The Role of the DNA Methyltransferase Family and the Therapeutic Potential of DNMT Inhibitors in Tumor Treatment
Abstract
1. Introduction
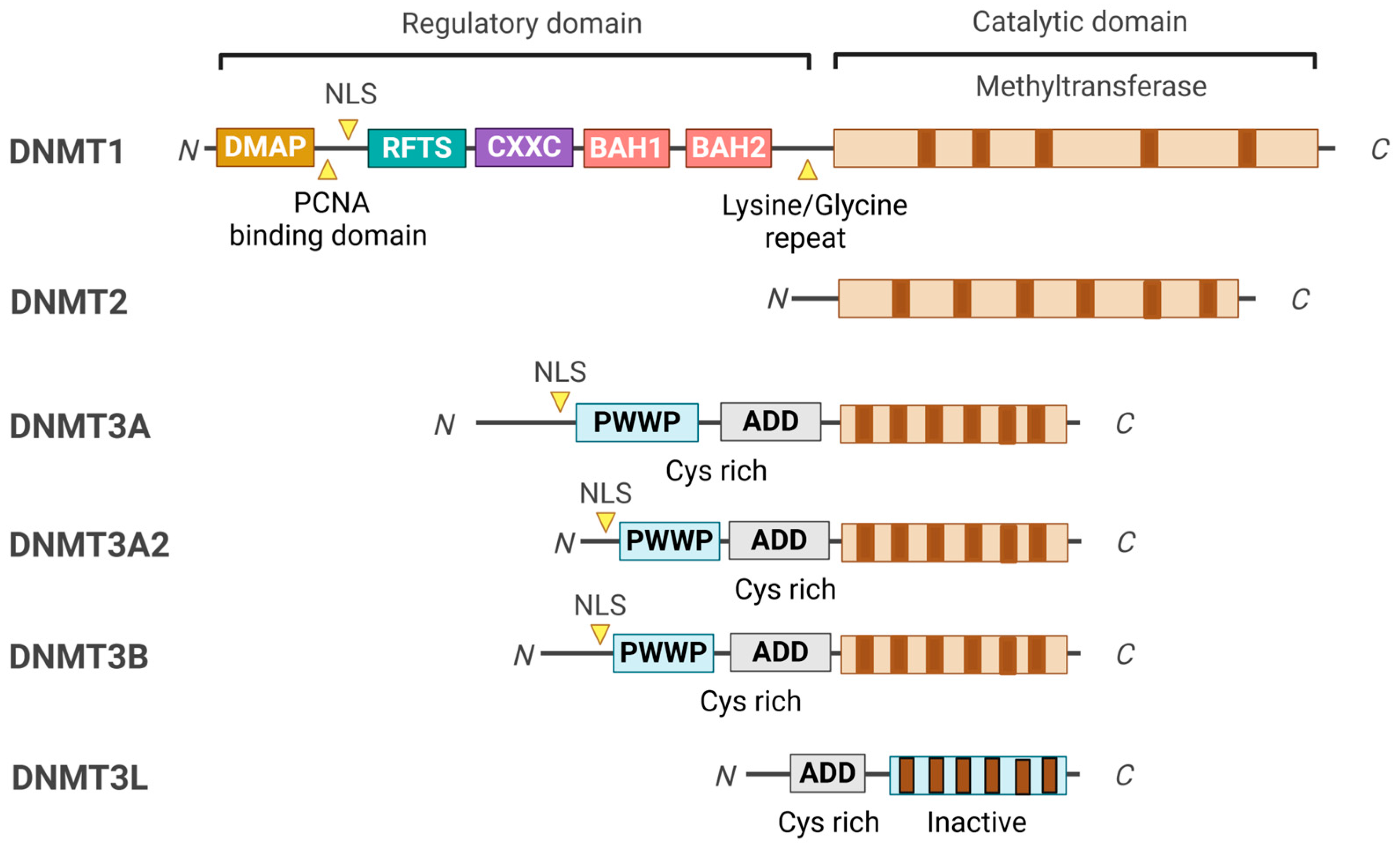
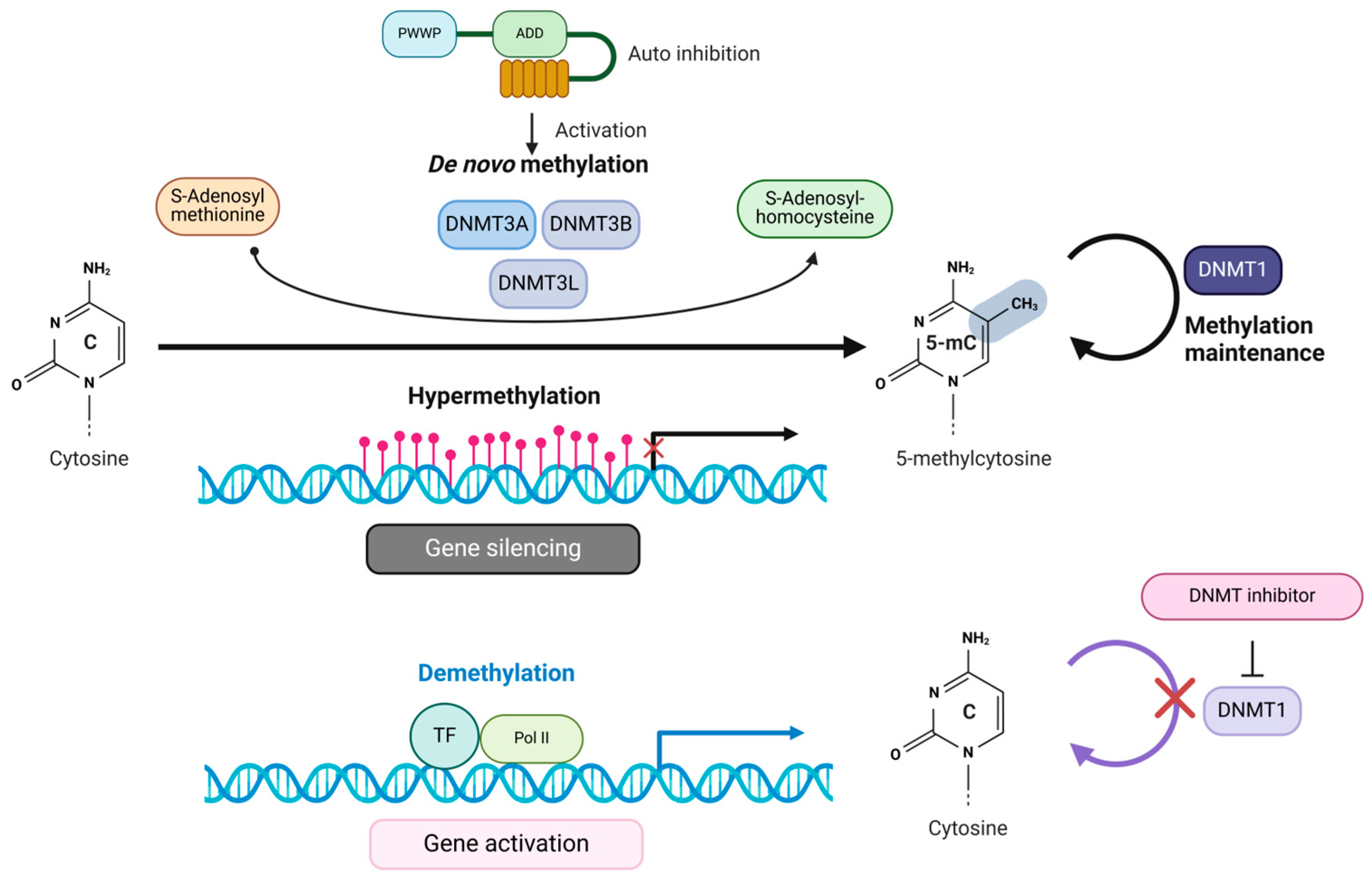
2. DNMT Domain Organizations and Their Functions
3. Recruitment of DNMTs to Gene Promoters
4. DNMT1 Binding Partners
- (1)
- MBDs
- (2)
- CFP1
- (3)
- DMAP1 (DNMT-associated protein1)
- (4)
- UHRF1
- (5)
- Sp1 (Specificity protein 1)
- (6)
- PCNA (Proliferating Cell Nuclear Antigen)
- (7)
- G9a/GLP (Histone Methyltransferases)
- (8)
- RUNX1-MTG8
- (9)
- HESX1 (HESX homeobox 1)
- (10)
- DAXX
- (11)
- CHAF1A
5. DNMT1 Regulators
- (1)
- MEK/ERK
- (2)
- STAT3
- (3)
- MicroRNAs
- (4)
- Circular RNAs
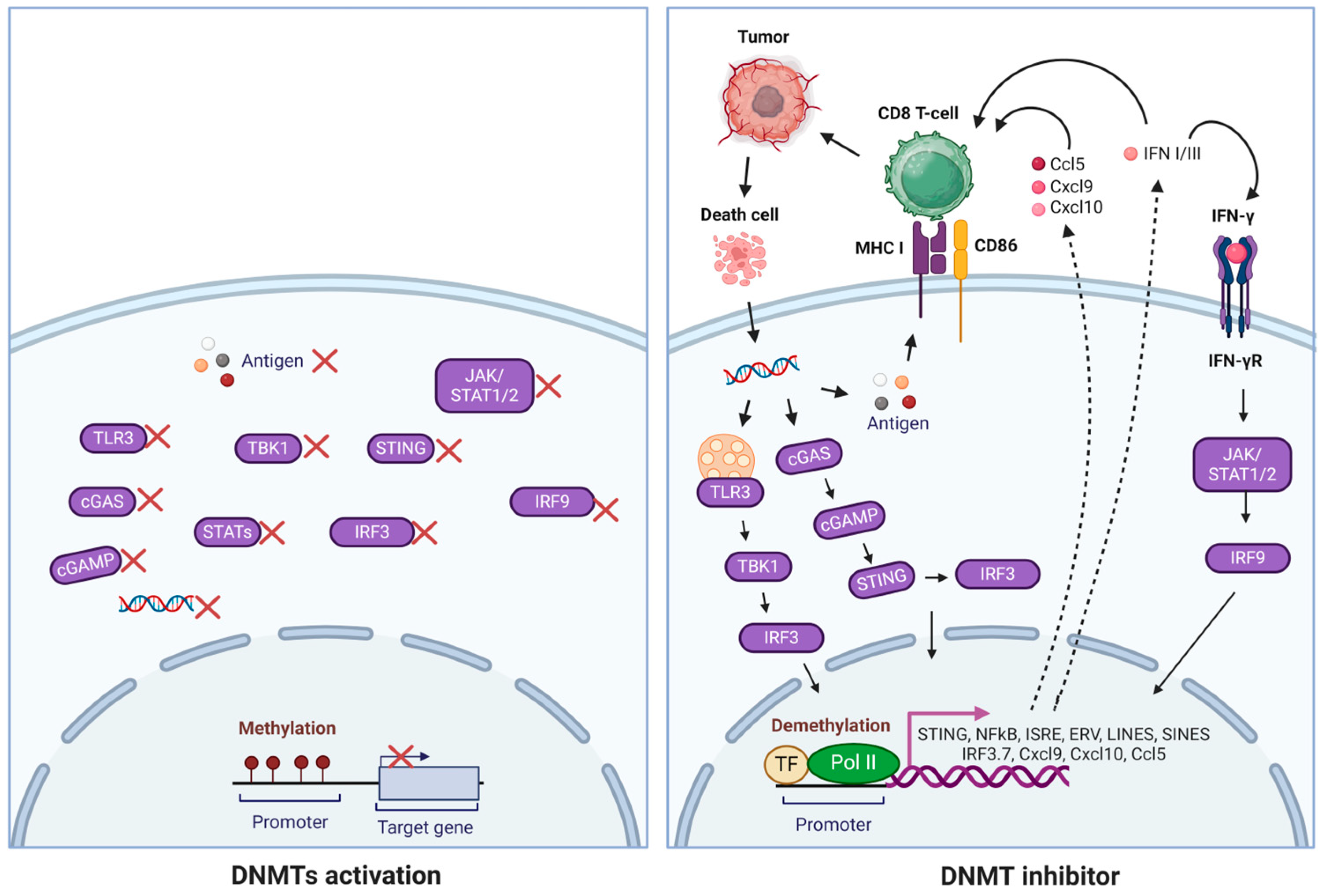
6. DNMT1 Inhibitor
7. DNMT Family and Cancer
8. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Sulewska, A.; Niklinska, W.; Kozlowski, M.; Minarowski, L.; Naumnik, W.; Niklinski, J.; Dabrowska, K.; Chyczewski, L. DNA methylation in states of cell physiology and pathology. Folia Histochem. Cytobiol. 2007, 45, 149–158. [Google Scholar] [PubMed]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Karnik, R.; Gu, H.; Ziller, M.J.; Clement, K.; Tsankov, A.M.; Akopian, V.; Gifford, C.A.; Donaghey, J.; Galonska, C.; et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015, 47, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2017, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Klimasauskas, S.; Kumar, S.; Roberts, R.J.; Cheng, X. Hhal methyltransferase flips its target base out of the DNA helix. Cell 1994, 76, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Zhang, X.; Chen, P.-Y.; Bostick, M.; Goll, M.G.; Hetzel, J.; Jain, J.; Strauss, S.H.; Halpern, M.E.; et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA 2010, 107, 8689–8694. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA Modification Mechanisms and Gene Activity During Development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Riggs, A. X inactivation, differentiation, and DNA methylation. Cytogenet. Genome Res. 1975, 14, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Gröne, H.; et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNA Asp by the DNA Methyltransferase Homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Tsumura, A.; Hayakawa, T.; Kumaki, Y.; Takebayashi, S.; Sakaue, M.; Matsuoka, C.; Shimotohno, K.; Ishikawa, F.; Li, E.; Ueda, H.R.; et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006, 11, 805–814. [Google Scholar] [CrossRef]
- Lei, H.; Oh, S.P.; Okano, M.; Jüttermann, R.; Goss, K.A.; Jaenisch, R.; Li, E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996, 122, 3195–3205. [Google Scholar] [CrossRef] [PubMed]
- Pechalrieu, D.; Etievant, C.; Arimondo, P.B. DNA methyltransferase inhibitors in cancer: From pharmacology to translational studies. Biochem. Pharmacol. 2017, 129, 1–13. [Google Scholar] [CrossRef]
- Gifford, C.A.; Ziller, M.J.; Gu, H.; Trapnell, C.; Donaghey, J.; Tsankov, A.; Shalek, A.K.; Kelley, D.R.; Shishkin, A.A.; Issner, R.; et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell 2013, 153, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Poppe, D.; Buckberry, S.; Pflueger, J.; Albertin, C.B.; Daish, T.; Bertrand, S.; de la Calle-Mustienes, E.; Gómez-Skarmeta, J.L.; Nery, J.R.; et al. The emergence of the brain non-CpG methylation system in vertebrates. Nat. Ecol. Evol. 2021, 5, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global Epigenomic Reconfiguration During Mammalian Brain Development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [PubMed]
- Gabel, H.W.; Kinde, B.; Stroud, H.; Gilbert, C.S.; Harmin, D.A.; Kastan, N.R.; Hemberg, M.; Ebert, D.H.; Greenberg, M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522, 89–93. [Google Scholar] [CrossRef]
- Pappalardo, X.G.; Barra, V. Losing DNA methylation at repetitive elements and breaking bad. Epigenetics Chromatin 2021, 14, 1–21. [Google Scholar] [CrossRef]
- Pacaud, R.; Brocard, E.; Lalier, L.; Hervouet, E.; Vallette, F.M.; Cartron, P.-F. The DNMT1/PCNA/UHRF1 disruption induces tumorigenesis characterized by similar genetic and epigenetic signatures. Sci. Rep. 2014, 4, 4230. [Google Scholar] [CrossRef]
- Gowher, H.; Liebert, K.; Hermann, A.; Xu, G.; Jeltsch, A. Mechanism of Stimulation of Catalytic Activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005, 280, 13341–13348. [Google Scholar] [CrossRef]
- Chédin, F. Chapter 7—The DNMT3 Family of Mammalian De Novo DNA Methyltransferases. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Academic Press: Cambridge, MA, USA, 2011; p. 255. [Google Scholar]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.; Chang, H.; Xu, G.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2008, 41, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Estève, P.-O.; Chin, H.G.; Benner, J.; Feehery, G.R.; Samaranayake, M.; Horwitz, G.A.; Jacobsen, S.E.; Pradhan, S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 2009, 106, 5076–5081. [Google Scholar] [CrossRef]
- Deplus, R.; Blanchon, L.; Rajavelu, A.; Boukaba, A.; Defrance, M.; Luciani, J.; Rothé, F.; Dedeurwaerder, S.; Denis, H.; Brinkman, A.B.; et al. Regulation of DNA Methylation Patterns by CK2-Mediated Phosphorylation of Dnmt3a. Cell Rep. 2014, 8, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Lin, Z.-J.; Li, C.-C.; Lin, X.; Shan, S.-K.; Guo, B.; Zheng, M.-H.; Li, F.; Yuan, L.-Q.; Li, Z.-H. Epigenetic regulation in metabolic diseases: Mechanisms and advances in clinical study. Signal Transduct. Target. Ther. 2023, 8, 98. [Google Scholar] [CrossRef]
