Machine Learning Model Based on Radiomic Features for Differentiation between COVID-19 and Pneumonia on Chest X-ray
Abstract
:1. Introduction
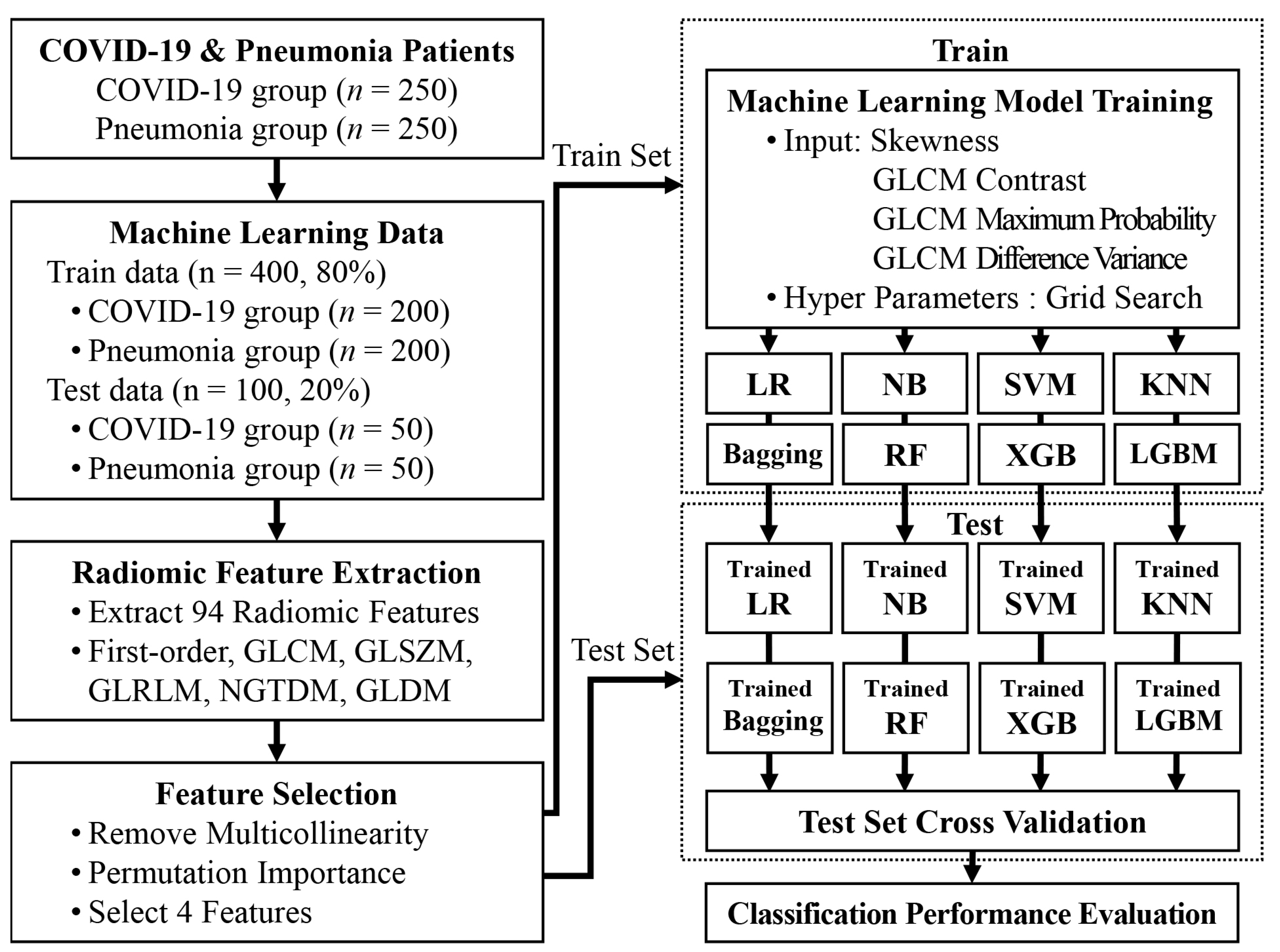
2. Materials and Methods
2.1. Ethics Statement
2.2. Data Collection
2.3. Region of Interest
2.4. Extraction of Radiomic Features
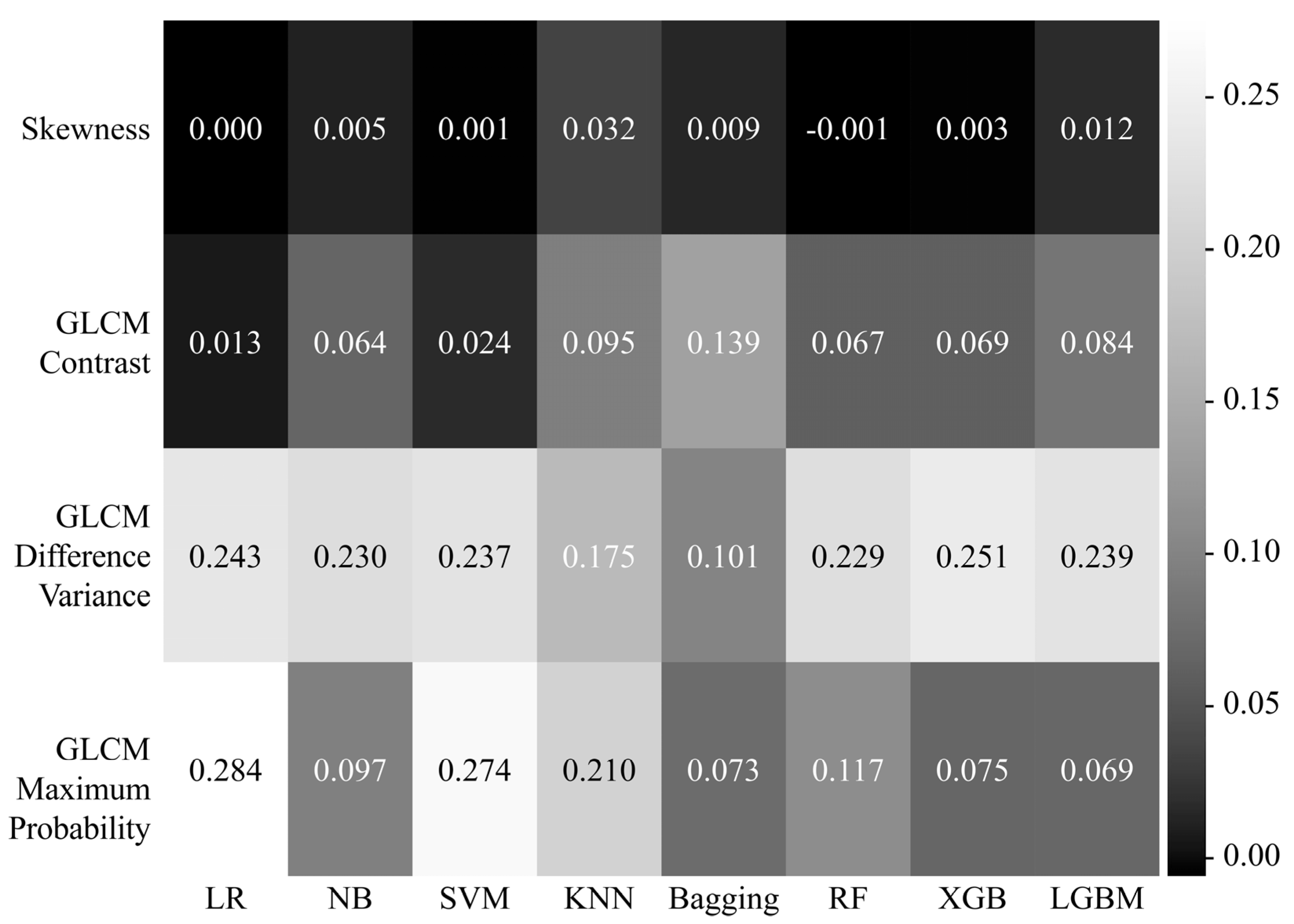
2.5. Feature Selection
2.6. Machine Learning Models to differentiate between COVID-19 and Pneumonia
2.7. Validation and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rai, P.; Kumar, B.K.; Deekshit, V.K.; Karunasagar, I.; Karunasagar, I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021, 105, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Naderi Ghale-Noie, Z.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef] [PubMed]
- Akl, E.A.; Blazic, I.; Yaacoub, S.; Frija, G.; Chou, R.; Appiah, J.A.; Fatehi, M.; Flor, N.; Hitti, E.; Jafri, H.; et al. Use of Chest Imaging in the Diagnosis and Management of COVID-19: A WHO Rapid Advice Guide. Radiology 2021, 298, E63–E69. [Google Scholar] [CrossRef]
- Cozzi, D.; Albanesi, M.; Cavigli, E.; Moroni, C.; Bindi, A.; Luvara, S.; Lucarini, S.; Busoni, S.; Mazzoni, L.N.; Miele, V. Chest X-ray in new Coronavirus Disease 2019 (COVID-19) infection: Findings and correlation with clinical outcome. Radiol. Med. 2020, 125, 730–737. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Hani, C.; Trieu, N.H.; Saab, I.; Dangeard, S.; Bennani, S.; Chassagnon, G.; Revel, M.P. COVID-19 pneumonia: A review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging 2020, 101, 263–268. [Google Scholar] [CrossRef]
- Barragan-Montero, A.; Javaid, U.; Valdes, G.; Nguyen, D.; Desbordes, P.; Macq, B.; Willems, S.; Vandewinckele, L.; Holmstrom, M.; Lofman, F.; et al. Artificial intelligence and machine learning for medical imaging: A technology review. Phys. Med. 2021, 83, 242–256. [Google Scholar] [CrossRef]
