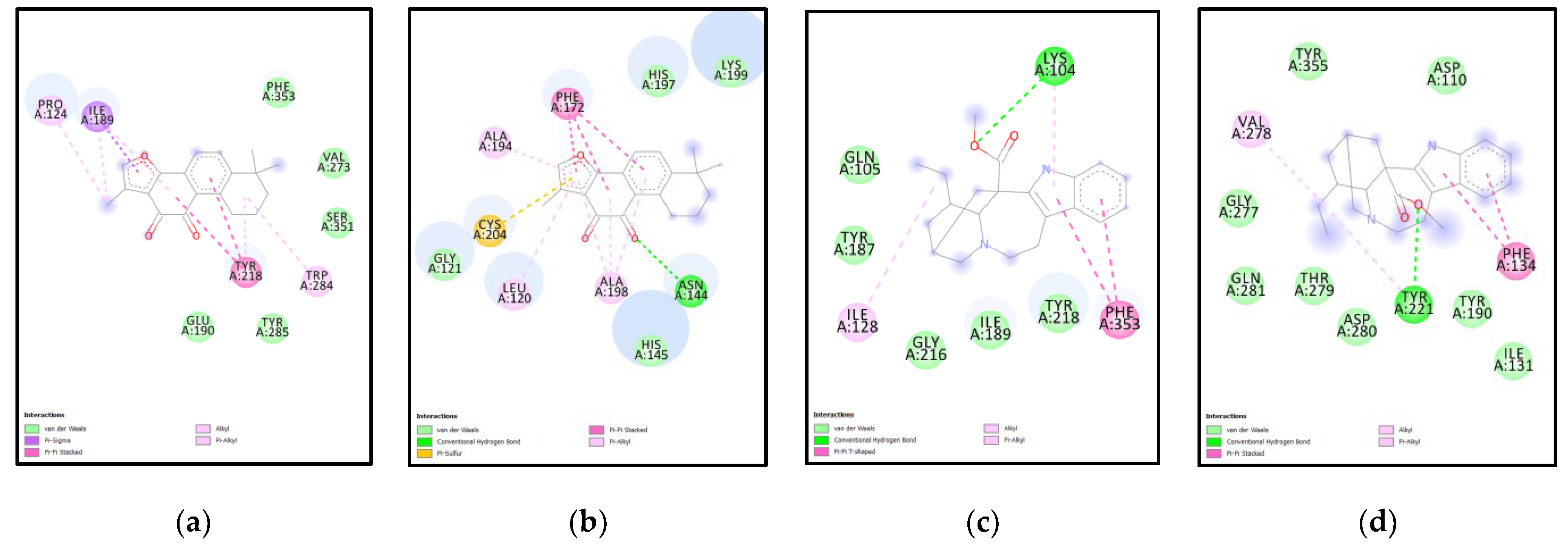
| Rank | XM vs. XP vs. Yeast | Binding Affinities (XM, XP, YEAST) Kcal/mol | XM vs. XP | Binding Affinities (XM, XP) Kcal/mol |
|---|---|---|---|---|
| 1. | Solamargine (PCID:73611) | −9.4, −10.6, −9.8 | Solamargine (PCID:73611) | −9.4, −10.6 |
| 2. | Wilforlide A (PCID:158477) | −9, −10.1, −8.4 | Obacunone (PCID:119041) | −9.2, −10.5 |
| 3. | Obacunone (PCID:119041) | −9.2, −10.5, −8.1 | Wilforlide A (PCID:158477) | −9, −10.1 |
| 4. | Cyclopamine (PCID:442972) | −8.6, −10.3, −8 | Cyclopamine [(PCID:442972) | −8.6, −10.3 |
| 5. | Ginsenoside Ro (PCID:11815492) | −8.5, −9.5, −8.5 | Tanshinone 2 (PCID:164676) | −8.6, −9.6 |
| 6. | Estradiol cypionate (PCID:9403) | −8.5, −9.6, −8 | Ginsinoside Ro (PCID:11815492) | −8.5, −9.5 |
| 7. | Natamycin (PCID:5284447) | −8.5, −9.4, −8.3 | Estradiol Cypionate (PCID:9403) | −8.5, −9.6 |
| 8. | Tiliroside (PCID:5320686) | −8.7, −8.3, −8.1 | Camptothecin (PCID:2360) | −8.6, −9 |
| 9. | Etoposide (PCID:36462) | −8.2, −9.1, −7.9 | Natamycin (PCID:5284447) | −8.5, −9.4 |
| 10. | Myricitrin (PCID:5281673) | −8, −8.1, −8.5 | Epothilone A | −8.3, −9.1 |
| 11. | Tigecycline (PCID:54686904) | −8, −8, −8.2 | Etoposide (PCID:36462) | −8.2, −9.1 |
| 12. | Chelerythrine (PCID:2703) | −7.8, −7.6, −7.9 | Timosaponin B 2 (PCID:44575945) | −8.3, −8.7 |
| 13. | Luteolin (PCID:5280445) | −7.9, −8.6, −7.2 | Tiliroside (PCID:5320686) | −8.7, −8.3 |
| 14. | Berberine (PCID:2353) | −7.8, −8.3, −7.3 | Beta−carotene (PCID:5280489) | −8.5, −8.3 |
| 15. | Butein (PCID:5281222) | −6.6, −6.7, −8 | Catharanthine (PCID:5458190) | −8, −8.4 |
| 16. | Clotrimazole (PCID:2812) | −8, −8.9 |
| Rank | XM vs. XP vs. Yeast | XM vs. XP |
|---|---|---|
| 1. | Cyclopamine (PCID:442972) | Cyclopamine (PCID:442972) |
| 2. | Chelerythrine (PCID:2703) | Tanshinone 2 (PCID:164676) |
| 3. | Berberine (PCID:2353) | Catharanthine (PCID:5458190) |
| Ligand | Molecular Weight (g/mol) | Lipophilicity (Log Po/w (XLOGP3)) | Hydrogen Bond Acceptors, Donors | Water Solubility (Log S (ESOL)) |
|---|---|---|---|---|
| Cyclopamine | 411.62 | 3.52 | 3, 2 | −4.61, Moderately soluble |
| Chelerythrine | 348.37 | 4.58 | 4, 0 | −5.27, Poorly soluble |
| Berberine | 336.36 | 3.62 | 4, 0 | −4.55, Moderately soluble |
| Tanshinone 2 A | 294.34 | 4.33 | 3, 0 | −4.76, Poorly soluble |
| Catharanthine | 336.43 | 2.80 | 3, 1 | −3.76, Moderately soluble |
| Ligand | Druglikeness Violations (Lipinski, Ghose, Veber, Egan, Muegge) | Bioavailability | Blood-Brain Barrier Crossing | GI Absorption | Skin Permeability (cm/s) |