- Park, S.Y.; Na Seo, A.; Jung, H.Y.; Gwak, J.M.; Jung, N.; Cho, N.-Y.; Kang, G.H. Alu and LINE-1 Hypomethylation Is Associated with HER2 Enriched Subtype of Breast Cancer. PLoS ONE 2014, 9, e100429. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M. DNA Hypomethylation, Cancer, the Immunodeficiency, Centromeric Region Instability, Facial Anomalies Syndrome and Chromosomal Rearrangements. J. Nutr. 2002, 132, 2424S–2429S. [Google Scholar] [CrossRef]
- Patra, S.K. Ras regulation of DNA-methylation and cancer. Exp. Cell Res. 2008, 314, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Pfundstein, G.; Nikonenko, A.G.; Sytnyk, V. Amyloid precursor protein (APP) and amyloid β (Aβ) interact with cell adhesion molecules: Implications in Alzheimer’s disease and normal physiology. Front. Cell Dev. Biol. 2022, 10, 969547. [Google Scholar] [CrossRef]
- de Mena, L.; Parés, G.; Garrido, A.; Pilco-Janeta, D.F.; Fernández, M.; Pérez, J.; Tolosa, E.; Cámara, A.; Valldeoriola, F.; Ezquerra, M.; et al. α-Synuclein Gene Hypomethylation in LRRK2 Parkinson’s Disease Patients. Mov. Disord. 2024. [Google Scholar] [CrossRef]
- Schmidt, C.S.; Bultmann, S.; Meilinger, D.; Zacher, B.; Tresch, A.; Maier, K.C.; Peter, C.; Martin, D.E.; Leonhardt, H.; Spada, F. Global DNA Hypomethylation Prevents Consolidation of Differentiation Programs and Allows Reversion to the Embryonic Stem Cell State. PLoS ONE 2012, 7, e52629. [Google Scholar] [CrossRef] [PubMed]
- Tobelaim, W.S.; Beaurivage, C.; Champagne, A.; Pomerleau, V.; Simoneau, A.; Chababi, W.; Yeganeh, M.; Thibault, P.; Klinck, R.; Carrier, J.C.; et al. Tumour-promoting role of SOCS1 in colorectal cancer cells. Sci. Rep. 2015, 5, 14301. [Google Scholar] [CrossRef]
- Zhao, R.; Choi, B.Y.; Lee, M.-H.; Bode, A.M.; Dong, Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16 INK4a) in Cancer. EBioMedicine 2016, 8, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wu, Q.; Cheng, J.; Qiu, X.; Zhang, J.; Fan, H. Depletion of DNMT3A Suppressed Cell Proliferation and Restored PTEN in Hepatocellular Carcinoma Cell. J. Biomed. Biotechnol. 2010, 2010, 737535. [Google Scholar] [CrossRef]
- Kientz, C.; Prieur, F.; Clemenson, A.; Joly, M.-O.; Stachowicz, M.-L.; Auclair, J.; Attignon, V.; Schiappa, R.; Wang, Q. MLH1 promoter hypermethylation: Are you absolutely sure about the absence of MLH1 germline mutation? About a new case. Fam. Cancer 2019, 19, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef]
- Bodis, G.; Toth, V.; Schwarting, A. Role of Human Leukocyte Antigens (HLA) in Autoimmune Diseases. Rheumatol. Ther. 2018, 5, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Guo, M. Synthetic lethality strategies: Beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci. 2020, 111, 3111–3121. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Pruitt, S.C.; Hershberger, P.A.; Witkiewicz, A.K.; Goodrich, D.W. Cell Cycle and Beyond: Exploiting New RB1 Controlled Mechanisms for Cancer Therapy. Trends Cancer 2019, 5, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Inoshita, S.; Terada, Y.; Nakashima, O.; Kuwahara, M.; Sasaki, S.; Marumo, F. Regulation of the G1/S transition phase in mesangial cells by E2F1. Kidney Int. 1999, 56, 1238–1241. [Google Scholar] [CrossRef]
- Pruitt, K.; Zinn, R.L.; Ohm, J.E.; McGarvey, K.M.; Kang, S.-H.L.; Watkins, D.N.; Herman, J.G.; Baylin, S.B. Inhibition of SIRT1 Reactivates Silenced Cancer Genes without Loss of Promoter DNA Hypermethylation. PLoS Genet. 2006, 2, e40. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-Q.; Tucker, K.L.; Parnell, L.D.; Adiconis, X.; García-Bailo, B.; Griffith, J.; Meydani, M.; Ordovás, J.M. PPARGC1A Variation Associated With DNA Damage, Diabetes, and Cardiovascular Diseases. Diabetes 2008, 57, 809–816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, J.; Yu, D.; Luo, C.; Bennett, C.; Jedrychowski, M.; Gygi, S.P.; Widlund, H.R.; Puigserver, P. Epigenetic suppression of PGC1α (PPARGC1A) causes collateral sensitivity to HMGCR-inhibitors within BRAF-treatment resistant melanomas. Nat. Commun. 2023, 14, 3251. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Lu, C.; Payne, D.; Paschall, A.V.; Klement, J.D.; Redd, P.S.; Ibrahim, M.L.; Yang, D.; Han, Q.; Liu, Z.; et al. Autocrine IL6-Mediated Activation of the STAT3–DNMT Axis Silences the TNFα–RIP1 Necroptosis Pathway to Sustain Survival and Accumulation of Myeloid-Derived Suppressor Cells. Cancer Res. 2020, 80, 3145–3156. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deuring, J.; Peppelenbosch, M.P.; Kuipers, E.J.; de Haar, C.; van der Woude, C.J. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinog. 2012, 33, 1889–1896. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Y.; Zhang, X.; Li, C.; Zhang, X.; Yang, D.; Liu, Y.; Li, L. DNA methylation of HOX genes and its clinical implications in cancer. Exp. Mol. Pathol. 2023, 134, 104871. [Google Scholar] [CrossRef] [PubMed]