- Hasoon, J.N.; Fadel, A.H.; Hameed, R.S.; Mostafa, S.A.; Khalaf, B.A.; Mohammed, M.A.; Nedoma, J. COVID-19 anomaly detection and classification method based on supervised machine learning of chest X-ray images. Results Phys. 2021, 31, 105045. [Google Scholar] [CrossRef]
- Kassania, S.H.; Kassanib, P.H.; Wesolowskic, M.J.; Schneidera, K.A.; Detersa, R. Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning Based Approach. Biocybern. Biomed. Eng. 2021, 41, 867–879. [Google Scholar] [CrossRef]
- Jain, R.; Gupta, M.; Taneja, S.; Hemanth, D.J. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell. 2021, 51, 1690–1700. [Google Scholar] [CrossRef]
- Sitaula, C.; Hossain, M.B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl. Intell. 2021, 51, 2850–2863. [Google Scholar] [CrossRef] [PubMed]
- von Eschenbach, W.J. Transparency and the Black Box Problem: Why We Do Not Trust AI. Philos. Technol. 2021, 34, 1607–1622. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Haggstrom, I.; Szczypinski, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Aggarwal, N.K.; Agrawal, R. First and Second Order Statistics Features for Classification of Magnetic Resonance Brain Images. J. Signal Inf. Process. 2012, 03, 146–153. [Google Scholar] [CrossRef]
- Avanzo, M.; Stancanello, J.; El Naqa, I. Beyond imaging: The promise of radiomics. Phys. Med. 2017, 38, 122–139. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kim, D.; Elgeti, T.; Steffen, I.G.; Hamm, B.; Nagel, S.N. Stability of Radiomic Features across Different Region of Interest Sizes-A CT and MR Phantom Study. Tomography 2021, 7, 238–252. [Google Scholar] [CrossRef]
- Garg, M.; Dhiman, G. A novel content-based image retrieval approach for classification using GLCM features and texture fused LBP variants. Neural Comput. Appl. 2020, 33, 1311–1328. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, L.F.; Zhang, X.; Nan, H.Y.; Hu, Y.C.; Han, Y.; Zhang, J.; Liu, Z.C.; Sun, Y.Z.; Tian, Q.; et al. Optimizing Texture Retrieving Model for Multimodal MR Image-Based Support Vector Machine for Classifying Glioma. J. Magn. Reason. Imaging 2019, 49, 1263–1274. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Akdemir, B. Application of Feature Extraction and Classification Methods for Histopathological Image using GLCM, LBP, LBGLCM, GLRLM and SFTA. Procedia Comput. Sci. 2018, 132, 40–46. [Google Scholar] [CrossRef]
