1. Introduction
Glioblastoma (GB) is the most common and the most aggressive brain tumor [
1]. The current standard of care involves maximal surgical resection followed by concurrent radiation therapy (RT) and temozolomide (TMZ), followed by adjuvant TMZ [
2]. There is currently a lack of clinical biomarkers in glioma that can be detected in minimally invasive biospecimens and are directly linked to progression or survival, with realistic prospects for implementation into clinical use [
3]. While MGMT methylation status is the single prognostic and predictive marker with MGMT-methylated patients having superior response to systemic management, improved progression, and overall survival [
4], and IDH mutation status has redefined GB classification with IDH-mutated tumors now reclassified given superior outcome [
5], both molecular features are detected in tumor tissue. This analysis can require several weeks to return results, which do not provide a distinct path towards harnessing tumor signaling with targeted agents or RT optimization. The exact mechanisms by which they exert their impact on the outcome are not fully understood. The paucity of data and, when available, the pervasive class imbalance in MGMT and IDH status annotation in public datasets pose a barrier to advancement in this direction for all biospecimens [
6,
7,
8]. From an imaging perspective, while patients are currently followed with MRI of the brain every 2–3 months following completion of up front CRT, MRI imaging of the brain remains difficult to interpret posttreatment indicating possible pseudoresponse or pseudoprogression and there is currently no biomarker that predicts for early progression and allows for early intervention or selection of patients for personalized management given evolving novel agents [
9,
10,
11]. The use of MRI as a data stream is limited by the complexity of the data and the restrictions on data sharing. The challenge with evolving biomarkers has been reproducibility across studies, matching biomarkers to outcomes as well as glioma subtypes, and correlation of markers across different biospecimens. Given the cost and practicality of acquisition, as well as the ability to rapidly grow large-scale data for computational analysis, blood is the most intuitive avenue as the noninvasive biospecimen of choice. Glioma patients already undergo weekly blood work to monitor temozolomide side effects as part of up-front management, providing an intuitive means of leveraging blood as the preferred biospecimen for biomarker development. The analysis of noninvasive biospecimen provides a compelling rationale for the advancement of both biological understanding of the disease and resistance to treatment as well as path to personalization of management given critical tumor heterogeneity [
12]. A number of studies are increasingly employing multichannel data to identify pivotal pathways [
13,
14,
15,
16,
17]; however, most studies are currently based on tumor tissue acquired at a single timepoint [
18,
19]. The present study involves serum as the biospecimen analyzed, obtained prior to and following completion of up-front CRT. A subset of patients also received valproic acid during the trial, and their analysis has been reported elsewhere [
20,
21]. In this study, we aimed to examine how differential alteration in proteomic and metabolomic expression pre- vs. post-completion of concurrent chemoirradiation (CRT) is associated with OS and PFS and molecular classification in GB.
4. Discussion
The present analysis involves serum biospecimens collected before and after completion of CRT in patients with pathologically proven GB. Four patients in this cohort were subsequently found to be IDH-mutated and would now be classified as astrocytoma IDH-mutated. The cohort spans 20 years during which first MGMT and then IDH became recognized molecular classifiers that were translated into the clinic, leading to data acquisition in some but not all patients, based on when they were diagnosed. We evaluated progression and overall survival, comparing MGMT-methylated patients with unmethylated patients, as well as IDH-mutated patients with IDH wild-type patients.