- De Kumar, B.; Darland, D.C. The Hox protein conundrum: The “specifics” of DNA binding for Hox proteins and their partners. Dev. Biol. 2021, 477, 284. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.J.; Shklovskaya, E.; Hersey, P. Epigenetic modulation in cancer immunotherapy. Curr. Opin. Pharmacol. 2017, 35, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, G.; Li, Y.; Lei, D.; Xiang, J.; Ouyang, L.; Wang, Y.; Yang, J. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 2022, 13, 1072651. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Anandh, S.; Null, J.L.; Przanowski, P.; Bhatnagar, S.; Kumar, P.; Shelton, S.E.; Grundy, E.E.; Chiappinelli, K.B.; Kamm, R.D.; et al. Priming a vascular-selective cytokine response permits CD8+ T-cell entry into tumors. Nat. Commun. 2023, 14, 2122. [Google Scholar] [CrossRef] [PubMed]
- Svedružić, Ž.M. Chapter 6—Dnmt1: Structure and Function. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Academic Press: Cambridge, MA, USA, 2011; p. 221. [Google Scholar]
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef]
- Leonhardt, H.; Page, A.W.; Weier, H.-U.; Bestor, T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 1992, 71, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Wijesinghe, S.; Wilson, C.; Halsall, J.; Liloglou, T.; Kanhere, A. A long intergenic non-coding RNA regulates nuclear localization of DNA methyl transferase-1. iScience 2021, 24, 102273. [Google Scholar] [CrossRef] [PubMed]
- Syeda, F.; Fagan, R.L.; Wean, M.; Avvakumov, G.V.; Walker, J.R.; Xue, S.; Dhe-Paganon, S.; Brenner, C. The Replication Focus Targeting Sequence (RFTS) Domain Is a DNA-competitive Inhibitor of Dnmt1. J. Biol. Chem. 2011, 286, 15344–15351. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Rechkoblit, O.; Bestor, T.H.; Patel, D.J. Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation. Science 2011, 331, 1036–1040. [Google Scholar] [CrossRef]
- Lai, J.; Chen, L.; Li, Q.; Zhao, G.; Li, X.; Guo, D.; Chen, Z.; Zhang, Y.; Fan, J.; Zhao, H.; et al. tRNA methyltransferase DNMT2 promotes hepatocellular carcinoma progression and enhances Bortezomib resistance through inhibiting TNFSF10. Cell. Signal. 2024, 127, 111533. [Google Scholar] [CrossRef]
- Arand, J.; Spieler, D.; Karius, T.; Branco, M.R.; Meilinger, D.; Meissner, A.; Jenuwein, T.; Xu, G.; Leonhardt, H.; Wolf, V.; et al. In Vivo Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases. PLoS Genet. 2012, 8, e1002750. [Google Scholar] [CrossRef]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3AMutations in Acute Myeloid Leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef]
- Childhood Overgrowth Consortium; Tatton-Brown, K.; Seal, S.; Ruark, E.; Harmer, J.; Ramsay, E.; Duarte, S.d.V.; Zachariou, A.; Hanks, S.; O’Brien, E.; et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014, 46, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Chen, C.-W.; Sundaramurthy, V.; Słabicki, M.; Hao, D.; Watson, C.J.; Tovy, A.; Reyes, J.M.; Dakhova, O.; Crovetti, B.R.; et al. Systematic Profiling of DNMT3A Variants Reveals Protein Instability Mediated by the DCAF8 E3 Ubiquitin Ligase Adaptor. Cancer Discov. 2021, 12, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Moarefi, A.H.; Chédin, F. ICF Syndrome Mutations Cause a Broad Spectrum of Biochemical Defects in DNMT3B-Mediated De Novo DNA Methylation. J. Mol. Biol. 2011, 409, 758–772. [Google Scholar] [CrossRef]
- Kubo, N.; Uehara, R.; Uemura, S.; Ohishi, H.; Shirane, K.; Sasaki, H. Combined and differential roles of ADD domains of DNMT3A and DNMT3L on DNA methylation landscapes in mouse germ cells. Nat. Commun. 2024, 15, 3266. [Google Scholar] [CrossRef]
- Xu, G.-L.; Bestor, T.H.; Bourc’His, D.; Hsieh, C.-L.; Tommerup, N.; Bugge, M.; Hulten, M.; Qu, X.; Russo, J.J.; Viegas-Péquignot, E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999, 402, 187–191. [Google Scholar] [CrossRef]
- Thijssen, P.E.; Ito, Y.; Grillo, G.; Wang, J.; Velasco, G.; Nitta, H.; Unoki, M.; Yoshihara, M.; Suyama, M.; Sun, Y.; et al. Mutations in CDCA7 and HELLS cause immunodeficiency–centromeric instability–facial anomalies syndrome. Nat. Commun. 2015, 6, 7870. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Hérault, Y.; Guillou, F.; Bourc’his, D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Duymich, C.E.; Charlet, J.; Yang, X.; Jones, P.A.; Liang, G. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 2016, 7, 11453. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, H.M.; Mohammad, H.P.; Baylin, S.B. Double Strand Breaks Can Initiate Gene Silencing and SIRT1-Dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 2008, 4, e1000155. [Google Scholar] [CrossRef]
- O’Hagan, H.M.; Wang, W.; Sen, S.; Shields, C.D.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Van Neste, L.; Easwaran, H.; et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands. Cancer Cell 2011, 20, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.Y.; Park, J.; Jang, E.-S.; Chi, S.W. 8-Oxoguanine: From oxidative damage to epigenetic and epitranscriptional modification. Exp. Mol. Med. 2022, 54, 1626–1642. [Google Scholar] [CrossRef]
- Burri, N.; Shaw, P.; Bouzourene, H.; Sordat, I.; Sordat, B.; Gillet, M.; Schorderet, D.; Bosman, F.T.; Chaubert, P. Methylation Silencing and Mutations of the p14ARF and p16INK4a Genes in Colon Cancer. Mod. Pathol. 2001, 81, 217–229. [Google Scholar] [CrossRef]
- Gao, P.; Yang, X.; Xue, Y.; Zhang, X.; Wang, Y.; Liu, W.; Wu, X. Promoter methylation of glutathione S-transferase π1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma and its correlation with DNA methyltransferase 1 expression. Cancer 2009, 115, 3222–3232. [Google Scholar] [CrossRef]
