Abstract
Chinese narcissus (Narcissus tazetta var. chinensis) was introduced to China 1300–1400 years ago, and has grown naturally in southeastern China. It is a popular Chinese traditional potted flower and a well-known flowering bulb cultivated worldwide with only two white-tepal triploid cultivars, ‘Jinzhan Yintai’ and ‘Yulinglong’. Recently, a mutant with yellow tepals was observed and promptly became popular. To clarify the key pigment for color difference and its molecular mechanism of accumulation, we conducted pigment metabolite analysis and comparative transcriptome analysis on ‘Jinzhan Yintai’ and the yellow-flowered mutant. The results showed that there was no significant difference in total flavonoid content between the mutant and ‘Jinzhan Yintai’, whereas the total carotenoid content of the mutant was more than 10-fold higher than that of ‘Jinzhan Yintai’. Based on the RNA-sequencing results, sixty-four unigenes, corresponding to 29 enzymes associated with the carotenoid biosynthesis pathway, were analyzed in detail. A comparative KEGG pathway enrichment analysis, in conjunction with quantitative real-time PCR data, revealed the opposite gene expression mode of the carotenoid biosynthesis pathway. Compared with ‘Jinzhan Yintai ’, PSY and PDS were up-regulated in the three mid-flowering stages of the mutant, whereas NCED genes were strongly down-regulated, which likely contributed to carotenoid accumulation in chromoplasts of the tepals in the mutant.
1. Introduction
Chinese narcissus (Narcissus tazetta var. chinensis) is a perennial bulbous plant belonging to the Amaryllidaceae that is widely cultivated in East Asia and China. More than 20 million bulbs are grown annually owing to its highly fragrant flowers and the flowering time overlapping with the Chinese Lunar New Year. As one of ten traditional flowers, it has been grown for more than 1000 years in China. However, only two major cultivars of Chinese narcissus are grown. The cultivar ‘Jinzhan Yintai’ produces a flower with six white petal-like tepals surmounted by a yellow cup-shaped corona. The other cultivar is a double-flowered form called ‘Yulinglong’, which produces flowers with 12 clustered white tepals without an obvious corona. Owing to its triploid chromosome complement (2n = 3x = 30), traditional cross breeding of Chinese narcissus is difficult. Consequently, important ornamental characters, especially the flower color, have yet to be improved [1].
In nature, flower color is a primary characteristic for plants to interact with pollinators for reproduction. Flower color is also the important commercial and quality trait for ornamental plants. The mechanisms of floral pigment biosynthesis have been studied extensively in the past three decades [2]. In most cases, flower color is determined by three classes of pigments: flavonoids, carotenoids, and alkaloids [2,3]. Among these compounds, the composition and content of carotenoids are regarded as the primary factors that influence the flora color of N. pseudonarcissus [4,5]. Ten carotenoids have been isolated from the perianth and corona of 15 N. pseudonarcissus cultivars [6].
Carotenoids, as light-harvesting pigments and structural components of photosystems, are crucial and participate in a variety of biological processes in plants, including photoprotection and the regulation of growth and development [7]. In addition, carotenoids are considered as antioxidants and a few of them could be precursors of provitamin A, which are of vital importance in the human diet [8]. Carotenoid metabolism in plants has been lucubrated in recent years since its importance both for plants and humans [9,10,11,12]. Carotenoid biosynthesis and storage primarily occurred in plastids, which strongly differ in their ability and capacity to accumulate carotenoids, ranging from extremely small quantities in etioplasts to massive amounts in chromoplasts [11,13].
There are usually five core steps in carotenoid biosynthesis pathway, involving condensation, desaturation/isomerization, hydroxylation, oxidation, and epoxidation to generate a variety of carotenes and xanthophylls [9,10,14]. These steps are catalyzed by numerous upstream enzymes, such as phytoene synthase (PSY), phytoene dehydrogenase (PDS), 15-cis-ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS), carotenoid isomerase (CRTISO), lycopene-ε-cyclase (LCYE), lycopene β-cyclase (LCYB), β-carotene hydroxylase 1 and 2 (BCH1/2), cytochrome P450 hydroxylases (CYP97A and CYP97C), violaxanthin de-epoxidase (VDE), zeaxanthin cyclooxygenase (ZEP), and neoxanthin synthase (NXS). Among these proteins, PSY is the first and important rate-limiting enzyme in the carotenoid biosynthesis [11]. Accumulation of carotenoids is a dynamic procedure of metabolic equilibrium between biosynthesis and degradation [9,15]. The primary factor of carotenoid biodegradation is carotenoid cleavage deoxygenases (CCDs), which degrade β-carotene and downstream carotenoids to generate apocarotenoids, abscisic acid (ABA), strigolactones, and other volatile compounds [16], Then 9-cis-Epoxycarotenoid dioxygenases (NCEDs) specifically cleave 9-cis-violaxanthin and 9-cis-neoxanthin to form xanthoxin for ABA production [17,18]. Zaxinone synthase (ZAS), a recent found new CCD subfamily in most land plants, cleaves zeaxanthin to produce zaxinone, and further regulates ABA and strigolactone biosynthesis [19,20]. In N. pseudonarcissus, CCD genes mediate carotenoid turnover and are crucial factors responsible for the low carotenoid content and floral color fading of certain cultivars [21]. However, to date, no NCED genes have been identified in narcissus species.
Transcription factors (TFs) are pivotal in regulating the transcription of carotenoid biosynthesis genes. Recent progress has been achieved in the identification and validation of TFs that positively or negatively regulate the expression of carotenoid structural genes in plants [14,22,23]. Numerous TFs members, including the MYB, WRKY, bHLH, and MADS families, regulate genes in single or multiple pathways in the leaf, flower, or fruit [7]. Moreover, as general regulators in multiple processes of plant growth and development, many TFs may indirectly affect carotenoid metabolism [22].
Recently, a mutant of Chinese narcissus with yellow tepals was discovered and has quickly gained popularity. However, the main kind of different flower pigments and its molecular basis for the difference in tepal color of Chinese narcissus and the yellow mutant have not been investigated. In the present study, RNA-sequencing (RNA-Seq) analysis of the tepal transcriptome of the two color forms of Chinese narcissus was performed using an Illumina sequencing platform. After microscopic observation of the tepals and total pigment metabolite analysis at different stages of flower development and pigmentation, we investigated the carotenoid metabolic pathways and compared the critical structural genes that are involved in tepal pigmentation in ‘Jingzhanyintai’ and the yellow mutant. The results provide a valuable theoretical basis for potential innovation of floral pigmentation in Chinese narcissus in the future.
2. Materials and Methods
2.1. Plant Materials
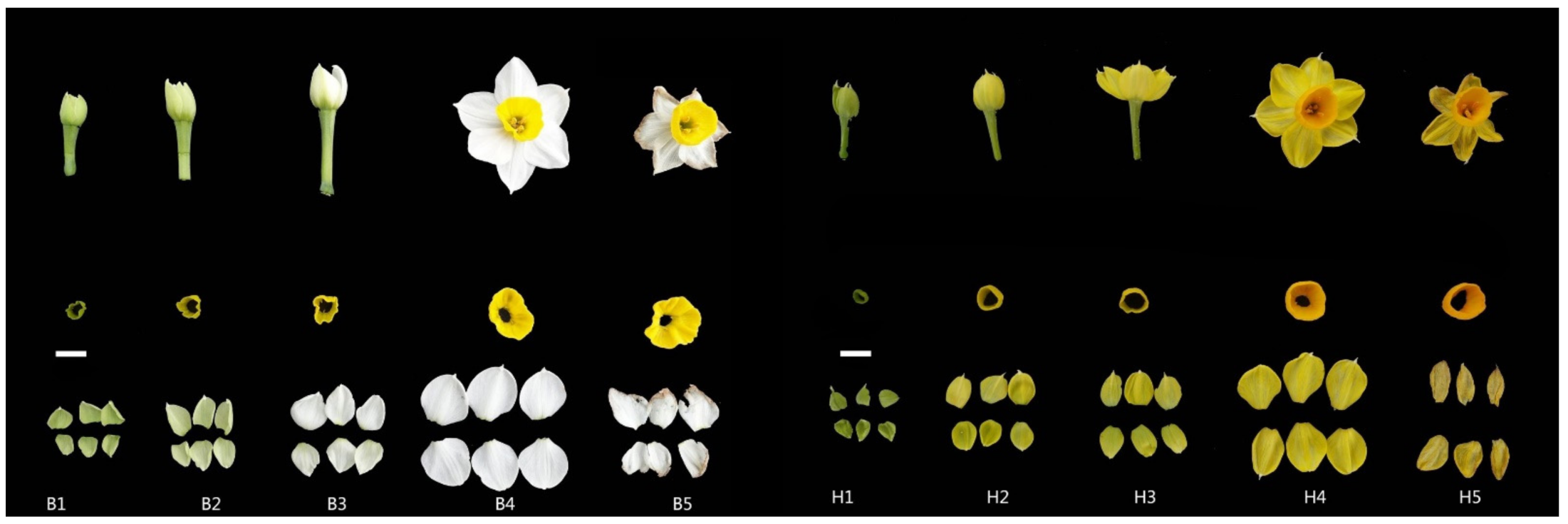
Chinese narcissus cultivar ‘Jinzhan Yintai’ and its yellow-tepal mutant were cultivated in the experimental field of the Research Institute of Narcissus, Zhangzhou, Fujian, China. Under natural temperature and light conditions, corolla and tepal tissues of two materials were collected from plants at bud stage (S1), initial flowering stage (S2), full bloom stage (S3), full expansion stage (S4), and decay stage (S5), respectively. We mainly concerned the pigment differences in two narcissus tepals. For RNA sequencing, the white tepal tissues were named B1, B2, B3, B4, B5 and separated for the S1 (T01 and T02), S2 (T03 and T04), S3 (T05 and T06), S4 (T07 and T08), and S5 (T09 and T10). The yellow tepals were marked as H1, H2, H3, H4, H5; at S1 (T11 and T12), S2 (T13 and T14), S3 (T15 and T16), S4 (T17 and T18) and S5 (T19 and T20), respectively (Figure 1). The collected tissues were frozen in liquid nitrogen and stored at −80 °C for RNA isolation.