- Chen, S.; Harmon, S.; Perk, T.; Li, X.; Chen, M.; Li, Y.; Jeraj, R. Using neighborhood gray tone difference matrix texture features on dual time point PET/CT images to differentiate malignant from benign FDG-avid solitary pulmonary nodules. Cancer Imaging 2019, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, N.; Akbarizadeh, G. Iris tissue recognition based on GLDM feature extraction and hybrid MLPNN-ICA classifier. Neural Comput. Appl. 2018, 32, 2267–2281. [Google Scholar] [CrossRef]
- Akinwande, M.O.; Dikko, H.G.; Samson, A. Variance Inflation Factor: As a Condition for the Inclusion of Suppressor Variable(s) in Regression Analysis. Open J. Stat. 2015, 5, 754–767. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Alonso-Betanzos, A. Ensembles for feature selection: A review and future trends. Inf. Fusion 2019, 52, 1–12. [Google Scholar] [CrossRef]
- Konig, G.; Molnar, C.; Bischl, B.; Grosse-Wentrup, M. Relative Feature Importance. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 9318–9325. [Google Scholar]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef]
- Abraham, R.; Simha, J.B.; Iyengar, S.S. Medical Datamining with a New Algorithm for Feature Selection and Naive Bayesian Classifier. In Proceedings of the 10th International Conference on Information Technology (ICIT 2007), Rourkela, India, 17–20 December 2007; pp. 44–49. [Google Scholar]
- Gaye, B.; Zhang, D.; Wulamu, A.; Tsai, S.-B. Improvement of Support Vector Machine Algorithm in Big Data Background. Math. Probl. Eng. 2021, 2021, 5594899. [Google Scholar] [CrossRef]
- Mittal, K.; Aggarwal, G.; Mahajan, P. Performance study of K-nearest neighbor classifier and K-means clustering for predicting the diagnostic accuracy. Int. J. Inf. Technol. 2018, 11, 535–540. [Google Scholar] [CrossRef]