- Catteau, A.; Morris, J.R. BRCA1 methylation: A significant role in tumour development? Semin. Cancer Biol. 2002, 12, 359. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qin, B.; Moyer, A.M.; Nowsheen, S.; Liu, T.; Qin, S.; Zhuang, Y.; Liu, D.; Lu, S.W.; Kalari, K.R.; et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J. Clin. Investig. 2018, 128, 2376–2388. [Google Scholar] [CrossRef]
- Robertson, K.D.; Ait-Si-Ali, S.; Yokochi, T.; Wade, P.A.; Jones, P.L.; Wolffe, A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000, 25, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Liang, G.; Lenz, H.-J. DNA methylation aberrancies delineate clinically distinct subsets of colorectal cancer and provide novel targets for epigenetic therapies. Oncogene 2017, 37, 566–577. [Google Scholar] [CrossRef]
- Lian, Y.; Meng, L.; Ding, P.; Sang, M. Epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification, and non-coding RNAs. Clin. Epigenetics 2018, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Al Emran, A.; Chatterjee, A.; Rodger, E.J.; Tiffen, J.C.; Gallagher, S.J.; Eccles, M.R.; Hersey, P. Targeting DNA Methylation and EZH2 Activity to Overcome Melanoma Resistance to Immunotherapy. Trends Immunol. 2019, 40, 328–344. [Google Scholar] [CrossRef]
- Exposito, F.; Redrado, M.; Serrano, D.; Calabuig-Fariñas, S.; Bao-Caamano, A.; Gallach, S.; Jantus-Lewintre, E.; Diaz-Lagares, A.; Rodriguez-Casanova, A.; Sandoval, J.; et al. G9a/DNMT1 co-targeting inhibits non-small cell lung cancer growth and reprograms tumor cells to respond to cancer-drugs through SCARA5 and AOX1. Cell Death Dis. 2024, 15, 787. [Google Scholar] [CrossRef]
- Tatematsu, K.; Yamazaki, T.; Ishikawa, F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 2000, 5, 677–688. [Google Scholar] [CrossRef]
- Yu, F. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000, 28, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Shiota, K. Methyl-CpG-binding Protein, MeCP2, Is a Target Molecule for Maintenance DNA Methyltransferase, Dnmt1. J. Biol. Chem. 2003, 278, 4806–4812. [Google Scholar] [CrossRef]
- Butler, J.S.; Lee, J.-H.; Skalnik, D.G. CFP1 Interacts with DNMT1 Independently of Association with the Setd1 Histone H3K4 Methyltransferase Complexes. DNA Cell Biol. 2008, 27, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.E.; Kim, J.H.; Taylor, M.; Muller, M.T. DNA Methyltransferase 1-associated Protein (DMAP1) Is a Co-repressor That Stimulates DNA Methylation Globally and Locally at Sites of Double Strand Break Repair. J. Biol. Chem. 2010, 285, 37630–37640. [Google Scholar] [CrossRef]
- Liu, Z.; Fisher, R.A. RGS6 Interacts with DMAP1 and DNMT1 and Inhibits DMAP1 Transcriptional Repressor Activity. J. Biol. Chem. 2004, 279, 14120–14128. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Horton, J.R.; Zhang, X.; Bostick, M.; Jacobsen, S.E.; Cheng, X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008, 455, 826–829. [Google Scholar] [CrossRef]
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Kishikawa, S.; Murata, T.; Kimura, H.; Shiota, K.; Yokoyama, K.K. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur. J. Biochem. 2002, 269, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Horio, Y.; Sekido, Y.; Minna, J.D.; Shimokata, K.; Hasegawa, Y. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14ARF, p16INK4a and RASSF1A in human lung cancer cell lines. Oncogene 2002, 21, 4822–4829. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.-H.; Ian, H.-I.; Koh, T.-W.; Ng, H.-H.; Xu, G.; Li, B.F.L. Human DNA-(Cytosine-5) Methyltransferase-PCNA Complex as a Target for p21WAF1. Science 1997, 277, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Haemmer, A.; Spada, F.; Rösing, N.; Meilinger, D.; Rothbauer, U.; Cardoso, M.C.; Leonhardt, H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007, 35, 4301–4312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Q.; Li, P.; Liu, X.; Jia, Y.; Wu, W.; Li, J.; Dong, S.; Koseki, H.; Wong, J. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011, 21, 1723–1739. [Google Scholar] [CrossRef]
- Hervouet, E.; Peixoto, P.; Delage-Mourroux, R.; Boyer-Guittaut, M.; Cartron, P.-F. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenetics 2018, 10, 17. [Google Scholar] [CrossRef]
- Casciello, F.; Windloch, K.; Gannon, F.; Lee, J.S. Functional Role of G9a Histone Methyltransferase in Cancer. Front. Immunol. 2015, 6, 487. [Google Scholar] [CrossRef] [PubMed]
- Sajedi, E.; Gaston-Massuet, C.; Andoniadou, C.L.; Signore, M.; Hurd, P.J.; Dattani, M.; Martinez-Barbera, J.P. DNMT1 interacts with the developmental transcriptional repressor HESX1. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008, 1783, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, J.; Li, J.; Tian, D.; Gu, L.; Zhou, M. Methylation of RASSF1A gene promoter is regulated by p53 and DAXX. FASEB J. 2012, 27, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, H.; Liao, A.; Yang, W. Aberrant SPOP-CHAF1A ubiquitination axis triggers tumor autophagy that endows a therapeutical vulnerability in diffuse large B cell lymphoma. J. Transl. Med. 2022, 20, 296. [Google Scholar] [CrossRef]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 1619–1633. [Google Scholar] [CrossRef]
- Lu, R.; Wang, X.; Chen, Z.-F.; Sun, D.-F.; Tian, X.-Q.; Fang, J.-Y. Inhibition of the Extracellular Signal-regulated Kinase/Mitogen-activated Protein Kinase Pathway Decreases DNA Methylation in Colon Cancer Cells. J. Biol. Chem. 2007, 282, 12249–12259. [Google Scholar] [CrossRef]