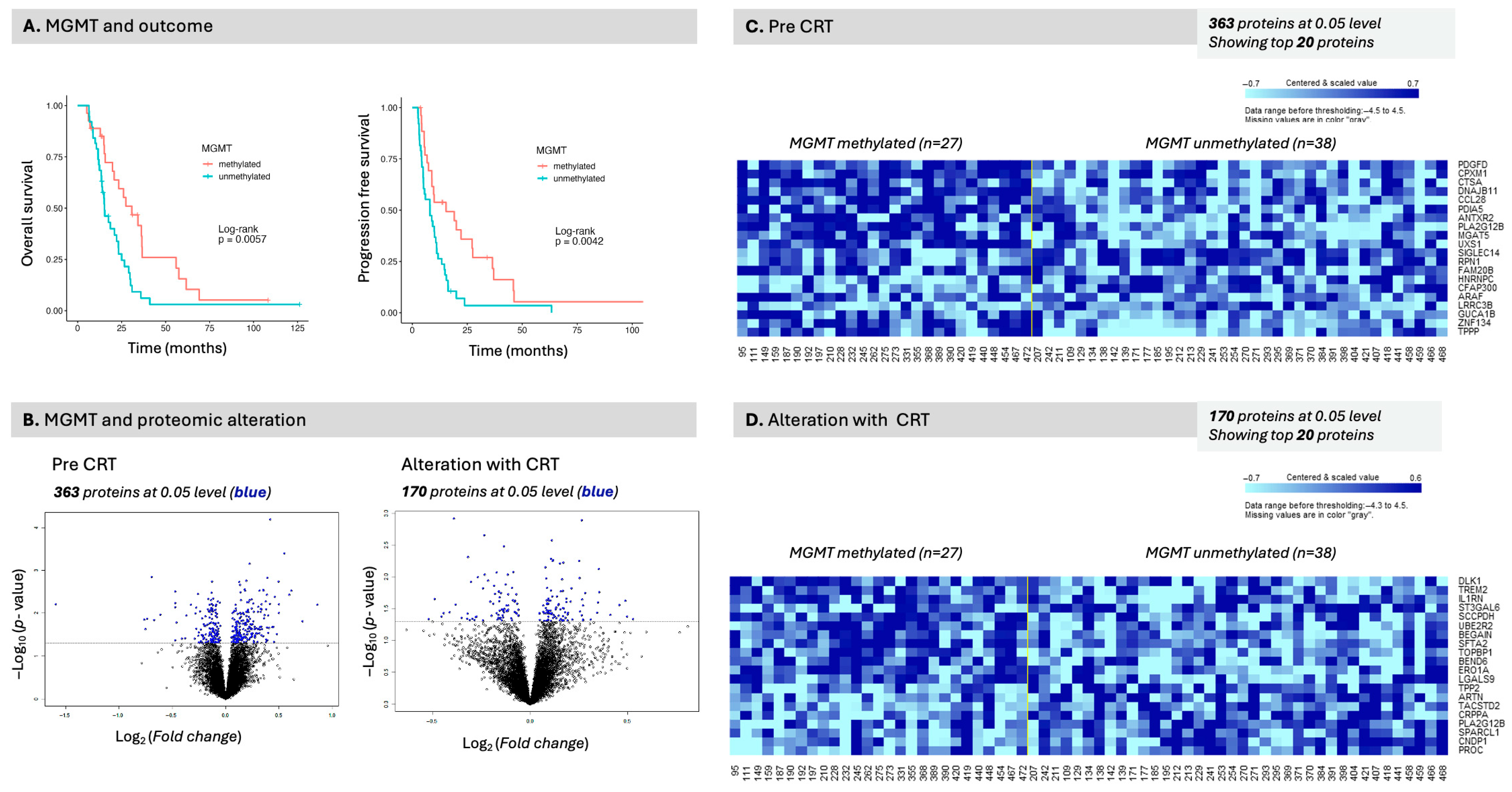
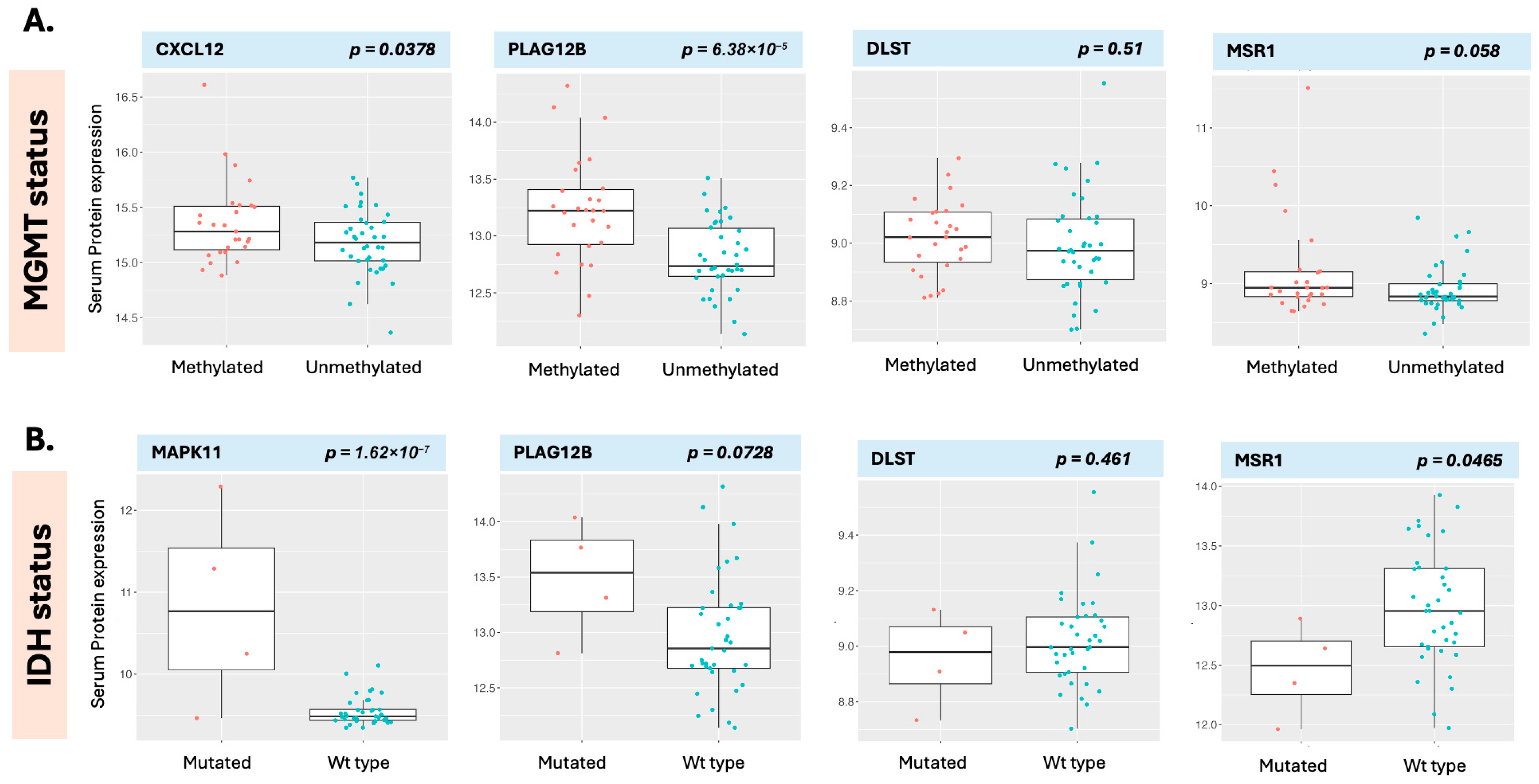
DLST was the only serum protein associated with overall survival to meet the significance threshold after adjustment for multiple comparisons. DLST (dihydrolipoamide S-succinyltransferase) is a component of the pyruvate dehydrogenase complex. In the present analysis, an elevated baseline serum DLST level was associated with decreased survival and was the only protein with a favorable FDR to do so. DLST is a mitochondrial critical component of the pyruvate dehydrogenase complex with significant implications for glucose metabolism (
Figure 9). We were unable to validate this finding using CPTAC tissue proteome data or TCGA genomic data. DLST was, however, significantly elevated in the serum of patients with GB compared with healthy individuals. Similarly, an increased MSR1 level emerged in baseline OS, PFS, and in its alteration with OS in the regression analysis and although it did not meet FDR threshold for significance, it was associated with OS in the survival analysis. MSR1 is an interesting molecule as we also noted that it has similar directionality in the CPTAC tissue proteome data, but not in the TCGA genomic data. MSR1 was, however, not significantly different in the serum of patients with GB as compared with healthy individuals. MSR1 (Macrophage Scavenger Receptor-A, aka CD204), has been found elevated in GB as compared to other cancers; its expression is associated with glioma progression [
32], and increased levels at the tissue level have been associated with poorer survival outcomes in several cancers, and the serum data in the present analysis support this. MSR1 is a membrane glycoprotein with complex functionality that intersects between lipid metabolism and immune regulation (
Figure 9) [
32,
33]. has been identified as an independent prognostic factor in TCGA and CGGA, and is associated with immune-associated gene sets [