- Tu, M.C.; Shin, D.; Shin, D. A Comparative Study of Medical Data Classification Methods Based on Decision Tree and Bagging Algorithms. In Proceedings of the 2009 Eighth IEEE International Conference on Dependable, Autonomic and Secure Computing, Washington, DC, USA, 12–14 December 2009; pp. 183–187. [Google Scholar]
- Schonlau, M.; Zou, R.Y. The random forest algorithm for statistical learning. Stata J. 2020, 20, 3–29. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Rufo, D.D.; Debelee, T.G.; Ibenthal, A.; Negera, W.G. Diagnosis of Diabetes Mellitus Using Gradient Boosting Machine (LightGBM). Diagnostics 2021, 11, 1714. [Google Scholar] [CrossRef]
- Bhat, P.C.; Prosper, H.B.; Sekmen, S.; Stewart, C. Optimizing event selection with the random grid search. Comput. Phys. Commun. 2018, 228, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.-T.; Yeh, P.-Y. Reliable Accuracy Estimates from k-Fold Cross Validation. IEEE Trans. Knowl. Data Eng. 2020, 32, 1586–1594. [Google Scholar] [CrossRef]
- Jin, H.; Ling, C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar]


| Characteristic | Number of Patients (%) | p Value | |
|---|---|---|---|
| COVID-19 (n = 250) | Pneumonia (n = 250) | ||
| Age (years) | 63.12 ± 19.59 | 71.11 ± 16.93 | p < 0.001 |
| Sex | |||
| - Males | 109 (43.6%) | 142 (56.8%) | p = 0.004 |
| - Females | 141 (56.4%) | 108 (43.2%) | p = 0.004 |
| Height (cm) | 162.30 ± 10.26 | 161.65 ± 9.96 | p = 0.497 |
| Weight (kg) | 63.59 ± 14.43 | 58.53 ± 13.49 | p < 0.001 |
| BMI (kg/m2) | 23.95 ± 3.81 | 22.49 ± 4.25 | p < 0.001 |
| HTN | 115 (46.0%) | 133 (53.2%) | p = 0.128 |
| DM | 63 (25.2%) | 82 (32.8%) | p = 0.076 |
| Smoking status | |||
| - None | 177 (70.8%) | 182 (72.8%) | p = 0.691 |
| - Former smoker | 15 (6.0%) | 7 (2.8%) | p = 0.127 |
| - Current smoker | 10 (4.0%) | 31 (12.4%) | p = 0.001 |
| Symptom | |||
| - Fever | 80 (32.1%) | 65 (26.0%) | p = 0.159 |
| - Cough | 85 (34.0%) | 34 (13.6%) | p < 0.001 |
| - Sputum | 45 (18.0%) | 43 (17.2%) | p = 0.907 |
