Proteomic Perspectives on KRAS-Driven Cancers and Emerging Therapeutic Approaches
Simple Summary
Abstract
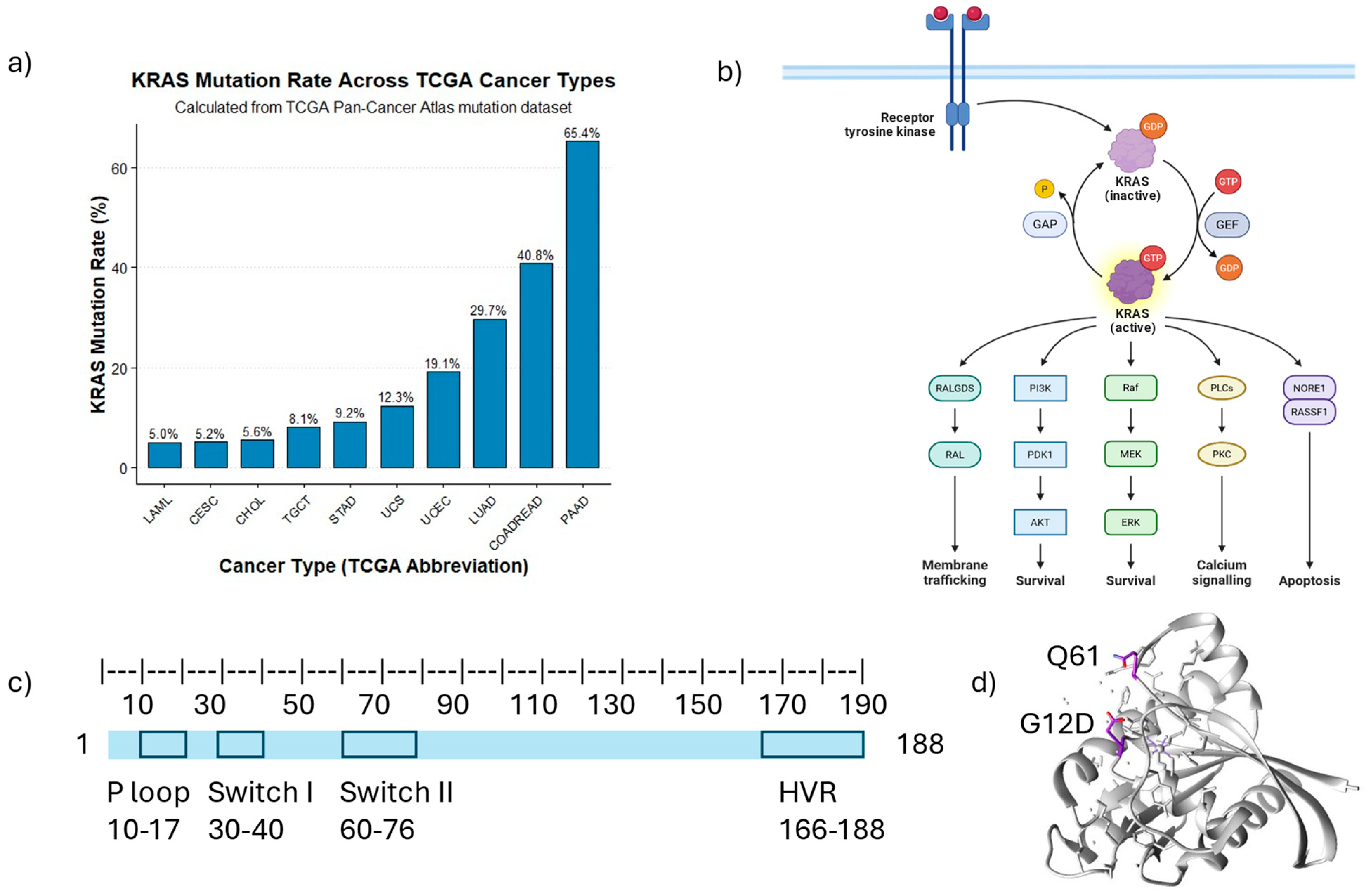
1. Introduction
2. Impact of KRAS Mutations on Cellular Proteome Homeostasis
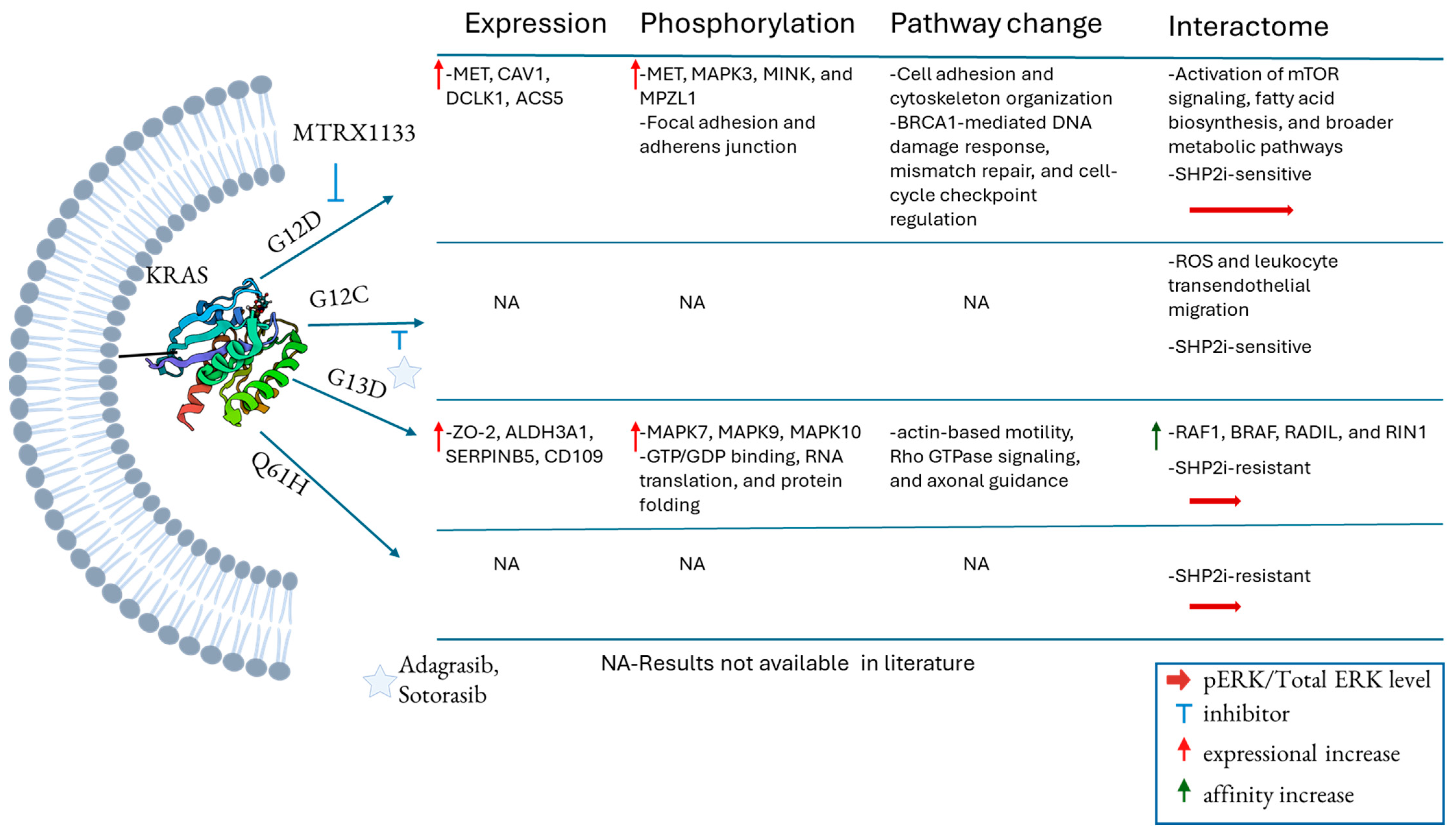
3. Distinct KRAS Mutations Associated with Various Functional Changes
4. Differential Interactomes Specific to KRAS Mutations
5. KRAS Conformational Changes Induced by Mutations
6. Post-Translational Modifications of KRAS and Their Involvement in Signaling
7. Current Progress in Inhibiting Different Mutations of KRAS
8. Resistance Mechanisms in Targeted Inhibition of KRAS
9. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, J.K.; Sivakumar, S.; Schrock, A.B.; Madison, R.; Fabrizio, D.; Gjoerup, O.; Ross, J.S.; Frampton, G.M.; Napalkov, P.; Montesion, M.; et al. Comprehensive pan-cancer genomic landscape of KRAS altered cancers and real-world outcomes in solid tumors. npj Precis. Oncol. 2022, 6, 91. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, W.; Wu, K.; Shi, L. The roles of KRAS in cancer metabolism, tumor microenvironment and clinical therapy. Mol. Cancer 2025, 24, 14. [Google Scholar] [CrossRef] [PubMed]
- Dias Carvalho, P.; Guimarães, C.F.; Cardoso, A.P.; Mendonça, S.; Costa, Â.M.; Oliveira, M.J.; Velho, S. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res. 2018, 78, 7–14. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef]
- Jarnuczak, A.F.; Najgebauer, H.; Barzine, M.; Kundu, D.J.; Ghavidel, F.; Perez-Riverol, Y.; Papatheodorou, I.; Brazma, A.; Vizcaíno, J.A. An integrated landscape of protein expression in human cancer. Sci. Data 2021, 8, 115. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R. Pathological implication of protein post-translational modifications in cancer. Mol. Asp. Med. 2022, 86, 101097. [Google Scholar] [CrossRef]
- Pan, S.; Brentnall, T.A.; Chen, R. Proteome alterations in pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 469, 429–436. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R.; Aebersold, R.; Brentnall, T.A. Mass Spectrometry Based Glycoproteomics—From a Proteomics Perspective. Mol. Cell. Proteom. 2011, 10, R110-003251. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Brentnall, T.A.; Chen, R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics 2015, 15, 2705–2715. [Google Scholar] [CrossRef]