33]. MSR1 is DLST and MSR1 emerged as prominent molecules in the present analysis but indicate that validation with independent datasets will be protein-dependent. A protein may be behaving similarly in serum and in tissue in relationship to the disease states (MSR1), while another may be providing specificity with altered detection in a disease state, as in this case of GB vs. healthy individuals (possible example DLST). Additional proteins of interest identified in the present analysis include PGAM and ATG5. While neither of these had a significant FDR in the initial analysis, both had pre-CRT levels that were significant for PFS, in contrast to other more prominent molecules such as MGMT and AKT1. This finding indicates that it is critical to examine signals by multiple different means (regression and survival analysis) to enhance detection of potentially hidden biomarkers. PGAM2 (Phosphoglycerate Mutase 2), for example converts 3-phosphoglycerate to 2-phosphoglycerate in the glycolytic pathway, a critical step that mediates glycolysis, the primary mode of energy generation in malignant conditions. PGAM2 is phosphorylated by PAK1, marking it for ubiquitin-mediated degradation [
34]. PGAM2 is upregulated in several cancers and can be critically increased by an alternative glycolytic pathway originating via a cancer-specific isoform of pyruvate kinase [
35]. PGAM2 activity is also linked to metabolic programming, including the regulation of phospholipid metabolism [
35]. Meanwhile, ATG5 (autophagy-related gene 5) is a proapoptotic molecule [
36] that has been demonstrated to be linked to the immune system and increased in the presence of hypoxia in GB and astrocytoma. It has also been linked to protective autophagy, a hypoxia-induced phenomenon in glioma. Studies have shown that knockdown of ATG5 decreases cell mobility and increases chemosensitivity in hypoxic conditions via the HIF1α/ATG5 axis [
37] and the CREB/ATG5 axis [
38]. ATG5 has been detected in serum and proposed as an early biomarker of malignant mesothelioma [