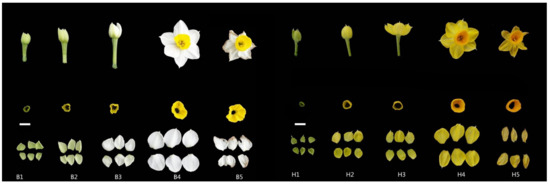
Figure 1.
Corolla and tepal tissue of Chinese narcissus. The materials harvested at bud stage, initial flowering stage, full bloom stage, full expansion stage, and decay stage. Among them, the left is ‘Jinzhan Yintai’ (B1–B5), and the right is the yellow mutant (H1–H5). Bar = 1 cm.
2.2. Pigment Semiquantification
Each sample (100 mg) was placed in a plug test tube, cooled in liquid nitrogen, and ground to powder. The samples were extracted with 3 mL petroleum:acetone ether (4:1, v/v) for 24 h. The total carotenoid content was determined by ultraviolet–visible (UV-Vis) spectrophotometry, at a wavelength of 400–500 nm. Total carotenoids were quantified using the method of the determination of carotenoid content in Manihot esculenta by Afonso T. et al. [24] (with slight modifications). For measurement of the total flavonoid content, standard solutions of rutin (Shanghai Yuanye Biotechnology Co., Ltd., Shanghai, China) of different concentrations were prepared and a standard curve generated. The samples were placed in test tubes with stoppers, cooled in liquid nitrogen, and ground to powder. The samples were extracted with 3 mL methanol:water:formic acid:trifluoroacetic acid (70:27:2:1, v/v/v/v) for 96 h. The absorbances of 200–450 nm wavelengths were determined by ultraviolet–visible detector. The quantification of total flavonoids adopted the method used by Cheng, B.X. et al. in the study of tetraploid rose [25] (with slight modifications).
2.3. Total RNA Extraction and RNA-Seq Analysis
Total RNA extraction and RNA-Seq analysis for ‘Jinzhan Yintai’ and the yellow mutant were performed by the Beijing Biomarker Company (Beijing, China). A Nanodrop spectrophotometer, Qubit 2.0 fluorometer, and Agilent 2100 bioanalyzer were used to detect the purity, concentration, and integrity of the RNA samples to ensure that the RNA quality was acceptable for transcriptome sequencing. The RNA library was constructed, and the concentration and fragment size of the library were determined using a Qubit 2.0 fluorometer and Agilent 2100 bioanalyzer. High-throughput sequencing was performed with an Illumina HiSeq system. Raw FASTQ reads (raw reads) are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and can be viewed under BioProject id: PRJNA855612. In the results, we measure the relative expression of genes by the number of fragments per kilobase of the exon model per million mapped fragments, the FPKM value.
2.4. Identification of Differentially Expressed Genes
The RNA-Seq reads were filtered and assembled to attain the transcripts. The transcripts were clustered and assembled into unigenes. The data then queried against the NCBI non-redundant (NR; https://ftp.ncbi.nih.gov/blast/db, Format: 24 July 2018), Pfam (http://pfam.xfam.org, Format: 24 July 2018), Clusters of Orthologous Groups of Proteins (KOG/COG/eggnog; https://www.mendeley.com/catalogue/cogclusters-orthologous-groups-proteins/, Format: 24 July 2018), Swiss-Prot (https://web.expasy.org/docs/swiss-prot_guideline.html, Format: 24 July 2018), Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html, Format: 27 July 2018), and Gene Ontology (GO; http://www.geneontology.org, Format: 24 July 2018) databases to obtain annotation information. Twenty tepal samples from ‘Jinzhan Yintai’ and the yellow-flowered mutant (the total number consisted of two biological replicates at each of the five flowering stages for each variety) were compared to identify up-regulated, down-regulated, or unchanged genes. Differentially expressed genes (DEGs) were defined as the up- or down-regulated unigenes with adjusted p-value ≤ 0.05 and |(log2 fold change) ≥ 1|.
2.5. Quantitative Real-Time PCR
The expression of six DEGs involved in carotenoid synthesis was measured by quantitative real-time PCR (qRT-PCR) using the Power SYBR® Green PCR Master Mix (Applied Biosystems) to verify the RNA-Seq data. Assays were conducted with a BIO-RAD CFX Connect™ Real-Time PCR Detection System. A three-step reaction was performed under the following conditions: initial denaturation at 95 °C for 3 min, followed by 40 cycles at 95 °C for 10 s and 55 °C for 20 s, and extended extension at 72 °C for 20 s and 75 °C for 5 s. Relative expression levels of the DEGs were calculated by normalization relative to the Actin gene using the 2−ΔΔCt method [26]. The expression patterns of DEGs were compared between samples at critical stages of carotenoid synthesis and decomposition (H3 and B3).
3. Results
3.1. Tepal Pigmentation Characteristics of Chinese Narcissus
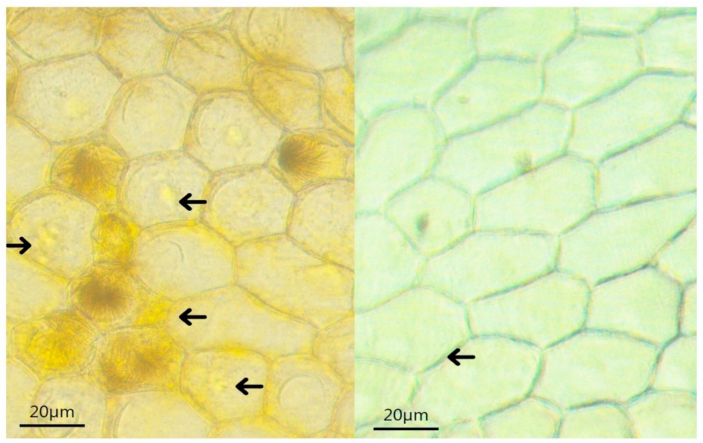
Microscopic observation of the tepals of the yellow mutant and ‘Jinzhan Yintai’ revealed that the tepal of the yellow mutant was thinner in transverse section, the epidermal cells contained a larger number of yellow liposomes, and each cell contained multiple spherical chromoplasts (Figure 2). The chromoplasts of the yellow mutant were more clearly visible and deeper yellow than those of ‘Jinzhan Yintai’.
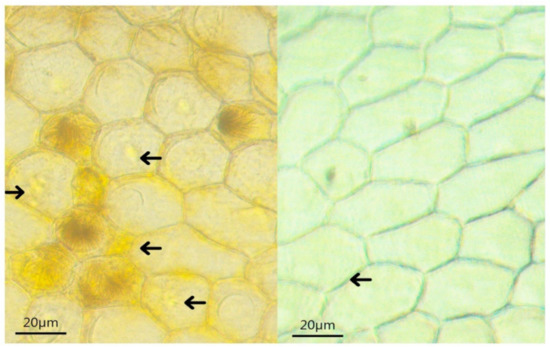
Figure 2.
Chromoplasts contained in epidermal cells of ‘Jinzhan Yintai’ and yellow mutant tepals. The arrows in the left picture point to yellow chromoplasts, and the arrows in the right picture point to white chromoplasts. Bar = 20 μm.
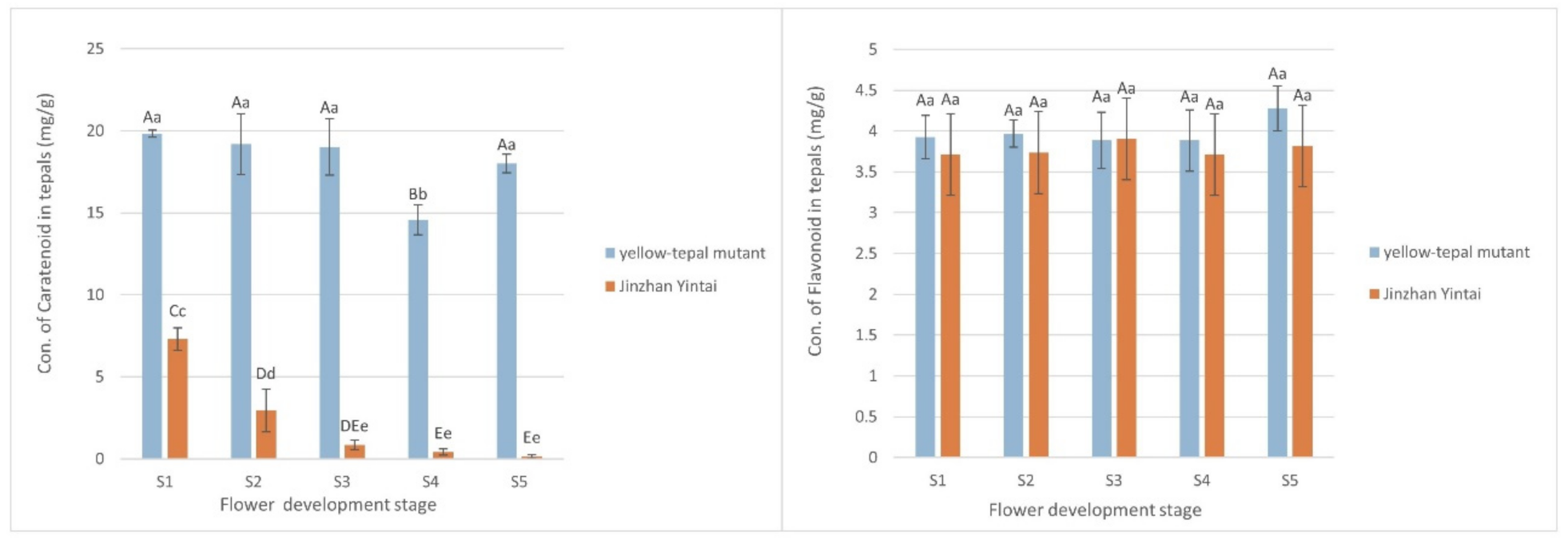
No significant difference in total flavonoid content was observed between the yellow mutant and ‘Jinzhan Yintai’, whereas the total carotenoid content in the yellow mutant was about ten-times higher than that of ‘Jinzhan Yintai’ at the five flowering stages. Although there was a turning point for decline at S3, the carotenoid content in the yellow mutant essentially remained high at all stages of flowering. The carotenoid content in ‘Jinzhan Yintai’ was less than half that of the yellow mutant at its highest level (S1), which then decreased over the course of flowering from fast to slow at the point of S3 (Figure 3). Thus, the carotenoid contents differed significantly between ‘Jinzhan Yintai’ and the yellow mutant before and after the S3 stage, which indicated carotenoids were likely to play a decisive role in the difference in flower color of the two varieties.