- Carrillo-Rodriguez, P.; Selheim, F.; Hernandez-Valladares, M. Mass Spectrometry-Based Proteomics Workflows in Cancer Research: The Relevance of Choosing the Right Steps. Cancers 2023, 15, 555. [Google Scholar] [CrossRef]
- Cui, M.; Deng, F.; Disis, M.L.; Cheng, C.; Zhang, L. Advances in the Clinical Application of High-throughput Proteomics. Explor. Res. Hypothesis Med. 2024, 9, 209–220. [Google Scholar] [CrossRef]
- Nolen, B.M.; Lokshin, A.E. Biomarker testing for ovarian cancer: Clinical utility of multiplex assays. Mol. Diagn. Ther. 2013, 17, 139–146. [Google Scholar] [CrossRef]
- Birhanu, A.G. Mass spectrometry-based proteomics as an emerging tool in clinical laboratories. Clin. Proteom. 2023, 20, 32. [Google Scholar] [CrossRef]
- Albrecht, V.; Müller-Reif, J.; Nordmann, T.M.; Mund, A.; Schweizer, L.; Geyer, P.E.; Niu, L.; Wang, J.; Post, F.; Oeller, M.; et al. Bridging the Gap from Proteomics Technology to Clinical Application: Highlights from the 68th Benzon Foundation Symposium. Mol. Cell. Proteom. 2024, 23, 100877. [Google Scholar] [CrossRef]
- Gillette, M.A.; Jimenez, C.R.; Carr, S.A. Clinical Proteomics: A Promise Becoming Reality. Mol. Cell. Proteom. 2024, 23, 100688. [Google Scholar] [CrossRef] [PubMed]
- Nedelkov, D. Mass spectrometry protein tests: Ready for prime time (or not). Expert Rev. Proteom. 2017, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cripps, C.; Gill, S.; Ahmed, S.; Colwell, B.; Dowden, S.; Kennecke, H.; Maroun, J.; Samson, B.; Thirlwell, M.; Wong, R. Consensus recommendations for the use of anti-EGFR therapies in metastatic colorectal cancer. Curr. Oncol. 2010, 17, 39. [Google Scholar] [CrossRef]
- Kim, Y.J.; Chambers, A.G.; Cecchi, F.; Hembrough, T. Targeted data-independent acquisition for mass spectrometric detection of RAS mutations in formalin-fixed, paraffin-embedded tumor biopsies. J. Proteom. 2018, 189, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.G.; Chain, D.C.; Sweet, S.M.; Song, Z.; Martin, P.L.; Ellis, M.J.; Rooney, C.; Kim, Y.J. Mass spectrometry quantifies target engagement for a KRASG12C inhibitor in FFPE tumor tissue. Clin. Proteom. 2023, 20, 47. [Google Scholar] [CrossRef]