- Gassenmaier, M.; Rentschler, M.; Fehrenbacher, B.; Eigentler, T.K.; Ikenberg, K.; Kosnopfel, C.; Sinnberg, T.; Niessner, H.; Bösmüller, H.; Wagner, N.B.; et al. Expression of DNA Methyltransferase 1 Is a Hallmark of Melanoma, Correlating with Proliferation and Response to B-Raf and Mitogen-Activated Protein Kinase Inhibition in Melanocytic Tumors. Am. J. Pathol. 2020, 190, 2155–2164. [Google Scholar] [CrossRef]
- Balada, E.; Ordi-Ros, J.; Vilardell-Tarrés, M. DNA Methylation and Systemic Lupus Erythematosus. Ann. N. Y. Acad. Sci. 2007, 1108, 127–136. [Google Scholar] [CrossRef]
- Sunahori, K.; Nagpal, K.; Hedrich, C.M.; Mizui, M.; Fitzgerald, L.M.; Tsokos, G.C. The Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Promotes DNA Hypomethylation by Suppressing the Phosphorylated Mitogen-activated Protein Kinase/Extracellular Signal-regulated Kinase (ERK) Kinase (MEK)/Phosphorylated ERK/DNMT1 Protein Pathway in T-cells from Controls and Systemic Lupus Erythematosus Patients. J. Biol. Chem. 2013, 288, 21936–21944. [Google Scholar] [CrossRef]
- Thomas, N.S.B. The STAT3-DNMT1 connection. JAK-STAT 2012, 1, 257–260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and MicroRNA-148a Contribute to DNA Hypomethylation in Lupus CD4+ T Cells by Directly and Indirectly Targeting DNA Methyltransferase 1. J. Immunol. 2010, 184, 6773–6781. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Wong, K.K.; Lawrie, C.H.; Green, T.M. Oncogenic Roles and Inhibitors of DNMT1, DNMT3A, and DNMT3B in Acute Myeloid Leukaemia. Biomark. Insights 2019, 14, 1177271919846454. [Google Scholar] [CrossRef]
- Mims, A.; Walker, A.R.; Huang, X.; Sun, J.; Wang, H.; Santhanam, R.; Dorrance, A.M.; Walker, C.; Hoellerbauer, P.; Tarighat, S.S.; et al. Increased anti-leukemic activity of decitabine via AR-42-induced upregulation of miR-29b: A novel epigenetic-targeting approach in acute myeloid leukemia. Leukemia 2012, 27, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Chen, W.; Wang, J. MicroRNA-20a Regulates Glioma Cell Proliferation, Invasion, and Apoptosis by Targeting CUGBP Elav-Like Family Member 2. World Neurosurg. 2019, 121, e519. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wu, X.; Xiang, S.; Qiao, M.; Cen, X.; Pan, X.; Huang, X.; Zhao, Z. Regulatory mechanism of miR-20a-5p expression in Cancer. Cell Death Discov. 2022, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Zhou, B.; Li, H.; He, L.; Wang, C.; Wang, Z.; Zhu, L.; Chen, M.; Gao, S. A novel miR-375-HOXB3-CDCA3/DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia. BMC Cancer 2018, 18, 182. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2023, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Spada, F.; Easwaran, H.P.; Zolghadr, K.; Margot, J.B.; Cardoso, M.C.; Leonhardt, H. Trapped in action: Direct visualization of DNA methyltransferase activity in living cells. Nat. Methods 2005, 2, 751–756. [Google Scholar] [CrossRef]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA Methyltransferases: A Novel Target for Prevention and Therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Laranjeira, A.B.A.; Hollingshead, M.G.; Nguyen, D.; Kinders, R.J.; Doroshow, J.H.; Yang, S.X. DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci. Rep. 2023, 13, 5964. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Robertson, K.D. DNA Methyltransferases, DNA Damage Repair, and Cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef]
- Wang, K.; He, Z.; Jin, G.; Jin, S.; Du, Y.; Yuan, S.; Zhang, J. Targeting DNA methyltransferases for cancer therapy. Bioorg. Chem. 2024, 151, 107652. [Google Scholar] [CrossRef]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Zhang, N.; van der Touw, W.; Ding, Y.; Ju, W.; Bottinger, E.P.; Reid, S.P.; Levy, D.E.; Bromberg, J.S. Epigenetic Regulation of Foxp3 Expression in Regulatory T Cells by DNA Methylation. J. Immunol. 2009, 182, 259–273. [Google Scholar] [CrossRef]
- Zhou, J.; Yao, Y.; Shen, Q.; Li, G.; Hu, L.; Zhang, X. Demethylating agent decitabine disrupts tumor-induced immune tolerance by depleting myeloid-derived suppressor cells. J. Cancer Res. Clin. Oncol. 2017, 143, 1371–1380. [Google Scholar] [CrossRef]
- Küçük, C.; Hu, X.; Gong, Q.; Jiang, B.; Cornish, A.; Gaulard, P.; McKeithan, T.; Chan, W.C. Diagnostic and Biological Significance of KIR Expression Profile Determined by RNA-Seq in Natural Killer/T-Cell Lymphoma. Am. J. Pathol. 2016, 186, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, P.; Lasagni, L.; Annunziato, F.; Serio, M.; Romagnani, S. CXC chemokines: The regulatory link between inflam-mation and angiogenesis. Trends Immunol. 2004, 25, 201. [Google Scholar] [CrossRef]
- Ahuja, N.; Easwaran, H.; Baylin, S.B. Harnessing the potential of epigenetic therapy to target solid tumors. J. Clin. Investig. 2014, 124, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Derissen, E.J.; Beijnen, J.H.; Schellens, J.H. Concise Drug Review: Azacitidine and Decitabine. Oncol. 2013, 18, 619–624. [Google Scholar] [CrossRef]
- Rius, M.; Stresemann, C.; Keller, D.; Brom, M.; Schirrmacher, E.; Keppler, D.; Lyko, F. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol. Cancer Ther. 2009, 8, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Momparler, R.L. Pharmacology of 5-Aza-2′-deoxycytidine (decitabine). Semin. Hematol. 2005, 42, S9–S16. [Google Scholar] [CrossRef] [PubMed]
- Kalac, M.; Scotto, L.; Marchi, E.; Amengual, J.; Seshan, V.E.; Bhagat, G.; Ulahannan, N.; Leshchenko, V.V.; Temkin, A.M.; Parekh, S.; et al. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood 2011, 118, 5506–5516. [Google Scholar] [CrossRef]