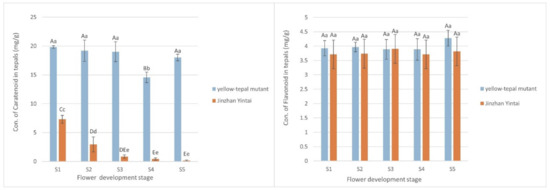
Figure 3.
Comparison of the total pigment content in ‘Jinzhan Yintai’ and yellow mutant tepals. The left one shows the comparison of the carotenoid content of the two samples, and the right one shows the comparison of the flavonoid content of the two samples. Different capital letters indicate significant differences at the p < 0.01 level, and different lowercase letters indicate significant differences at the p < 0.05 level; same letter means no significant difference. Error bars represent standard deviation.
3.2. Tepal Transcriptome Sequencing, Assembly and Annotation
Twenty cDNA libraries were constructed for the two varieties for transcriptome sequencing. In total, 148.85 Gb clean reads were obtained. The percentage of Q30 bases in each sample was not less than 85.59% and the GC content was 46.44%–48.90% (Table 1). After quality control, the filtered data were used for gene annotation and functional classification analysis. In total, 225,032 transcripts were assembled, with an average length of 1108 bases, of which 92,071 were unigenes. The N50 of the unigenes was 1543 and the assembly integrity was high.

Table 1.
Annotation and statistics of unigenes.
The 92,071 unigenes assembled de novo were aligned and annotated by means of a BLAST search against the NR, Pfam, KOG/COG/eggNOG, Swiss-Prot, KEGG, and GO databases. Altogether, 32,816 unigenes (35.64%) were annotated based on annotations extracted from the databases. In total, 8713 (26.55%), 18,219 (55.52%), 11,156 (34.00%), 18,928 (57.68%), 20,011 (60.98%), 19,326 (58.89%), 29,824 (90.88%), and 31,761 (96.79%) unigenes were annotated from the COG, GO, KEGG, KOG, Pfam, Swiss-Prot, eggNOG, and NR databases, respectively. Overall, 1055 (3.21%) unigenes were not annotated.
3.3. Expression Profiles of DEGs in Tepals
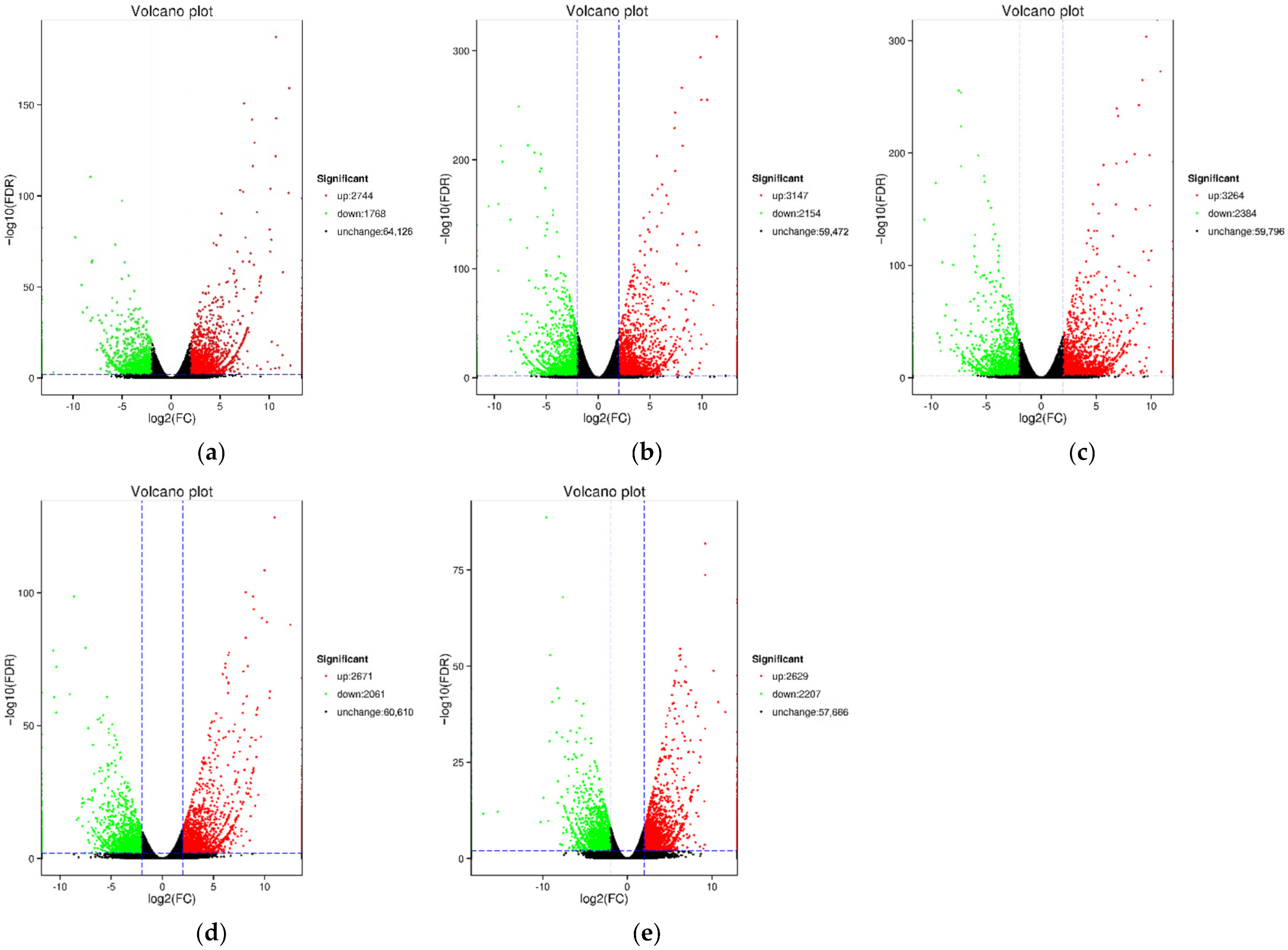
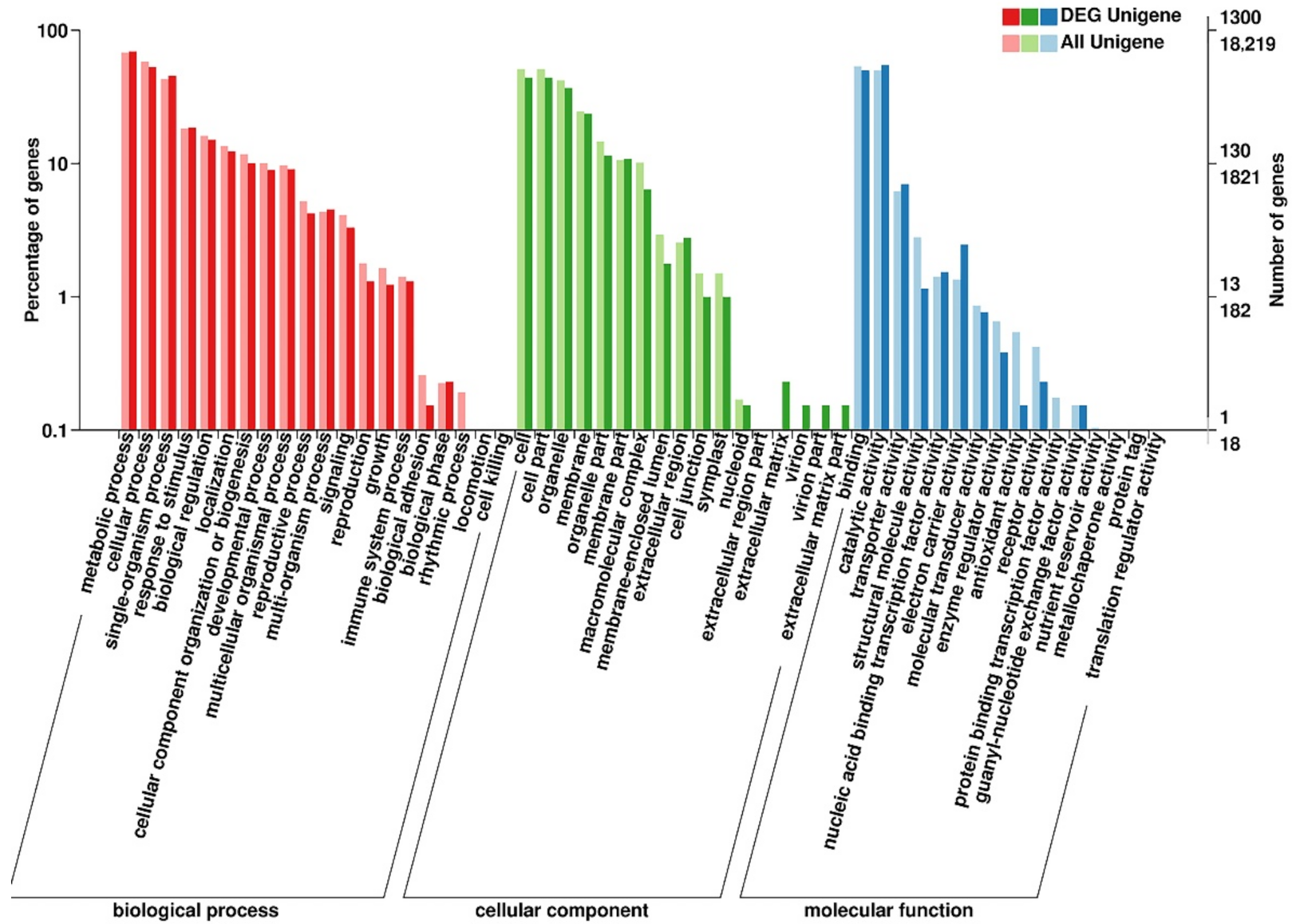
The highest number of DEGs was detected in the comparison between the B3 and H3 samples, combined with the former results of the total carotenoid content, we select S3 to analysis further (Table 2). In total, 5648 DEGs were detected in the B3 versus H3 comparison, of which 3264 DEGs were up-regulated and 2384 DEGs were down-regulated (Figure 4, Table 2). (Figure 5). In the biological process category, most DEGs were annotated as metabolic process, cellular process, and single-organism process. In the cellular component category, the majority of DEGs were annotated as cell, cell part, and organelle terms. In the molecular function category, the number of DEGs annotated as binding and catalytically active terms was obviously higher than others. The GO annotations indicated that the DEGs were involved in various metabolic and cellular processes.