|---|---|---|---|---|---|
| Cyclopamine | Lipinski (1, MLOGP > 4.15), Ghose (1, atoms > 70) violations | 0.55 | Yes | High | −6.31 |
| Chelerythrine | No | 0.55 | Yes | High | 5.17 |
| Berberine | No | 0.55 | Yes | High | −5.78 |
| Tanshinone 2 | No | 0.55 | Yes | High | −5.02 |
| Catharanthine | No | 0.55 | Yes | High | −6.36 |
| Protein | Amino Acid Sequence |
|---|---|
| YEAST SMT | MSETELRKRQAQFTRELHGDDIGKKTGLSALMSKNNSAQKEAV QKYLRNWDGRTDKDAEERRLEDYNEATHSYYNVVTDFYEYGW GSSFHFSRFYKGESFAASIARHEHYLAYKAGIQRGDLVLDVGCGV GGPAREIARFTGCNVIGLNNNDYQIAKAKYYAKKYNLSDQMDF VKGDFMKMDFEENTFDKVYAIEATCHAPKLEGVYSEIYKVLKPG GTFAVYEWVMTDKYDENNPEHRKIAYEIELGDGIPKMFHVDVA RKALKNCGFEVLVSEDLADNDDEIPWYYPLTGEWKYVQNLANL ATFFRTSYLGRQFTTAMVTVMEKLGLAPEGSKEVTAALENAAVG LVAGGKSKLFTPMMLFVARKPENAETPSQTSQEATQ |
| XM complete sequence * |
MSLFLLGVVSIVVIGFVVYLFKFKNQIKGYHLTNENVTGTYQDLF AQDTKDTHDKRKNAGWDVAGKYYDMVTDFYLYGWGRSFHFA PRHKKESMIESIQRHEYWLAKQMDLKKGMKCLDLGCGVMGPA TNISRFTGAHITGVNNHPYQSQQAKEYISQMGLSEQCQIVRGDFN NLDDNSDLPSESYDAAYTIEASCHAKDRPHCYKQIYNKLKPGAIF AGYEWVMISGKYDSKNEEHNKIKFDIMKGDGLPEILMDKEIDESL RKAGFEVIKTEDVGVTDQIHPVPWYQPIDNGGWDFTSWFQTSYG RFIVHKLVGILESVGLVPKSSQQAYEFLMAGASGLVAGGKTGIFTP CYFFMARKPLTAAE |
| XP complete sequence * | MFLIILTVIVGGFILYLFKFREQIKGSHLTDTDKTASYQELFATDNQ QTHEKRKKAGWDVAGKYYDMVTDFYLYGWGRSFHFATRHSRE SLIESILRHEYWLAKQLDLKPGMKCLDLGCGVMGPAVNIARFSG CNVTGVNNHPYQSERAKVFINEMGMDGRCNIVRGDFNNLDDN KDLPAESYDAAYAIEATCHAKDRPHCYKQIFNKLKPGAVFGGYE WVMITGKYDSKNEEHNKIKFDIMKGDGLPEILMDKEIDDALVKA GFEVIRTEDVAITDKINPIPWYQPLDNGGWELTNWFQTSYGRWV VHKLVGILEKIGLVPKTSQQAYEFLMAGAGLVGGGKTGIFTPSYF FLARKPLKN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abraham, J.; Chauhan, N.; Ray, S. Virtual Screening of Alkaloid and Terpenoid Inhibitors of SMT Expressed in Naegleria sp. Molecules 2022, 27, 5727. https://doi.org/10.3390/molecules27175727
Abraham J, Chauhan N, Ray S. Virtual Screening of Alkaloid and Terpenoid Inhibitors of SMT Expressed in Naegleria sp. Molecules. 2022; 27(17):5727. https://doi.org/10.3390/molecules27175727
Chicago/Turabian StyleAbraham, Jason, Neha Chauhan, and Supriyo Ray. 2022. "Virtual Screening of Alkaloid and Terpenoid Inhibitors of SMT Expressed in Naegleria sp." Molecules 27, no. 17: 5727. https://doi.org/10.3390/molecules27175727
APA StyleAbraham, J., Chauhan, N., & Ray, S. (2022). Virtual Screening of Alkaloid and Terpenoid Inhibitors of SMT Expressed in Naegleria sp. Molecules, 27(17), 5727. https://doi.org/10.3390/molecules27175727