- Wang, L.-X.; Mei, Z.-Y.; Zhou, J.-H.; Yao, Y.-S.; Li, Y.-H.; Xu, Y.-H.; Li, J.-X.; Gao, X.-N.; Zhou, M.-H.; Jiang, M.-M.; et al. Low Dose Decitabine Treatment Induces CD80 Expression in Cancer Cells and Stimulates Tumor Specific Cytotoxic T Lymphocyte Responses. PLoS ONE 2013, 8, e62924. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.A.; Cahill, K.; Saygin, C.; Odenike, O. Cedazuridine/decitabine: From preclinical to clinical development in myeloid malignancies. Blood Adv. 2021, 5, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Bouligny, I.M.; Mehta, V.; Isom, S.; Ellis, L.R.; Bhave, R.R.; Howard, D.S.; Lyerly, S.; Manuel, M.; Dralle, S.; Powell, B.L.; et al. Efficacy of 10-day decitabine in acute myeloid leukemia. Leuk. Res. 2021, 103, 106524. [Google Scholar] [CrossRef]
- Luker, A.J.; Graham, L.J.; Smith, T.M.; Camarena, C.; Zellner, M.P.; Gilmer, J.-J.S.; Damle, S.R.; Conrad, D.H.; Bear, H.D.; Martin, R.K. The DNA methyltransferase inhibitor, guadecitabine, targets tumor-induced myelopoiesis and recovers T cell activity to slow tumor growth in combination with adoptive immunotherapy in a mouse model of breast cancer. BMC Immunol. 2020, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: Mechanism and clinical application. Clin. Epigenetics 2021, 13, 166. [Google Scholar] [CrossRef]
- Uddin, M.G.; Fandy, T.E. Chapter Five—DNA methylation inhibitors: Retrospective and perspective view. In Advances in Cancer Research; Tew, K.D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2021; p. 205. [Google Scholar]
- Fenaux, P.; Gobbi, M.; Kropf, P.L.; Issa, J.-P.J.; Roboz, G.J.; Mayer, J.; Krauter, J.; Robak, T.; Kantarjian, H.M.; Novak, J.; et al. Guadecitabine vs treatment choice in newly diagnosed acute myeloid leukemia: A global phase 3 randomized study. Blood Adv. 2023, 7, 5027–5037. [Google Scholar] [CrossRef]
- Avendaño, C.; Menéndez, J.C. (Eds.) Chapter 8—Epigenetic therapy of cancer. In Medicinal Chemistry of Anticancer Drugs, 3rd ed.; Elsevier: Boston, MA, USA, 2023; p. 389. [Google Scholar]
- Dai, W.; Qiao, X.; Fang, Y.; Guo, R.; Bai, P.; Liu, S.; Li, T.; Jiang, Y.; Wei, S.; Na, Z.; et al. Epigenetics-targeted drugs: Current paradigms and future challenges. Signal Transduct. Target. Ther. 2024, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Noviello, T.M.R.; Di Giacomo, A.M.; Caruso, F.P.; Covre, A.; Mortarini, R.; Scala, G.; Costa, M.C.; Coral, S.; Fridman, W.H.; Sautès-Fridman, C.; et al. Guadecitabine plus ipilimumab in unresectable melanoma: Five-year follow-up and integrated multi-omic analysis in the phase 1b NIBIT-M4 trial. Nat. Commun. 2023, 14, 5914. [Google Scholar] [CrossRef]
- Thottassery, J.V.; Sambandam, V.; Allan, P.W.; Maddry, J.A.; Maxuitenko, Y.Y.; Tiwari, K.; Hollingshead, M.; Parker, W.B. Novel DNA methyltransferase-1 (DNMT1) depleting anticancer nucleosides, 4′-thio-2′-deoxycytidine and 5-aza-4′-thio-2′-deoxycytidine. Cancer Chemother. Pharmacol. 2014, 74, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.; Thottassery, J. 5-Aza-4′-thio-2′-deoxycytidine, a new orally bioavailable non-toxic “best-in-class” DNMT1 depleting agent in clinical development. J. Pharmacol. Exp. Ther. 2021, 379, JPET-MR. [Google Scholar] [CrossRef]
- Ehrlich, M. Dna Hypomethylation In Cancer Cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Spencer, D.H.; Russler-Germain, D.A.; Ketkar, S.; Helton, N.M.; Lamprecht, T.L.; Fulton, R.S.; Fronick, C.C.; O’laughlin, M.; Heath, S.E.; Shinawi, M.; et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell 2017, 168, 801–816.e13. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Schulze, I.; Rohde, C.; Scheller-Wendorff, M.; Bäumer, N.; Krause, A.; Herbst, F.; Riemke, P.; Hebestreit, K.; Tschanter, P.; Lin, Q.; et al. Increased DNA methylation of Dnmt3b targets impairs leukemogenesis. Blood 2016, 127, 1575–1586. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nakamura, M.; Nishikawa, J.; Saito, M.; Sakai, K.; Sasaki, S.; Hashimoto, S.; Okamoto, T.; Suehiro, Y.; Yamasaki, T.; Sakaida, I. Decitabine inhibits tumor cell proliferation and up-regulates e-cadherin expression in Epstein–Barr virus-associated gastric cancer. J. Med. Virol. 2016, 89, 508–517. [Google Scholar] [CrossRef]
- Kamal, K.M.; Ghazali, A.R.; Ab Mutalib, N.S.; Abu, N.; Chua, E.W.; Masre, S.F. The role of DNA methylation and DNA methyltransferases (DNMTs) as potential biomarker and therapeutic target in non-small cell lung cancer (NSCLC). Heliyon 2024, 10, e38663. [Google Scholar] [CrossRef] [PubMed]
- Tekpli, X.; Landvik, N.E.; Anmarkud, K.H.; Skaug, V.; Haugen, A.; Zienolddiny, S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol. Immunother. 2012, 62, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Han, J.; Shim, Y.M.; Park, J.; Kim, D. Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung carcinoma. Cancer 2005, 104, 1825–1833. [Google Scholar] [CrossRef]
- Gordian, E.; Ramachandran, K.; Singal, R. Methylation Mediated Silencing of TMS1 in Breast Cancer and its Potential Contribution to Docetaxel Cytotoxicity. Anticancer. Res. 2009, 29, 3207–3210. [Google Scholar]
- Vasanthakumar, A.; Lepore, J.B.; Zegarek, M.H.; Kocherginsky, M.; Singh, M.; Davis, E.M.; Link, P.A.; Anastasi, J.; Le Beau, M.M.; Karpf, A.R.; et al. Dnmt3b is a haploinsufficient tumor suppressor gene in Myc-induced lymphomagenesis. Blood 2013, 121, 2059–2063. [Google Scholar] [CrossRef]