36], with an elevated level associated with activated autophagy. This process may occur via the acetylation of PAK1 (p21-activated kinase 1), which leads to autophagy and tumor proliferation by phosphorylating ATG5, thereby protecting ATG5 from ubiquitination-dependent degradation [
39]. In the present study, elevated serum ATG5 and PGAM2 were both highly statistically significantly associated with improved progression-free survival, which indicates a possible role for these molecules as potential biomarkers with avenues for validation given that they both have a relationship to p21-activated kinase 1. PAK1 is not currently present in the 7K Somalogic panel and therefore could not be directly measured but with growing proteomic datasets, this may change. It is possible that ATG5 degradation was more prevalent in patients with superior tumor biology, leading to its detection in serum and observed improved outcome. It is also possible that both signals originate from a cellular population that is either less hypoxic or less driven by protective autophagy, or both, hence benefiting from an improved outcome independent of treatment. Elevated levels of serum PGAM2 and ATG5 were associated with improved progression-free survival in a similar manner. Since both are directly linked to p21-activated kinase 1, which is heavily implicated in cancer, this also leads to potential regulation with GSK3 and Akt1 [
40] and ubiquitination-dependent degradation, again potentially providing upstream avenues for validation. This process may, presumably, increase the ability for detection in serum, as observed in the present study. PAK1 is a p21-activated kinase associated with GB development, hypoxic conditions, and poorer survival, characterized by increased invasiveness, cellular proliferation, and autophagy [
39]. The postulated interaction (
Figure 9) links PAK1 and, via phosphorylation, ATG5 and PGAM2, subsequently driving autophagy and glycolysis, respectively (
Figure 9).