Table 2.
Number of DEGs identified in different comparisons between the yellow mutant and ‘Jinzhan Yintai’.
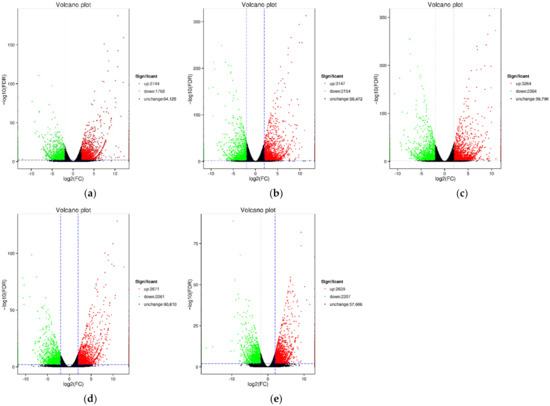
Figure 4.
Volcano plots of DEGs in different groups. (a) H1 vs. B1; (b) H2 vs. B2; (c) H3 vs. B3; (d) H4 vs. B4; (e) H5 vs. B5. Each point in the differential expression volcano plot represents a gene, and the abscissa represents the logarithm of the fold difference in expression of a certain gene between the two samples. The larger the absolute value, the greater the fold difference of the expression level between the two samples. The ordinate represents the negative logarithm of the false discovery rate. The larger the value, the more significant the differential expression, and the more reliable the differentially expressed genes obtained by screening. Red and green dots represent up-regulated and down-regulated DEGs, respectively. Black dots represent unchanged DEGs.
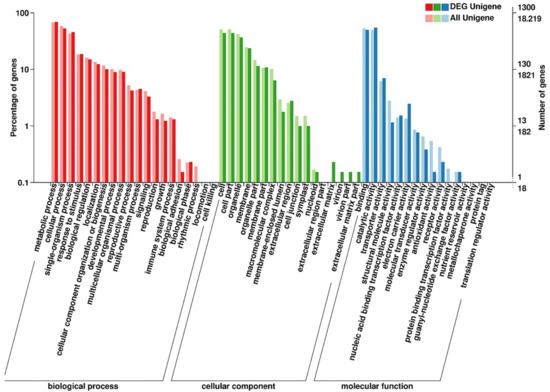
Figure 5.
GO annotation of DEGs identified in the comparison between H3 and B3. The horizontal axis is the GO classification, the left side of the vertical axis is the percentage of the number of genes, and the right side is the number of genes. Red columns represented biological processes, green columns represented cellular components, and blue columns represented molecular functions.
3.4. DEGs Involved in Carotenoid Biosynthesis
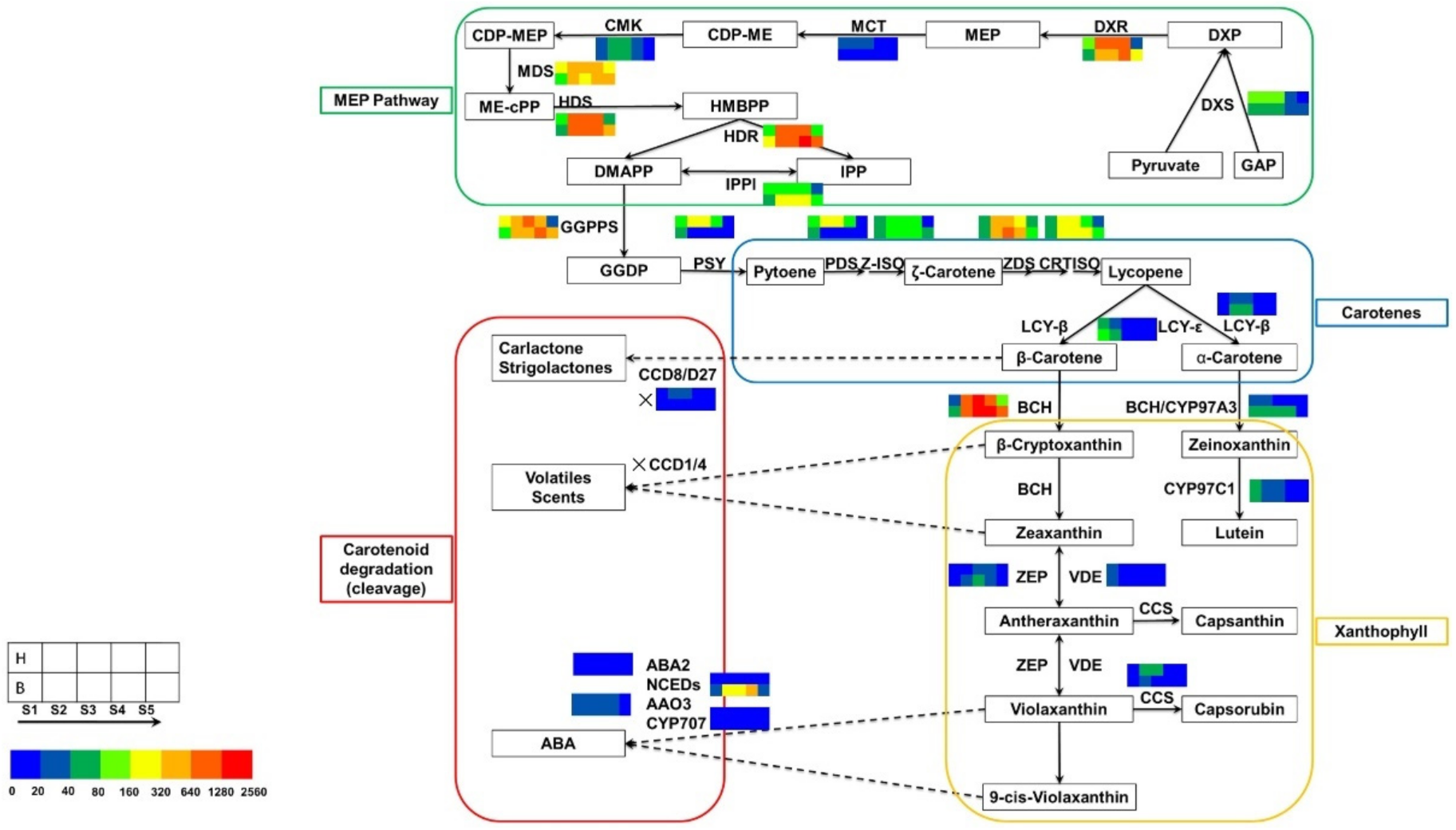
DEGs encoding proteins involved in the carotenoid metabolic pathway were analyzed. Sixty-four unigenes related to 29 enzymes were identified in the tepal transcriptome by mapping to KEGG reference pathways. Among these unigenes, 17 unigenes were associated with carotene precursor synthesis (the methylerythritol 4-phosphate [MEP] pathway), 28 unigenes were involved in carotenoid synthesis, and 19 unigenes were involved in carotenoid degradation/cleavage. Based on the FPKM values, the changes in expression of the 64 unigenes and the three branched pathway genes in tepals during tepal pigmentation were compared. These data showed that the DEGs were mainly involved in carotenoid synthesis and carotenoid degradation/cleavage (Figure 6, Table 3).