- Brunelli, L.; Caiola, E.; Marabese, M.; Broggini, M.; Pastorelli, R. Comparative metabolomics profiling of isogenic KRAS wild type and mutant NSCLC cells in vitro and in vivo. Sci. Rep. 2016, 6, 28398. [Google Scholar] [CrossRef]
- Hallin, J.; Bowcut, V.; Calinisan, A.; Briere, D.M.; Hargis, L.; Engstrom, L.D.; Laguer, J.; Medwid, J.; Vanderpool, D.; Lifset, E.; et al. Anti-tumor efficacy of a potent and selective non-covalent KRASG12D inhibitor. Nat. Med. 2022, 28, 2171–2182. [Google Scholar] [CrossRef] [PubMed]
- Chong, W.; Zhu, X.; Ren, H.; Ye, C.; Xu, K.; Wang, Z.; Jia, S.; Shang, L.; Li, L.; Chen, H. Integrated multi-omics characterization of KRAS mutant colorectal cancer. Theranostics 2022, 12, 5138. [Google Scholar] [CrossRef] [PubMed]
- Tahir, R.; Renuse, S.; Udainiya, S.; Madugundu, A.K.; Cutler, J.A.; Nirujogi, R.S.; Na, C.H.; Xu, Y.; Wu, X.; Pandey, A. Mutation-Specific and Common Phosphotyrosine Signatures of KRAS G12D and G13D Alleles. J. Proteome Res. 2021, 20, 670–683. [Google Scholar] [CrossRef]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.-J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic Analysis of Exosomes from Mutant KRAS Colon Cancer Cells Identifies Intercellular Transfer of Mutant KRAS. Mol. Cell. Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef]
- Baldelli, E.; El Gazzah, E.; Moran, J.C.; Hodge, K.A.; Manojlovic, Z.; Bassiouni, R.; Carpten, J.D.; Ludovini, V.; Baglivo, S.; Crinò, L.; et al. Wild-Type KRAS Allele Effects on Druggable Targets in KRAS Mutant Lung Adenocarcinomas. Genes 2021, 12, 1402. [Google Scholar] [CrossRef]
- Zhou, B.; Der, C.J.; Cox, A.D. The role of wild type RAS isoforms in cancer. Semin. Cell Dev. Biol. 2016, 58, 60–69. [Google Scholar] [CrossRef]
- Yan, H.; Yu, C.-C.; Fine, S.A.; Youssof, A.L.; Yang, Y.-R.; Yan, J.; Karg, D.C.; Cheung, E.C.; Friedman, R.A.; Ying, H.; et al. Loss of the wild-type KRAS allele promotes pancreatic cancer progression through functional activation of YAP1. Oncogene 2021, 40, 6759–6771. [Google Scholar] [CrossRef]
- Matallanas, D.; Romano, D.; Al-Mulla, F.; O’Neill, E.; Al-Ali, W.; Crespo, P.; Doyle, B.; Nixon, C.; Sansom, O.; Drosten, M.; et al. Mutant K-Ras Activation of the Proapoptotic MST2 Pathway Is Antagonized by Wild-Type K-Ras. Mol. Cell 2011, 44, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Y.; Qian, L.; Jiang, S.; Gai, X.; Ye, S.; Chen, Y.; Wang, X.; Zhai, L.; Xu, J.; et al. A proteomic and phosphoproteomic landscape of KRAS mutant cancers identifies combination therapies. Mol. Cell 2021, 81, 4076–4090.e4078. [Google Scholar] [CrossRef]