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Wang, P.; Parton, T.; Zhou, Y.; Chrysovergis, K.; Rockowitz, S.; Chen, W.-Y.; Abdel-Wahab, O.; Wade, P.A.; Zheng, D.; et al. Epigenetic Perturbations by Arg882-Mutated DNMT3A Potentiate Aberrant Stem Cell Gene-Expression Program and Acute Leukemia Development. Cancer Cell 2016, 30, 92–107. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.-Y.; Dai, Y.-J.; Zhang, W.; Zhang, W.-N.; Xiong, S.-M.; Gu, Z.-H.; Wang, K.-K.; Zeng, R.; Chen, Z.; et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc. Natl. Acad. Sci. USA 2014, 111, 2620–2625. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Ushijima, S.; Nakanishi, Y.; Sakamoto, M.; Hirohashi, S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003, 192, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zheng, L.; Zhang, Y.; Xue, M. Bioinformatics analysis of DNMT1 expression and its role in head and neck squamous cell carcinoma prognosis. Sci. Rep. 2021, 11, 2267. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, S.-I.; Chijiwa, T.; Okamura, T.; Akashi, K.; Fukumaki, Y.; Niho, Y.; Sasaki, H. Expression of DNA methyltransferases DNMT1,3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 2001, 97, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.L.; Hlady, R.A.; Opavska, J.; Klinkebiel, D.; Novakova, S.; Smith, L.M.; Lewis, R.E.; Karpf, A.R.; Simpson, M.A.; Wu, L.; et al. Essential Role for Dnmt1 in the Prevention and Maintenance of MYC-Induced T-Cell Lymphomas. Mol. Cell. Biol. 2013, 33, 4321–4333. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Ramachandran, S.; Elangovan, S.; Padia, R.; Yang, P.; Cinghu, S.; Veeranan-Karmegam, R.; Arjunan, P.; Gnana-Prakasam, J.P.; Sadanand, F.; et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat. Commun. 2015, 6, 6910. [Google Scholar] [CrossRef]
- Agoston, A.T.; Argani, P.; Yegnasubramanian, S.; De Marzo, A.M.; Ansari-Lari, M.A.; Hicks, J.L.; Davidson, N.E.; Nelson, W.G. Increased Protein Stability Causes DNA Methyltransferase 1 Dysregulation in Breast Cancer. J. Biol. Chem. 2005, 280, 18302–18310. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-J.; Xu, J.; Gu, Z.-H.; Pan, C.-M.; Lu, G.; Shen, Y.; Shi, J.-Y.; Zhu, Y.-M.; Tang, L.; Zhang, X.-W.; et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011, 43, 309–315. [Google Scholar] [CrossRef]
- Yamashita, Y.; Yuan, J.; Suetake, I.; Suzuki, H.; Ishikawa, Y.; Choi, Y.L.; Ueno, T.; Soda, M.; Hamada, T.; Haruta, H.; et al. Array-based genomic resequencing of human leukemia. Oncogene 2010, 29, 3723–3731. [Google Scholar] [CrossRef]
- Walter, M.J.; Ding, L.; Shen, D.; Shao, J.; Grillot, M.; McLellan, M.; Fulton, R.; Schmidt, H.; Kalicki-Veizer, J.; O’Laughlin, M.; et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 2011, 25, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-E.; Hou, H.-A.; Tsai, C.-H.; Wu, S.-J.; Kuo, Y.-Y.; Tseng, M.-H.; Liu, M.-C.; Liu, C.-W.; Chou, W.-C.; Chen, C.-Y.; et al. Dynamics of DNMT3A mutation and prognostic relevance in patients with primary myelodysplastic syndrome. Clin. Epigenetics 2018, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Heesch, S.; Schlee, C.; Schwartz, S.; Gökbuget, N.; Hoelzer, D.; Konstandin, N.P.; Ksienzyk, B.; Vosberg, S.; Graf, A.; et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood 2013, 121, 4749–4752. [Google Scholar] [CrossRef]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Pérez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A Critical Role for Dnmt1 and DNA Methylation in T Cell Development, Function, and Survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Alvarez, B.; Rodriguez, R.M.; Fraga, M.F.; López-Larrea, C. DNA methylation: A promising landscape for immune system-related diseases. Trends Genet. 2012, 28, 506–514. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Liu, D.; Yu, L.; Xue, B.; Shi, H. Epigenetic Regulation of Macrophage Polarization by DNA Methyltransferase 3b. Mol. Endocrinol. 2014, 28, 565–574. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, X.; Jia, L.; Mondal, A.K.; Diallo, A.; Hawkins, G.A.; Das, S.K.; Parks, J.S.; Yu, L.; Shi, H.; et al. Inhibiting DNA Methylation by 5-Aza-2′-deoxycytidine Ameliorates Atherosclerosis Through Suppressing Macrophage Inflammation. Endocrinology 2014, 155, 4925–4938. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Zeng, S.; Su, C.; Li, J.; Xuan, Y.; Lin, Y.; Xu, E.; Fan, Q. The interaction between DNA methylation and tumor immune microenvironment: From the laboratory to clinical applications. Clin. Epigenetics 2024, 16, 24. [Google Scholar] [CrossRef]
- Hey, J.; Halperin, C.; Hartmann, M.; Mayer, S.; Schönung, M.; Lipka, D.B.; Scherz-Shouval, R.; Plass, C. DNA methylation landscape of tumor-associated macrophages reveals pathways, transcription factors and prognostic value relevant to triple-negative breast cancer patients. Int. J. Cancer 2022, 152, 1226–1242. [Google Scholar] [CrossRef] [PubMed]
- Elkoshi, Z. TGF-β, IL-1β, IL-6 levels and TGF-β/Smad pathway reactivity regulate the link between allergic diseases, cancer risk, and metabolic dysregulations. Front. Immunol. 2024, 15, 1371753. [Google Scholar] [CrossRef]
- Goyal, A.; Bauer, J.; Hey, J.; Papageorgiou, D.N.; Stepanova, E.; Daskalakis, M.; Scheid, J.; Dubbelaar, M.; Klimovich, B.; Schwarz, D.; et al. DNMT and HDAC inhibition induces immunogenic neoantigens from human endogenous retroviral element-derived transcripts. Nat. Commun. 2023, 14, 6731. [Google Scholar] [CrossRef]