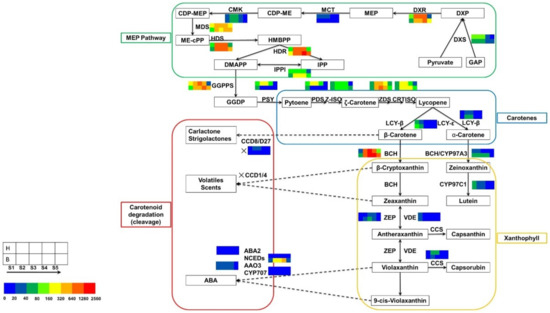
Figure 6.
Expression patterns of DEGs related to the carotenoid biosynthesis pathway in tepals during flower development. Enzyme names and expression patterns are indicated at the side of each step. Color boxes from left to right represent unigenes showing lower or higher expression level in bud stage, initial flowering stage, full bloom stage, full expansion stage, and decay stage, respectively. The first row represents the yellow mutant, while the second row represents ‘Jinzhan Yintai’. Different colors represent magnitude of log2 (expression ratio) for each unigenes, with the FPKM values being grouped in 0–20, 20–40, 40–80, 80–160, 160–320, 320–640,640–1280, 1280–2560, respectively.

Table 3.
The number of unigenes related to carotenoids metabolic pathway in Chinese narcissus.
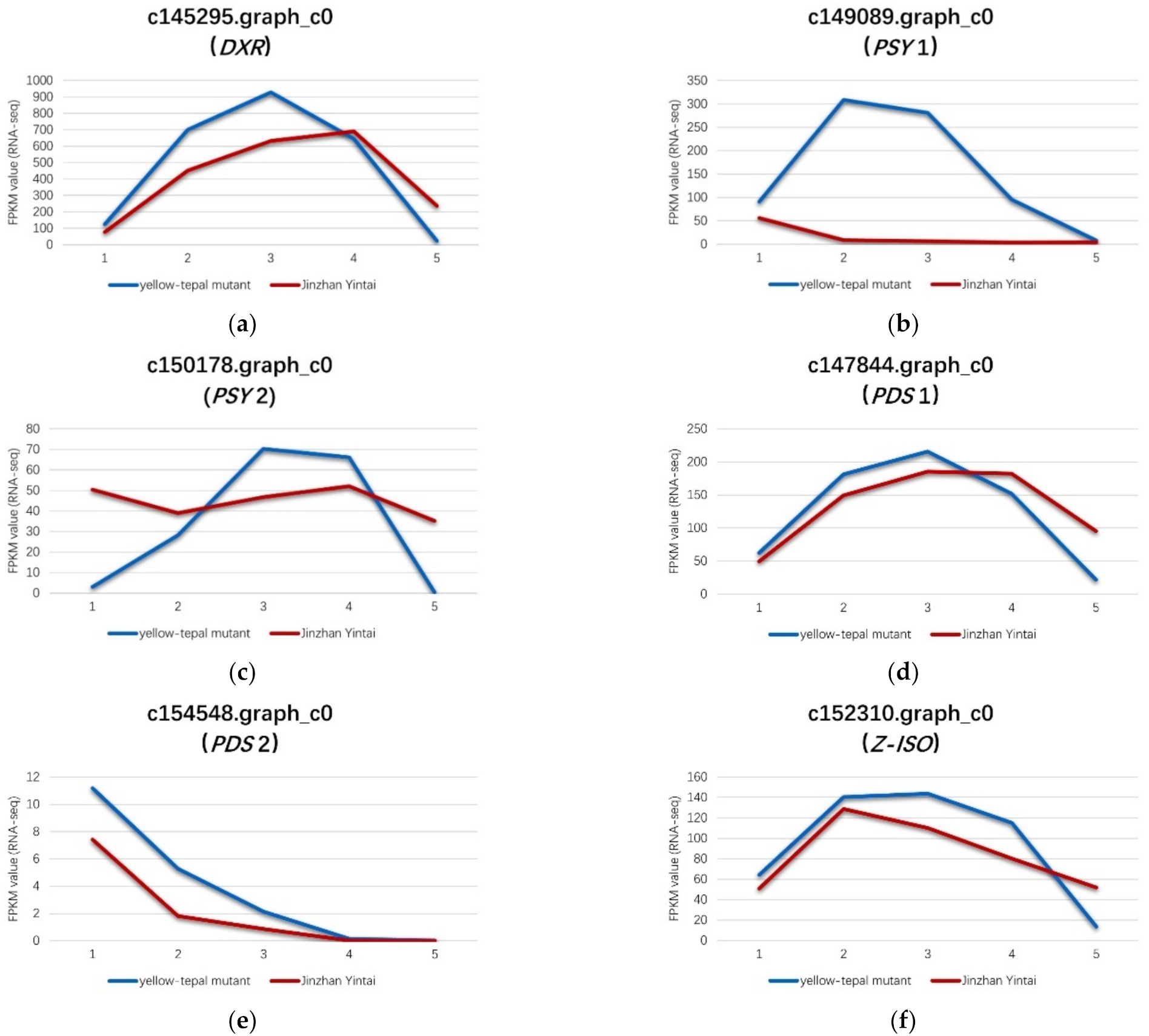
In this study, eight genes were differentially expressed in the critical period of carotenoid synthesis and degradation. Of these eight genes, six (DXR, PSY1, PSY2, PDS1, PDS2 and Z-ISO) were up-regulated and two (NCED1 and NCED2) were down-regulated in the yellow mutant, and were associated with carotenoid synthesis and carotenoid degradation, respectively. Among the genes involved in carotenoid synthesis, the expression levels of genes encoding four types of enzymes differed significantly between the yellow mutant and ‘Jinzhan Yintai’. DXR is an important enzyme involved in carotene precursor synthesis (the MEP pathway) [27]. The expression level of DXR gene (c145295.graph_c0) in the present study was significantly higher in the yellow mutant than in ‘Jinzhan Yintai’ (Figure 7).
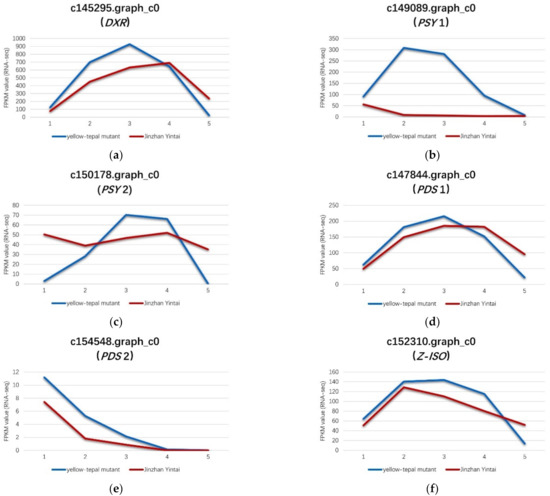
Figure 7.
Expression levels of six key genes in ‘Jinzhan Yintai’ and its yellow mutant. (a) c145295.graph_c0, (b) c149089.graph_c0, (c) c150178.graph_c0, (d) c147844.graph_c0, (e) c154548.graph_c0, (f) c152310.graph_c0. The blue line represents the expression level of the gene in the yellow mutant, while the red line represents the expression level of the gene in ‘Jinzhan Yintai’.
A floral PSY gene was reported to play a possible regulatory role in color formation during flower development in N. pseudonarcissus [28]. PSY is a significant plastidial enzyme, catalyzing the conversion of two molecules of geranylgeranyl pyrophosphate (GGPP) to phytoene during carotenoid biosynthesis [29]. In the current study, we detected two PSY genes (c149089.graph_c0 and c150178.graph_c0) that were up-regulated in the yellow mutant compared with ‘Jinzhan Yintai’ (Figure 7).
Among these genes, the expression level of one PSY gene (c149089.graph_c0) in the yellow mutant was significantly higher than that in ‘Jinzhan Yintai’ at the bud, initial flowering, full bloom, and full development stages. Another PSY gene (c150178.graph_c0) was more highly expressed in the yellow mutant at the full bloom and full development stages. These results indicated that PSY genes were more positively expressed in the tepals of the yellow mutant than in ‘Jinzhan Yintai’.
Through the catalytic activity of PDS, 15-cis-phytoene is reduced to 9,15-cis-phytoene, which is subsequently reduced to 9,15,9′-trans-cis-ζ-carotene. This process is an important step in carotenoid synthesis. Two PDS genes (c154548.graph_c0 and c147844.graph_c0) were up-regulated in the yellow mutant (Figure 7). The expression of one PDS gene (c154548.graph_c0) was slightly higher in the yellow mutant overall, whereas the expression level of the other PDS gene (c147844.graph_c0) was significantly higher in the yellow mutant at the bud, initial flowering, and full flowering stages compared with ‘Jinzhan Yintai’ tepals at the same stages.
The present data revealed that the Z-ISO gene (c152310.graph_c0) was up-regulated in the yellow mutant. The Z-ISO enzyme catalyzes the conversion of 9,15,9′-trans-cis-ζ-carotene to 9,9′-di-cis-ζ-carotene (Figure 7).