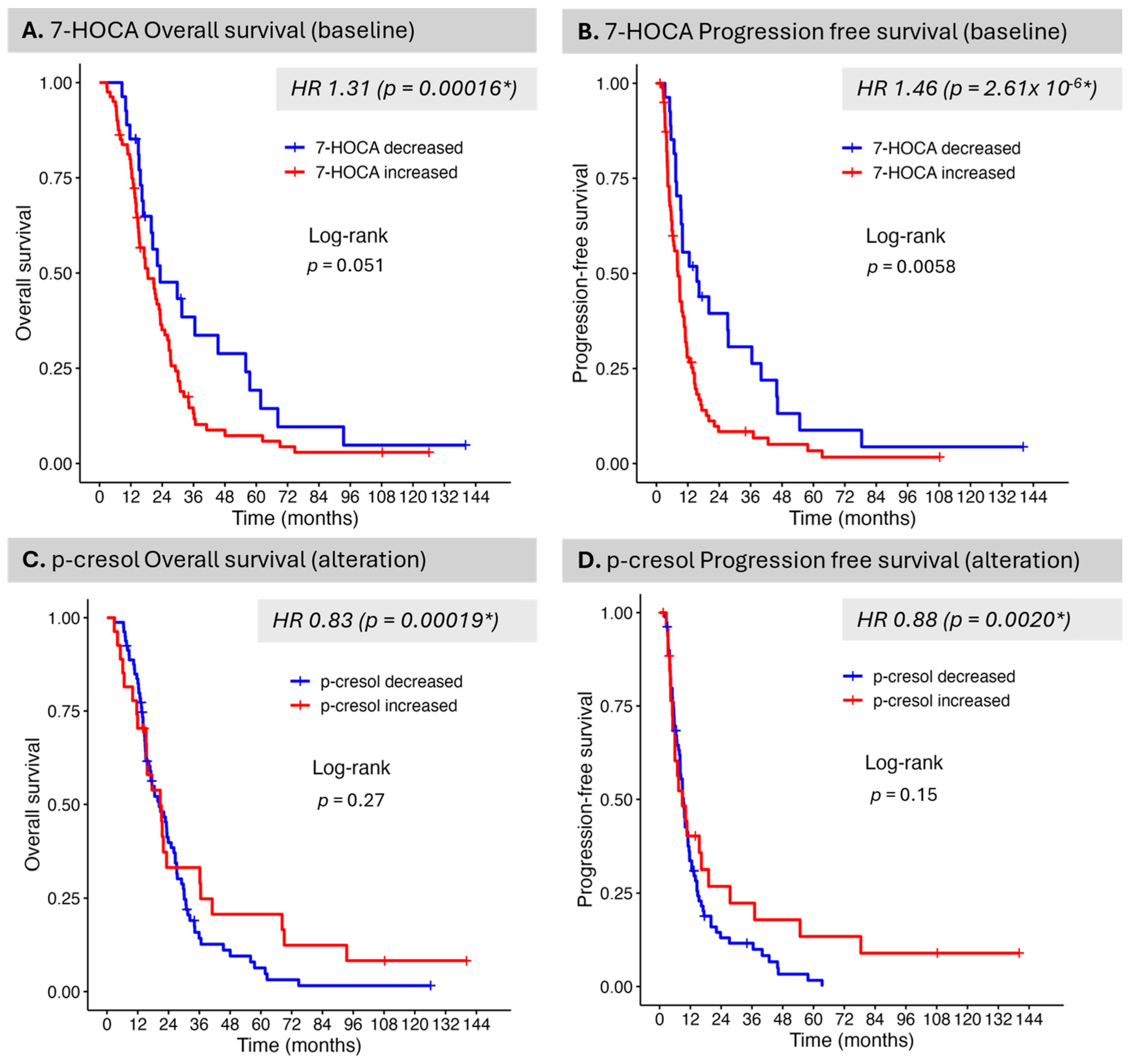
Despite a relatively much smaller set of measured metabolites that are biologically annotated and KEGG-linked, several were identified as critical in connection with OS, PFS, and IDH mutation status. 7-alpha-hydroxy-3-oxo-4-cholestenoate (7-HOCA) is a bile acid intermediate and cholesterol metabolite implicated in the classical pathway of bile acid synthesis occurring in the liver. Recent evidence has shown that 7-HOCA may represent a promising biomarker in GB. It was detected in both CSF and plasma with 7-HOCA levels found elevated in both biospecimen type, in patients with GB as compared to healthy individuals. 7-HOCA levels were associated with an increased risk of GB [
41]. This finding is supported by serum data in the present study with increased levels associated with detriment in OS and PFS. Previous research has also shown that 7-HOCA can be employed as a marker of blood–brain barrier dysfunction, with data supporting the flux of 7-HOCA from the brain against a concentration gradient, given its preferential binding to albumin [
42]. 7-HOCA is converted to CDCA (chenodeoxycholic acid) and cholic acid, metabolites that were also identified as associated with PFS and OS in the present analysis. The observed hazard ratios were greater than 1 for these metabolites, similar to 7-HOCA, before CRT and less than 1 after CRT, indicating that CRT administration impacts the bile acid signaling pathway and its various enzymatic steps, resulting in an improvement in outcome when these metabolites are decreased. The present study revealed primary bile acid biosynthesis, the biosynthesis of unsaturated fatty acids, and arachidonic acid metabolism across several analyses based on metabolomic serum signals. From a proteomic signal standpoint, PLA2G12B, a critical enzyme in arachidonic acid metabolism, was differentially expressed between MGMT-methylated patients, with increased levels observed in these patients. This finding links arachidonic acid metabolism to both outcome and molecular classification in GB. The brain is highly dependent on cholesterol for its function, accounting for approximately 20% of the body’s total cholesterol, and uniquely synthesizes cholesterol de novo. Under conditions of hypoxia, enzymatic activity leads to the biosynthesis of fatty acids and cholesterol [
41].