- Rowe, H.M.; Friedli, M.; Offner, S.; Verp, S.; Mesnard, D.; Marquis, J.; Aktas, T.; Trono, D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 2013, 140, 519–529. [Google Scholar] [CrossRef]
- Falahat, R.; Berglund, A.; Putney, R.M.; Perez-Villarroel, P.; Aoyama, S.; Pilon-Thomas, S.; Barber, G.N.; Mulé, J.J. Epigenetic reprogramming of tumor cell–intrinsic STING function sculpts antigenicity and T cell recognition of melanoma. Proc. Natl. Acad. Sci. USA 2021, 118, e2013598118. [Google Scholar] [CrossRef]
- Etcheverry, A.; Aubry, M.; de Tayrac, M.; Vauleon, E.; Boniface, R.; Guenot, F.; Saikali, S.; Hamlat, A.; Riffaud, L.; Menei, P.; et al. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genom. 2010, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Wee, C.W.; Kim, J.H.; Kim, H.J.; Kang, H.-C.; Suh, S.Y.; Shin, B.S.; Ma, E.; Kim, I.H. Radiosensitization of Glioblastoma Cells by a Novel DNA Methyltransferase-inhibiting Phthalimido-Alkanamide Derivative. Anticancer. Res. 2019, 39, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hu, S.; Hou, P.; Jiang, D.; Condouris, S.; Xing, M. Suppression of BRAF/MEK/MAP Kinase Pathway Restores Expression of Iodide-Metabolizing Genes in Thyroid Cells Expressing the V600E BRAF Mutant. Clin. Cancer Res. 2007, 13, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.V.; Nestler, K.A.; Chiappinelli, K.B. Therapeutic targeting of DNA methylation alterations in cancer. Pharmacol. Ther. 2024, 258, 108640. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Wishka, D.G.; Lopez, O.D.; Rudchenko, V.; Huang, G.; Hoffman, S.N.; Borgel, S.; Georgius, K.; Carter, J.; Stotler, H.; et al. F-aza-T-dCyd (NSC801845), a Novel Cytidine Analog, in Comparative Cell Culture and Xenograft Studies with the Clinical Candidates T-dCyd, F-T-dCyd, and Aza-T-dCyd. Mol. Cancer Ther. 2021, 20, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Kini, A.G. Engineering and Application of Zinc Finger Proteins and TALEs for Biomedical Research. Mol. Cells 2017, 40, 533–541. [Google Scholar] [CrossRef]
- Werner, E.W.; Mei, T.-S.; Burckle, A.J.; Sigman, M.S. Enantioselective Heck Arylations of Acyclic Alkenyl Alcohols Using a Redox-Relay Strategy. Science 2012, 338, 1455–1458. [Google Scholar] [CrossRef]
- Bernstein, D.L.; Le Lay, J.E.; Ruano, E.G.; Kaestner, K.H. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J. Clin. Investig. 2015, 125, 1998–2006. [Google Scholar] [CrossRef]
| Mouse/Cell | Survival | Development | Reference | Etc | |
|---|---|---|---|---|---|
| Dnmt1 | Mouse | Embryonically lethal | Defect | [17] | Global loss of DNA methylation |
| Dnmt2 | Mouse/Fly | Viable and fertile | No defect | [12,13] | Aberrant hematopoiesis |
| Dnmt3 | Mouse | Embryonically lethal | Impaired postnatal development | [10] | |
| Dnmt3b | Mouse | Embryonically lethal | Defect | [11] | |
| Dnmt1 | Mouse ES cell | [16] | Died upon the induction of differentiation | ||
| Dnmt1 | Human ES cell | [18,19] | Died upon the induction of differentiation | ||
| Triple knockout (Dnmt1, Dnmt3a, Dnmt3b) | Cell | [15] | Proliferation |
| Mutation | Location | Effects | Cancer Association | Ref | |
|---|---|---|---|---|---|
| DNMT1 | Somatic Mutations | Mutations in coding exons of the DNMT1 gene | Genomic instability, loss of DNA methylation control | Colorectal cancers Lung cancer | [154,162] |
| DNMT1 | Catalytic Domain Mutations | Catalytic domain | Loss of function, global hypomethylation, oncogenic activation | Hematologic malignancies, Solid tumors | [163,164] |
| DNMT1 | Splice-Site Mutations | Splice sites | Abnormal splicing, leading to truncated proteins with altered function | AML, MDS, Leukemia | [165] |
| DNMT1 | Deletion/Overexpression | Coding exons | Significantly suppressed tumor formation/Tumorigenesis | Mammary tumor, Breast cancer, Pancreas/Liver | [162,166,167] |
| DNMT2 | Deletion | tRNA methyltransferase domain | Promotes the proliferation, colony formation, and metastasis of hepatocellular carcinoma cells | Hepatocellular carcinoma | [67] |
| DNMT3A | R882H, R882C, R882P, R882S | Catalytic domain | Dominant-negative inhibition, genome-wide hypomethylation, AML association | Acute Myeloid Leukemia (AML)/MDS | [168,169,170] |
| DNMT3A | Catalytic domain mutations | C-terminal domain | Loss of function, reduced enzymatic activity, hypomethylation of TSGs | AML, Myelodysplastic Syndromes (MDS) | [160,171] |
| DNMT3B | Frameshift mutations | Various coding regions | Protein truncation, loss of enzymatic function, genomic instability | Lymphoma | [72,172] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.J. The Role of the DNA Methyltransferase Family and the Therapeutic Potential of DNMT Inhibitors in Tumor Treatment. Curr. Oncol. 2025, 32, 88. https://doi.org/10.3390/curroncol32020088
Kim DJ. The Role of the DNA Methyltransferase Family and the Therapeutic Potential of DNMT Inhibitors in Tumor Treatment. Current Oncology. 2025; 32(2):88. https://doi.org/10.3390/curroncol32020088
Chicago/Turabian StyleKim, Dae Joong. 2025. "The Role of the DNA Methyltransferase Family and the Therapeutic Potential of DNMT Inhibitors in Tumor Treatment" Current Oncology 32, no. 2: 88. https://doi.org/10.3390/curroncol32020088
APA StyleKim, D. J. (2025). The Role of the DNA Methyltransferase Family and the Therapeutic Potential of DNMT Inhibitors in Tumor Treatment. Current Oncology, 32(2), 88. https://doi.org/10.3390/curroncol32020088