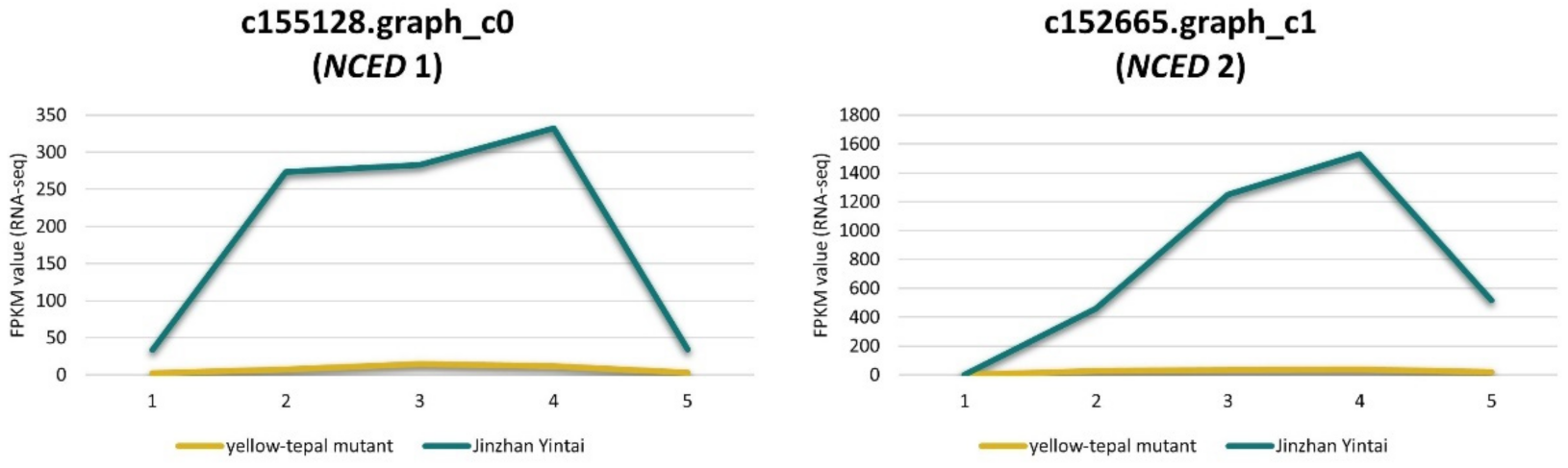
In plants, NCED is an important enzyme involved in carotenoid decomposition and ABA synthesis. We found that two NCED genes (c155128.graph_c0 and c152665.graph_c1) were significantly down-regulated in the yellow mutant, whereas their expression was up-regulated in ‘Jinzhan Yintai’ (Figure 8). This result suggested that low-expressed NCEDs may inhibit carotenoid degradation in tepals of the yellow mutant, which contributed to the accumulation of carotenoids in tepals. For ‘Jinzhan Yintai’, the limited amount of carotenoids synthesized in the early stages of flower development were likely to be decomposed quickly under the high activity of NCED, which furtherly activated the ABA production for uncertain reasons.
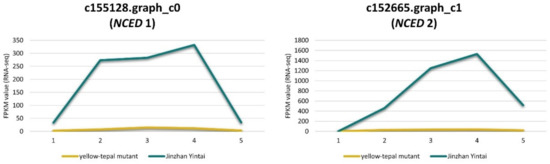
Figure 8.
Expression levels of NCED genes in ‘Jinzhan Yintai’ and its yellow mutant. The yellow line represents the expression level of the gene in the yellow mutant, while the green line represents the expression level of the gene in ‘Jinzhan Yintai’.
3.5. Transcription Factors Involved in Narcissus Tepal Pigmentation
In plants, transcription factors usually play key roles in regulating diverse developmental processes, and biotic and abiotic stress responses [30,31,32]. In the comparisons of ‘Jinzhan Yintai’ and the yellow mutant, a total of 200 putative DEGs encoding TFs were identified, including 36 encoding WRKY TFs, 35 encoding ethylene-related TFs, 34 encoding bHLH TFs, 32 encoding MYB and MYB-related TFs, 11 encoding GATA TFs, and 10 encoding NAC TFs (Table 4). These TF DEGs likely participated in the synthesis of floral pigment metabolites in the tepals. Their potential function and interaction either with structural genes or with other TFs would be further testified in future experiments.

Table 4.
List of 200 putative TFs differentially expressed between ‘Jinzhan Yintai’ and the yellow mutant tepals.
3.6. Verification of Gene Expression by qRT-PCR
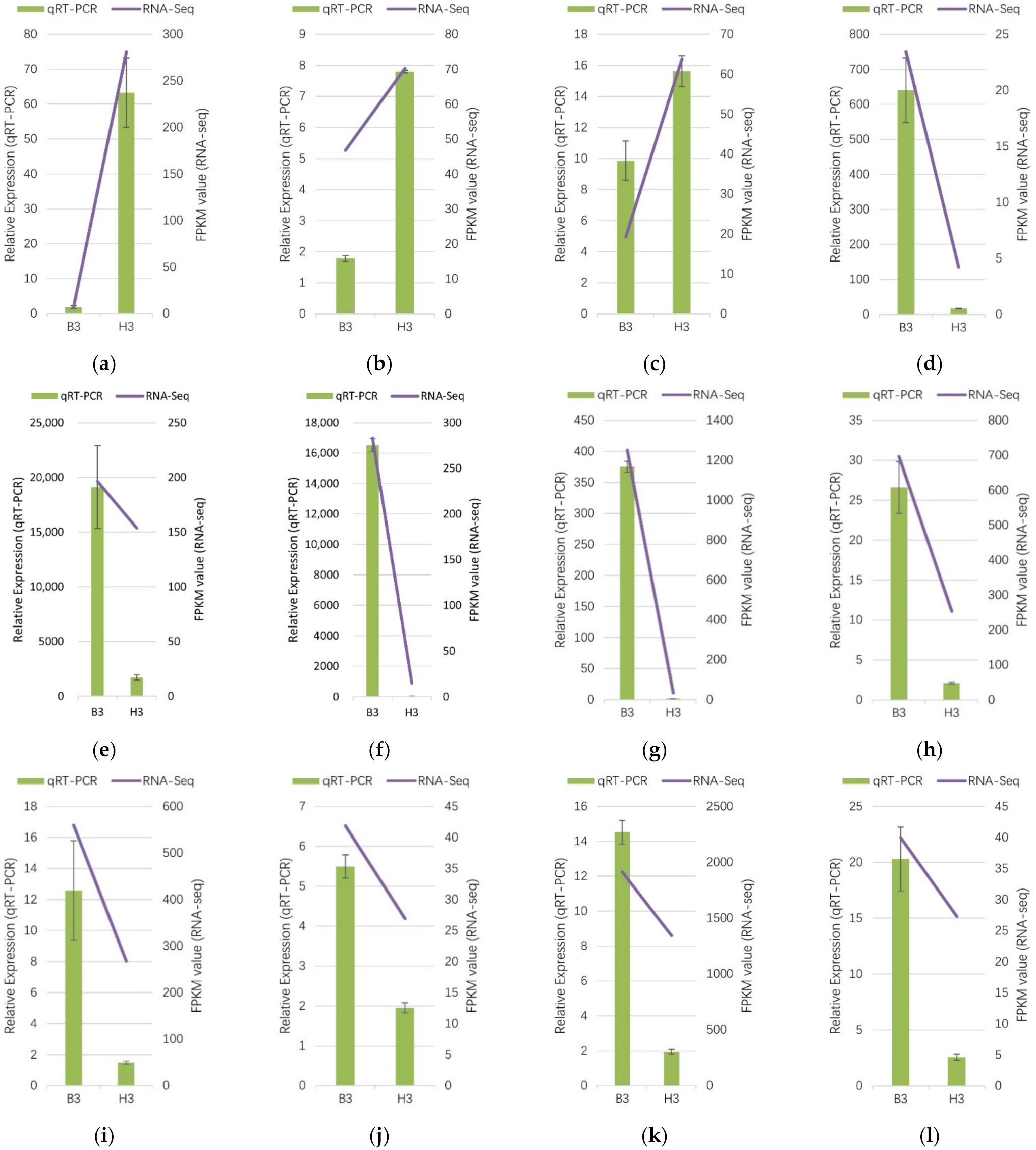
To verify the RNA-Seq data, in addition to the key genes PSY (c149089.graph_c0 and c150178.graph_c0) and NCED (c155128.graph_c0 and c152665.graph_c1), eight other floral pigment-related DEGs were selected for qRT-PCR assays. All of the selected twelve genes encoded enzymes involved in carotenoid synthesis and degradation, namely PSY1, PSY2, CCS, DXS1, DXS2, NCED1, NCED2, GGPPS1, GGPPS2, LCYB, crtZ and ZEP. According to RNA-Seq analysis, three DEGs (c149089.graph_c0, c150178.graph_c0 and c147078.graph_c0,) were up-regulated in the yellow mutant, whereas nine genes (c118364.graph_c0, c153078.graph_c1, c155128.graph_c0, c152665.graph_c1, c144484.graph_c3, c140360.graph_c1, c145169.graph_c0, c151028.graph_c0, and c140017.graph_c0) were down-regulated. The result showed that the expression patterns of these twelve DEGs were consistent between the qRT–PCR and RNA-seq results (Figure 9), which indicated the RNA-Seq data were highly reliable for future genetic experiments.
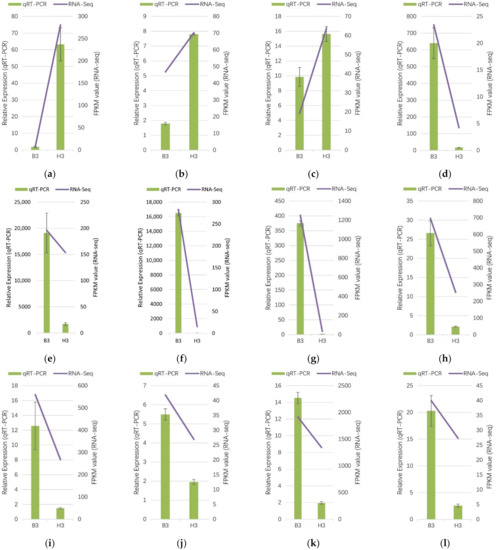
Figure 9.