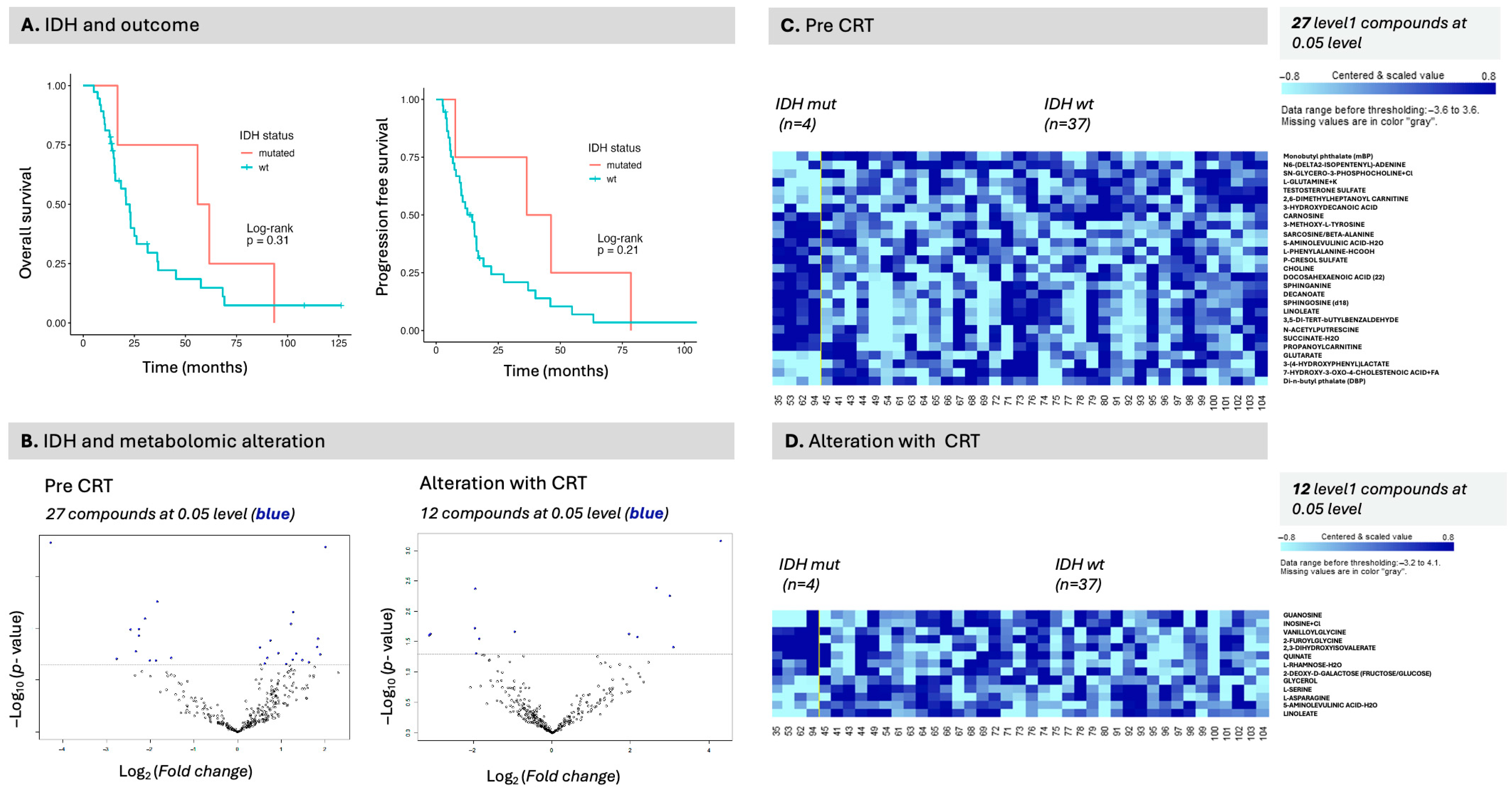
We identified serum p-cresol as being associated with outcome, as well as IDH status, in the present study. P-Cresol has also been previously identified as a potential marker of IDH status in CSF [
43]. The class comparison analysis for IDH status, while exploratory, nonetheless provides valuable insights into differences based on IDH status. Similar to the work of Möhn et al., p-cresol levels were decreased in patients with an IDH mutation compared to patients with an IDH wild-type, providing the literature validation. We note that IDH status is not present in many large glioma databases with the exception of TCGA [
44] and also not present in either RTOG0525 [
45] or RTOG0825 [
46]. When present, a relatively small number of patients are IDH-mutated [
47], which can render data analysis in this space and in serum particularly valuable. In upstream signaling (
Figure 9), S1P (sphingosine-1-phosphate) has been implicated in several cancers, including GB [
48]. In the present study, it has been associated with PFS, as has SPHK1 (Sphingosine Kinase 1), which is responsible for rendering S1P a bioactive lipid. Elevations in S1P and SPHK1 were both associated with a higher risk of progression in the analysis; however, this association was not statistically significant in the Kaplan–Meier survival analysis. SPHK1 and S1P provide an instrumental connection to lipid metabolism, as well as MSR1, the metabolites p-cresol and 7-HOCA, and a means of validation in other biospecimens and datasets.
These multi-omic analyses indicate that CRT produces measurable, clinically relevant shifts in GB biology, with GB linked biomarkers tracking patient outcome. Although only one pre-CRT protein, DLST, remained significant for OS after multiple-testing correction (higher baseline DLST predicting markedly worse OS), convergent Kaplan–Meier findings reinforce a treatment effect: lower baseline DLST and MSR1 aligned with improved OS, and higher baseline PGAM2 and ATG5 associated with longer PFS. The metabolomics further support CRT impact on systemic metabolism. P-cresol was the top pre- vs. post-CRT altered compound and showed a benefit (HR < 1 for both OS and PFS), while decreased baseline 7-HOCA favored better PFS (and trended for OS). S1P also tracked with outcome (lower baseline levels associating with improved PFS), and several analytes exhibited percentile dependent significance, consistent