- Nolan, A.; Raso, C.; Kolch, W.; Kriegsheim, A.v.; Wynne, K.; Matallanas, D. Proteomic Mapping of the Interactome of KRAS Mutants Identifies New Features of RAS Signalling Networks and the Mechanism of Action of Sotorasib. Cancers 2023, 15, 4141. [Google Scholar] [CrossRef]
- Hutton, J.E.; Wang, X.; Zimmerman, L.J.; Slebos, R.J.C.; Trenary, I.A.; Young, J.D.; Li, M.; Liebler, D.C. Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer. Mol. Cell. Proteom. 2016, 15, 2924–2938. [Google Scholar] [CrossRef] [PubMed]
- Saliakoura, M.; Konstantinidou, G. Lipid Metabolic Alterations in KRAS Mutant Tumors: Unmasking New Vulnerabilities for Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 1793. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.M.; Waters, A.M.; Klomp, J.E.; Javaid, S.; Bryant, K.L.; Stalnecker, C.A.; Drizyte-Miller, K.; Papke, B.; Yang, R.; Amparo, A.M.; et al. Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-Mutant Pancreatic Cancer. Cancer Res. 2023, 83, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.P.; Jamieson, N.B.; Karim, S.A.; Athineos, D.; Ridgway, R.A.; Nixon, C.; McKay, C.J.; Carter, R.; Brunton, V.G.; Frame, M.C.; et al. LKB1 Haploinsufficiency Cooperates with KRAS to Promote Pancreatic Cancer Through Suppression of p21-Dependent Growth Arrest. Gastroenterology 2010, 139, 586–597.e586. [Google Scholar] [CrossRef]
- Gastl, B.; Klotz-Noack, K.; Klinger, B.; Ispasanie, S.; Salib, K.H.F.; Zuber, J.; Mamlouk, S.; Bublitz, N.; Blüthgen, N.; Horst, D.; et al. Reduced replication origin licensing selectively kills KRAS-mutant colorectal cancer cells via mitotic catastrophe. Cell Death Dis. 2020, 11, 499. [Google Scholar] [CrossRef]
- Kong, Y.; Allison, D.B.; Zhang, Q.; He, D.; Li, Y.; Mao, F.; Li, C.; Li, Z.; Zhang, Y.; Wang, J.; et al. The kinase PLK1 promotes the development of Kras/Tp53-mutant lung adenocarcinoma through transcriptional activation of the receptor RET. Sci. Signal. 2022, 15, eabj4009. [Google Scholar] [CrossRef]