Verification of RNA-Seq data of carotenoid biosynthesis related DEGs by qRT-PCR. (a) c149089.graph_c0 (PSY1); (b) c150178.graph_c0 (PSY2); (c) c147078.graph_c0 (CCS); (d) c118364.graph_c0 (DXS1); (e) c153078.graph_c1 (DXS2); (f) c155128.graph_c0 (NCED1); (g) c152665.graph_c1 (NCED2); (h) c144484.graph_c3 (GGPPS1); (i) c140360.graph_c1 (GGPPS2); (j) c145169.graph_c0 (LCYB); (k) c151028.graph_c0 (crtZ); (l) c140017.graph_c0 (ZEP). Histograms represent the fold change in gene expression determined by qRT-PCR using the 2−ΔΔCт method. Line plots represent gene expression determined according to the log2 ratio (FPKM of H1 sample relative to B1 sample [H1/B1]) determined by RNA-Seq. All genes selected for qRT-PCR analysis were analysed in three replicates. Error bars represent standard deviation.
4. Discussion
In nature, the color of flowers can provide ornamental value and use value related to plant reproduction. Exploring the regulatory mechanism of floral color differentiation is of great value for understanding the evolution of plants and cultivating new ornamental lines [33]. Floral color differentiation is controlled by multiple factors, including environmental factors and endogenous factors under natural conditions [34,35]. The Chinese narcissus cultivar ‘Jinzhan Yintai’ and the yellow-tepal mutant produce flowers with a central corona and six tepals. The difference in flower color between the two varieties is mainly reflected in the tepals. The tepals of both varieties are yellow at the bud and initial flowering stages, and those of the yellow mutant are more strongly pigmented. During the full bloom, full expansion, and decay stages, the tepals of ‘Jinzhan Yintai’ are less pigmented and appear white, whereas the tepals of the yellow mutant consistently retain a more saturated yellow color.
The appearance of flower color is mainly affected by abundant pigments synthesized by plants, among which flavonoids and carotenoids are the main components of yellow pigments in flowers [25]. In the present study, UV-Vis spectrophotometry was used to estimate the total flavonoid and total carotenoid contents in the tepals of ‘Jinzhan Yintai’ and the yellow mutant. The difference in tepal color was due to the difference in carotenoid content. The content of flavonoids was similar in both materials, and the total flavonoid content of both materials was substantially less than the total carotenoid content. Therefore, it was likely that the detected DEGs associated with carotenoid metabolism played a crucial role in the phenotypic difference of the two genotypes.
In this study, we compared the differential expression of key genes in the carotenoid metabolic pathway in ‘Jinzhan Yintai’ and the yellow mutant and identified eight major DEGs. Phytochrome-interacting factor 1 (PIF1) and other TFs in the phytochrome-interacting factor (PIF) family down-regulate carotenoid production by specifically repressing PSY expression [36]. Therefore, PSY is important for accumulation of carotenoid in plants. The DEGs c149089.graph_c0 and c150178.graph_c0 were annotated to encode PSY proteins, and were up-regulated in the comparisons of B3 versus H3 and B4 versus H4, and in the case of c149089.graph_c0, also in the comparisons B1 versus H1 and B2 versus H2. Previous data have shown that overexpression of PDS significantly enhances lycopene biosynthesis and accumulation in transgenic tomato fruit [37]. In the present study, the DEGs c147844.graph_c0 and c154548.graph_c0 were annotated to encode PDS proteins and were up-regulated in the comparisons B1 versus H1, B2 versus H2, and B3 versus H3. The up-regulation of PSY and PDS genes in the yellow mutant resulted in a significantly.
Increased carotenoid synthesis compared with that in ‘Jinzhan Yintai’. In addition, the expression of PSY and PDS was also found to be significantly correlated with the accumulation of one or more carotenoid compounds in studies on persimmon fruit development [38]. The DEG c152310.graph_c0 was annotated to encode a carotene isomerase, which presumably enables carotenoid biosynthesis to occur in the dark and in nonphotosynthetic tissues [39]. The elevated expression of Z-ISO in the yellow mutant was important for the yellow pigmentation of the tepal. Thus, we identified six DEGs associated with carotenoid synthesis that were significantly up-regulated in the yellow mutant, and each encode enzymes that catalyze the conversion of substrates to carotenoid-accumulating secondary metabolites.
In addition, among the identified DEGs associated with flower color development, two were significantly down-regulated in the yellow mutant, namely c155128.graph_c0 and c152665.graph_c1. These two DEGs were annotated to encode NCED proteins. In the advanced steps of the carotenoid metabolic pathway, NCED plays an important role in the breakdown of carotenoids. We speculate that NCED is an important factor in the difference in tepal color of the two narcissus accessions.
In conclusion, compared with ‘Jinzhan Yintai’, the expression of the PSY, PDS, and Z-ISO genes (involved in carotenoid synthesis) were up-regulated in the yellow mutant, whereas the expression of NCED (involved in carotenoid decomposition) was down-regulated. The accumulation of carotenoids in tepals of the yellow mutant was much stronger than that in tepals of ‘Jinzhan Yintai’. The present data suggested that the differential expression of these eight DEGs was mainly responsible for the difference in tepal color between ‘Jinzhan Yintai’ and the yellow mutant.
An additional eight genes were differentially expressed in ‘Jinzhan Yintai’ and the yellow mutant, and were also involved in the carotenoid metabolic pathway. In the MEP pathway, the first step catalyzed by DXS is the rate-determining step of the pathway, thus DXS is considered to be the most crucial enzyme of the pathway. Geranylgeranyl diphosphate synthase catalyzes the synthesis of its C20 precursor, isoprenoid geranylgeranyl diphosphate [40]. Lycopene β-cyclase is a critical enzyme directly involved in the synthesis of α-carotene and β-carotene through the cyclization of lycopene [41]. Beta-carotene hydroxylase (crtZ) is a rate-limiting enzyme in carotenoid synthesis [42]. Zeaxanthin cyclooxygenase (ZEP) affects the production of lutein-like pigments downstream of the carotenoid metabolic pathway [43]. The DXS gene (c118364.graph_c0 and c153078.graph_c1), GGPPS (c144484.graph_c3 and c140360.graph_c1), LCYB (c145169.graph_c0), crtZ (c151028.graph_c0), and ZEP (c140017.graph_c0) were all down-regulated in the yellow mutant. Capsanthin-capsorubin synthase (Ccs) is a catalytic enzyme that catalyzes the synthesis of capsanthin from epoxy zeaxanthin, which plays a role in carotenoid pigment biosynthesis [44]. This gene (c147078.graph_c0) was up-regulated in the yellow mutant and likely promotes the accumulation of carotenoid pigments in the yellow mutant.
We identified a total of 200 DEGs that encode TFs, mainly from the WRKY, ethylene-related, bHLH, and MYB/MYB-related TF families. Thirty-six differentially expressed unigenes were annotated as encoding WRKY TFs, which were the largest percentage of TFs DEGs (18.00%), demonstrating that these TFs may critically influence narcissus tepal pigmentation. The WRKY TFs were reported participate in carbohydrate and secondary metabolite biosynthesis, and plant development [45]. Ethylene affects the anthocyanin content of flowers, which affects their brightness, redness, and chroma [46]. The second highest number of TFs identified in the present study was 35 DEGs encoding ethylene-related TFs (17.50%), followed by bHLH TFs (34 DEGs, 17.00%) and MYB/MYB-related TFs (32 DEGs, 16.00%). The interaction between bHLH and MYB TFs regulates anthocyanin biosynthesis, thereby affecting flower pigmentation [47,48]. Transcription factors of these four families accounted for 68.50% of the total TFs. Elucidation of their roles in regulating the biosynthesis of pigment-related metabolites in narcissus tepals requires further investigation.
RNA-Seq is among the most profitable and practical method for studying genes in plant species without model, especially those that lack genomic data, such as Narcissus [32]. The data obtained in the present study provided information on the relative levels of gene expression, which may reflect the regulatory state of genes in the tepal of the two varieties. It will be beneficial to explore the mechanism of yellow pigment accumulation in the tepal of the yellow mutant by predicting the functions of the DEGs. The current study provides a foundation for further research on the mechanism of pigment development in Narcissus. Elucidating the roles of these genes in narcissus floral color development is a topic for future research.
5. Conclusions
The total carotenoid content and transcriptomic profiles differ between the narcissus cultivar ‘Jinzhan Yintai’ and its yellow mutant. The difference in tepal color of the two narcissus accessions was mainly caused by the different balance mode of carotenoid biosynthesis pathway. The combo operations of ‘pushing gas’ in synthesis (up-regulation of carotenoid synthesis genes) and ‘braking’ in degradation (the down-regulation of degradation genes) resulted in the high content of carotenoids in the yellow mutant. The reverse operation consumed the few carotenoids in ‘Jinzhan Yintai’. This study would enrich the molecular database for Chinese narcissus carotenoids biosynthesis and provide the potential framework of flower color genetic modification.
Author Contributions
Conceptualization, M.C.; formal analysis, Y.Z. (Yiming Zhang); investigation, Y.Z. (Yi Zhou); resources, J.C., Y.Z. (Yiqiang Zhang), J.S., S.W. and W.L.; data curation, Y.Z. (Yiming Zhang); writing—original draft preparation, Y.Z. (Yiming Zhang); writing—review and editing, Y.W., T.C. and Q.Z.; visualization, Y.Z. (Yiming Zhang); supervision, M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported financially by the Fundamental Research Funds for the Central Universities (YX2013-05), the National Natural Science Foundation of China (31500574) and Zhangzhou Science and Technology Project (2016).