with heterogeneous therapy responses. Together, these patterns suggest that CRT not only selects prognostic molecular states present at baseline possibly via purine and fatty acid metabolism but also drives post-treatment metabolic reprogramming detectable in circulation which may at least partly be mediated by lipid metabolism. The present analysis builds further rationale for dynamic monitoring of serum samples in larger cohorts of patients.
To translate the findings from this study, we propose a clinical validation pathway integrating multi-omic, imaging, and longitudinal sampling. Initial steps include analytical validation of a serum assay panel (p-cresol, S1P, 7-HOCA, DLST, MSR1, PGAM2, and ATG5) as well as known hallmarks of cancer proteins with standardized pre-analytics and central IDH and MGMT testing. We also propose parallel analysis of IDH status for all patients where tissue is available and blood collection at multiple timepoints on existing trials. A prospective observational study would aim to collect serum at multiple timepoints (pre-CRT, during, post-CRT, and follow-up) and fuse metabolite trajectories with MRI radiomics to track therapy response and progression. Parallel mechanistic studies can correlate serum S1P and SPHK1 activity with tumor tissue and CSF to confirm biological linkage. Subsequent biomarker-guided adaptive trials could test whether monitoring of metabolites, such as p-cresol, are feasible and can be utilized to alter management and outcomes. Addressing data gaps, particularly the limited IDH-mutant representation requires central genotyping, targeted accrual, and Bayesian modeling to ensure robust subgroup inference. To move proteomic and metabolic markers from retrospective discovery toward clinically actionable tools for dynamic glioma monitoring, integrating omics with imaging and molecular profiling is critical and should form a cornerstone of ongoing and upcoming trials in glioma.
Limitations of the current study include its retrospective nature and the period covered, which spans over 20 years during which diagnostic standards, including the prevalence of MGMT and IDH testing, have changed. MGMT and IDH status were missing in 40% and 59% of the cohort, potentially limiting conclusions that can be made without further validation. Only four patients were IDH-mutated, which may also limit the conclusions that can be drawn regarding the IDH class comparison.