| - Dyspnea | 28 (11.2%) | 85 (34.0%) | p < 0.001 |
| - Myalgia | 33 (13.2%) | 0 (0.0%) | p < 0.001 |
| - Sore throat | 37 (14.8%) | 1 (0.4%) | p < 0.001 |
| Model | Hyperparameters |
|---|---|
| LR | C: 997.265, penalty: L2, random state: 50 |
| NB | var smoothing: 1 × 10−9 |
| SVM | C: 998.196, kernel: linear, probability: true, random state: 50 |
| KNN | leaf size: 20, n neighbors: 2, weights: distance |
| Bagging | max features: 3, max samples: 5, n estimators: 100, random state: 50 |
| RF | max depth: 190, n estimators: 709, random state: 50 |
| XGB | learning rate: 0.068, max depth: 11, n estimators: 100, random state: 50 |
| LGBM | learning rate: 0.05, max depth: 12, min child samples: 10, num leaves: 40, random state: 50 |
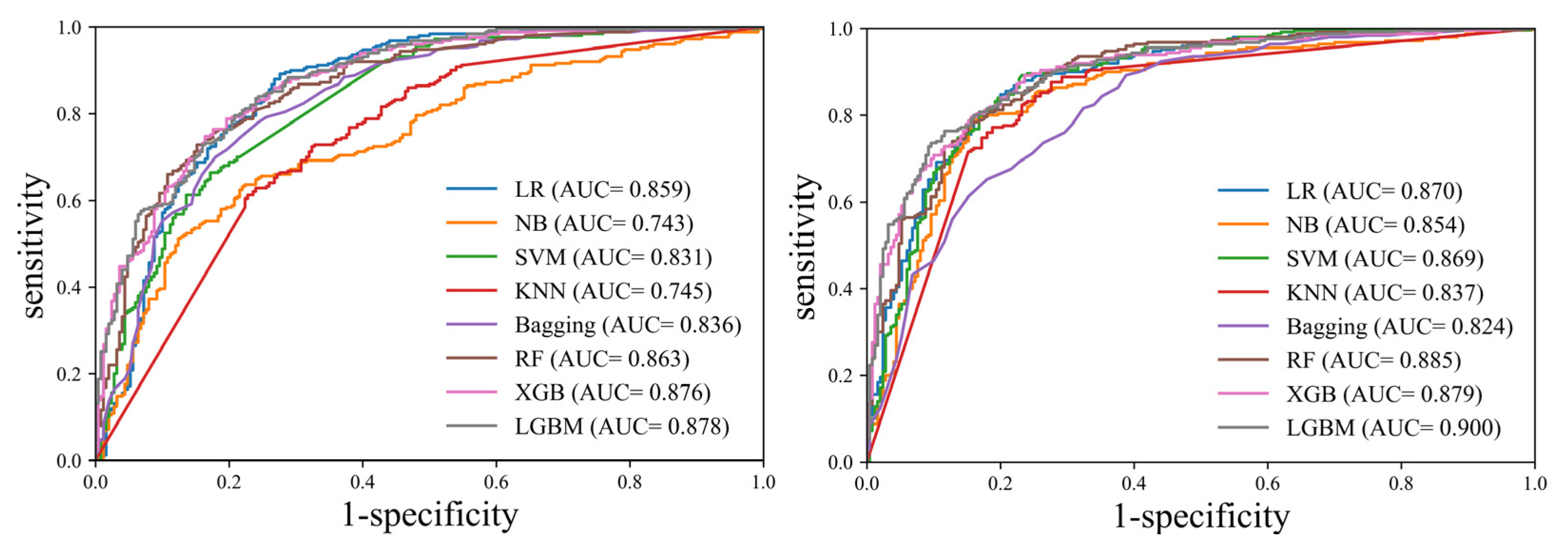
| AUC (95% CI) | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|---|---|---|---|---|---|---|
| (Without Feature Selection) | ||||||
| LR | 0.859 (0.825–0.888) | 0.746 (0.706–0.784) | 0.980 (0.954–0.994) | 0.512 (0.448–0.576) | 0.668 (0.639–0.695) | 0.962 (0.914–0.984) |
| NB | 0.743 (0.703–0.781) | 0.560 (0.515–0.604) | 0.960 (0.928–0.981) | 0.160 (0.117–0.211) | 0.533 (0.518–0.548) | 0.800 (0.672–0.887) |
| SVM | 0.831 (0.795–0.863) | 0.532 (0.487–0.576) | 0.996 (0.978–0.999) | 0.068 (0.040–0.107) | 0.517 (0.508–0.525) | 0.944 (0.695–0.992) |
| KNN | 0.745 (0.704–0.782) | 0.682 (0.639–0.723) | 0.912 (0.870–0.944) | 0.452 (0.389–0.516) | 0.625 (0.596–0.652) | 0.837 (0.771–0.887) |
| Bagging | 0.836 (0.801–0.868) | 0.716 (0.674–0.755) | 0.984 (0.960–0.996) | 0.448 (0.385–0.512) | 0.641 (0.614–0.666) | 0.966 (0.913–0.987) |
| RF | 0.863 (0.830–0.892) | 0.720 (0.678–0.759) | 0.948 (0.913–0.972) | 0.492 (0.428–0.556) | 0.651 (0.622–0.679) | 0.904 (0.846–0.942) |
| XGB | 0.876 (0.843–0.903) | 0.792 (0.754–0.827) | 0.880 (0.833–0.918) | 0.704 (0.643–0.760) | 0.748 (0.710–0.784) | 0.854 (0.806–0.892) |
| LGBM | 0.878 (0.846–0.906) | 0.784 (0.745–0.819) | 0.900 (0.856–0.934) | 0.668 (0.606–0.726) | 0.731 (0.694–0.765) | 0.870 (0.820–0.907) |