Data Availability Statement
In this research, all samples were collected in compliance with relevant guidelines and legislation. The raw transcriptomic data is deposited under BioProject id PRJNA855612 and can be accessed from the NCBI Sequence Read Archive (SRA) platform via accession numbers SRR19993659(T01), SRR19993658(T02), SRR19993647(T03), SRR19993646(T04), SRR19993645(T05), SRR19993644(T06), SRR19993643(T07), SRR19993642(T08), SRR19993641(T09), SRR19993640(T10), SRR19993657(T11), SRR19993656(T12), SRR19993655(T13), SRR19993654(T14), SRR19993653(T15), SRR19993652(T16), SRR19993651(T17), SRR19993650(T18), SRR19993649(T19), SRR19993648(T20).
Acknowledgments
We thank Robert McKenzie, PhD for editing the English text of a draft of this manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Ren, Y.; Liu, C.; Li, X. Comparison of karyotype between univalve Chinese narcissus and multiple-valve Chinese narcissus. J. Agriculyural Univ. Hebei 2010, 33, 9–12. [Google Scholar]
- Liu, Y.; Song, X.; Zhang, D.; Zhou, F.; Wang, D.; Wei, Y. Blueberry anthocyanins: Protection against ageing and light-induced damage in retinal pigment epithelial cells. Br. J. Nutr. 2012, 108, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Alix, K.; Damerval, C. The evolution of flower development: Current understanding and future challenges. Ann. Bot. 2011, 107, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Valadon, L.R.; Mummery, R.S. Carotenoids in floral parts of a narcissus, a daffodil and a tulip. Biochem. J. 1968, 106, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Beyer, P. Carotene biosynthesis in daffodil chromoplasts: On the membrane-integral desaturation and cyclization reactions. Curr. Top. Plant Physiol. 1989, 2, 157–170. [Google Scholar]
- Li, X.; Lu, M.; Tang, D.; Shi, Y. Composition of Carotenoids and Flavonoids in Narcissus Cultivars and their Relationship with Flower Color. PLoS ONE 2015, 10, e0142074. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Krinsky, N.I.; Johnson, E.J. Carotenoid actions and their relation to health and disease. Mol. Asp. Med. 2005, 26, 459–516. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant. 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, E.T. Changing Form and Function through Carotenoids and Synthetic Biology. Plant Physiol. 2019, 179, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yuan, H.; Zeng, Y.; Xu, Q. Plastids and Carotenoid Accumulation. Carotenoids Nat. 2016, 79, 273–293. [Google Scholar]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for Carotenoid Biosynthesis, Degradation, and Storage. In Plant and Food Carotenoids; Rodríguez-Concepción, M., Welsch, R., Eds.; Humana: New York, NY, USA, 2020; Volume 2083, pp. 3–23. [Google Scholar]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Molnár, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Wang, J.Y.; Haider, I.; Jamil, M.; Fiorilli, V.; Saito, Y.; Mi, J. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 2019, 10, 810. [Google Scholar] [CrossRef]
- Ablazov, A.; Mi, J.; Jamil, M.; Jia, K.P.; Wang, J.Y.; Feng, Q. The Apocarotenoid Zaxinone Is a Positive Regulator of Strigolactone and Abscisic Acid Biosynthesis in Arabidopsis Roots. Front. Plant Sci. 2020, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, D.; Du, H.; Shi, Y.H. Transcriptome sequencing and biochemical analysis of perianths and coronas reveal flower color formation in narcissus pseudonarcissus. Int. J. Mol. Sci. 2018, 19, 4006. [Google Scholar] [CrossRef] [PubMed]
- Stanley, L.; Yuan, Y.W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, A.S.; Zhou, X.S.; Xu, Q.; Tadmor, Y.; Li, L. Carotenoid Pigment Accumulation in Horticultural Plants. Hortic. Plant J. 2020, 6, 343–360. [Google Scholar] [CrossRef]
- Afonso, T.; Moresco, R.; Uarrota, V.G.; Navarro, B.B.; Nunes, E.D.C.; Maraschin, M.; Rocha, M. UV-Vis and CIELAB Based Chemometric Characterization of Manihot esculenta Carotenoid Contents. J. Integr. Bioinform. 2017, 14, 20170056. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.X.; Wan, H.H.; Han, Y.; Yu, C.; Luo, L.; Pan, H.T.; Zhang, Q.X. Identification and QTL Analysis of Flavonoids and Carotenoids in Tetraploid Roses Based on an Ultra-High-Density Genetic Map. Front. Plant Sci. 2021, 12, 1071. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Morikawa, T.; Uraguchi, Y.; Sanda, S. Overexpression of DnaJ-Like Chaperone Enhances Carotenoid Synthesis in Chlamydomonas reinhardtii. Appl. Biochem. Biotechnol. 2017, 184, 80–91. [Google Scholar] [CrossRef]
- Schledz, M.; Al-Babili, S.; von Lintig, J.; Haubruck, H.; Rabbani, S.; Kleinig, H.; Beyer, P. Phytoene synthase from Narcissus pseudonarcissus: Functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J. Cell Mol. Biol. 1996, 10, 781–792. [Google Scholar] [CrossRef]
- Welsch, R.; Zhou, X.; Yuan, H.; Alvarez, D.; Sun, T.; Schlossarek, D.; Li, L. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in arabidopsis. Mol. Plant 2018, 11, 149–162. [Google Scholar] [CrossRef]
- Xu, M.; Liu, C.L.; Luo, J.; Qi, Z.; Yan, Z.; Fu, Y.; Tang, H. Transcriptomic de novo analysis of pitaya (Hylocereus polyrhizus) canker disease caused by Neoscytalidium dimidiatum. BMC Genom. 2019, 20, 10. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Lutova, L.A.; Dodueva, I.E.; Lebedeva, M.A.; Tvorogova, V.E. Transcription factors in developmental genetics and the evolution of higher plants. Genetika 2015, 51, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Chen, L.J.; Yang, Q.L.; Hu, Z.L.; Guo, P.Y.; Xie, Q.L.; Chen, G.P. New insight into the pigment composition and molecular mechanism of flower coloration in tulip (Tulipa gesneriana L.) cultivars with various petal colors. Plant Sci. 2022, 317, 111193. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xu, M.; Li, R. Transcriptomic investigation of the basis of Corona and petal colour in Chinese narcissus. J. Hortic. Sci. Biotechnol. 2020, 95, 565–577. [Google Scholar] [CrossRef]
- Yan, J.; Wang, M.; Zhang, L. Light induces petal color change in Quisqualis indica (Combretaceae). Plant Divers. 2018, 40, 28–34. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Zou, L.P.; Gao, H.P.; Zhong, Y.Q. Construction of Overexpression Vector for Phytoene Dehydrogenase Gene and Its Expression Identification in Tomato. Hubei Agric. Sci. 2012, 51, 393–395. [Google Scholar]
- Qi, Y.W.; Wu, H.X.; Liu, J.; Chen, L.J.; Jiang, Z.T.; Zhang, Y.J.; Tian, X.T.; Li, R.; Yang, Y.; Ren, X.L. Lycopene β-cyclase plays a critical role in carotenoid biosynthesis during persimmon fruit development and postharvest ripening. Sci. Hortic. 2021, 287, 110265. [Google Scholar] [CrossRef]
- Isaacson, T.; Ronen, G.; Hirschberg, Z.J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 2002, 14, 333–342. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, X.; Song, X. Photoperiodic Responses and Characterization of the Cmvvd Gene Encoding a Blue Light Photoreceptor from the Medicinal Caterpillar Fungus Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms 2017, 19, 163. [Google Scholar] [CrossRef]
- Moreno, J.C.; Pizarro, L.; Fuentes, P. Levels of Lycopene β-Cyclase 1 Modulate Carotenoid Gene Expression and Accumulation in Daucus carota. PLoS ONE 2013, 8, e58144. [Google Scholar]
- Wu, X.; Zhu, L.; Li, O. Less Dynamic Sphingomonas Bacteria crtZ Cell Gene, crtG Gene and Its Application. CN Patent CN102286495 A, 2011. [Google Scholar]
- Petry, F.C.; Nadai, F.D.; Cristofani-Yaly, M. Carotenoid biosynthesis and quality characteristics of new hybrids between tangor (Citrus reticulata × C. sinensis) cv. ‘Murcott’ and sweet orange (C. sinensis) cv. ‘Pêra’. Food Res. Int. 2019, 122, 461–470. [Google Scholar] [CrossRef]
- Arumingtyas, E.L.; Fuadati, A.Z.; Dwinianti, E.F. Study on the Profile of Capsanthin-Capsurobin Synthase (Ccs) Gene responsible for Carotenoid Synthesis in Chili Pepper (Capsicum frutescens L.) Mutants G1M6 M2 Generation. IOP Conf. Ser. Earth Environ. Sci. 2019, 391, 012019. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, H.; Liu, H.; Yang, J.; Hu, Y. Extensive transcriptome changes underlying the flower c intensity variation in Paeonia ostii. Front. Plant Sci. 2015, 6, 1205. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Sun, S.; Chen, X.; Wu, S.; Wang, D.; Chen, X. PyMYB10 and PyMYB10.1 Interact with bHLH to enhance anthocyanin accumulation in Pears. PLoS ONE 2015, 10, e0142112. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Verweij, W.; Kroon, A.; Spelt, C.; Mol, J.; Koes, R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 2006, 18, 1274–1291. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).