| (With Feature Selection) | ||||||
| LR | 0.870 (0.837–0.898) | 0.796 (0.758–0.831) | 0.860 (0.811–0.901) | 0.732 (0.673–0.786) | 0.762 (0.722–0.799) | 0.839 (0.792–0.877) |
| NB | 0.854 (0.820–0.883) | 0.696 (0.654–0.736) | 0.952 (0.918–0.975) | 0.440 (0.378–0.504) | 0.630 (0.603–0.656) | 0.902 (0.838–0.942) |
| SVM | 0.869 (0.836–0.897) | 0.790 (0.752–0.825) | 0.876 (0.829–0.914) | 0.704 (0.643–0.760) | 0.747 (0.709–0.783) | 0.850 (0.802–0.889) |
| KNN | 0.837 (0.802–0.869) | 0.682 (0.639–0.723) | 0.912 (0.870–0.944) | 0.452 (0.389–0.516) | 0.625 (0.596–0.652) | 0.837 (0.771–0.887) |
| Bagging | 0.824 (0.787–0.856) | 0.742 (0.701–0.780) | 0.924 (0.884–0.954) | 0.560 (0.496–0.623) | 0.677 (0.645–0.708) | 0.881 (0.825–0.920) |
| RF | 0.885 (0.854–0.912) | 0.808 (0.771–0.842) | 0.816 (0.762–0.862) | 0.800 (0.745–0.848) | 0.803 (0.760–0.840) | 0.813 (0.769–0.850) |
| XGB | 0.879 (0.848–0.907) | 0.790 (0.752–0.825) | 0.804 (0.749–0.851) | 0.776 (0.719–0.826) | 0.782 (0.739–0.820) | 0.798 (0.753–0.837) |
| LGBM | 0.900 (0.870–0.925) | 0.796 (0.758–0.831) | 0.916 (0.875–0.947) | 0.676 (0.614–0.734) | 0.739 (0.702–0.773) | 0.890 (0.841–0.924) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.J. Machine Learning Model Based on Radiomic Features for Differentiation between COVID-19 and Pneumonia on Chest X-ray. Sensors 2022, 22, 6709. https://doi.org/10.3390/s22176709
Kim YJ. Machine Learning Model Based on Radiomic Features for Differentiation between COVID-19 and Pneumonia on Chest X-ray. Sensors. 2022; 22(17):6709. https://doi.org/10.3390/s22176709
Chicago/Turabian StyleKim, Young Jae. 2022. "Machine Learning Model Based on Radiomic Features for Differentiation between COVID-19 and Pneumonia on Chest X-ray" Sensors 22, no. 17: 6709. https://doi.org/10.3390/s22176709
APA StyleKim, Y. J. (2022). Machine Learning Model Based on Radiomic Features for Differentiation between COVID-19 and Pneumonia on Chest X-ray. Sensors, 22(17), 6709. https://doi.org/10.3390/s22176709