- Azoitei, N.; Hoffmann, C.M.; Ellegast, J.M.; Ball, C.R.; Obermayer, K.; Gößele, U.; Koch, B.; Faber, K.; Genze, F.; Schrader, M.; et al. Targeting of KRAS mutant tumors by HSP90 inhibitors involves degradation of STK33. J. Exp. Med. 2012, 209, 697–711. [Google Scholar] [CrossRef]
- Shirazi, F.; Jones, R.J.; Singh, R.K.; Zou, J.; Kuiatse, I.; Berkova, Z.; Wang, H.; Lee, H.C.; Hong, S.; Dick, L.; et al. Activating KRAS, NRAS, and BRAF mutants enhance proteasome capacity and reduce endoplasmic reticulum stress in multiple myeloma. Proc. Natl. Acad. Sci. USA 2020, 117, 20004–20014. [Google Scholar] [CrossRef]
- Alves, S.; Castro, L.; Fernandes, M.S.; Francisco, R.; Castro, P.; Priault, M.; Chaves, S.R.; Moyer, M.P.; Oliveira, C.; Seruca, R.; et al. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget 2015, 6, 30787–30802. [Google Scholar] [CrossRef]
- Rangel, D.F.; Dubeau, L.; Park, R.; Chan, P.; Ha, D.P.; Pulido, M.A.; Mullen, D.J.; Vorobyova, I.; Zhou, B.; Borok, Z.; et al. Endoplasmic reticulum chaperone GRP78/BiP is critical for mutant Kras-driven lung tumorigenesis. Oncogene 2021, 40, 3624–3632. [Google Scholar] [CrossRef] [PubMed]
- Hamarsheh, S.a.; Groß, O.; Brummer, T.; Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020, 11, 5439. [Google Scholar] [CrossRef]
- Shang, A.; Gu, C.; Zhou, C.; Yang, Y.; Chen, C.; Zeng, B.; Wu, J.; Lu, W.; Wang, W.; Sun, Z.; et al. Exosomal KRAS mutation promotes the formation of tumor-associated neutrophil extracellular traps and causes deterioration of colorectal cancer by inducing IL-8 expression. Cell Commun. Signal. 2020, 18, 52. [Google Scholar] [CrossRef]
- Dias Carvalho, P.; Martins, F.; Carvalho, J.; Oliveira, M.J.; Velho, S. Mutant KRAS-Associated Proteome Is Mainly Controlled by Exogenous Factors. Cells 2022, 11, 1988. [Google Scholar] [CrossRef]
- Haigis, K.M. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017, 3, 686–697. [Google Scholar] [CrossRef]
- Hammond, D.E.; Mageean, C.J.; Rusilowicz, E.V.; Wickenden, J.A.; Clague, M.J.; Prior, I.A. Differential Reprogramming of Isogenic Colorectal Cancer Cells by Distinct Activating KRAS Mutations. J. Proteome Res. 2015, 14, 1535–1546. [Google Scholar] [CrossRef]
- Preto, A.; Figueiredo, J.; Velho, S.; Ribeiro, A.S.; Soares, P.; Oliveira, C.; Seruca, R. BRAF provides proliferation and survival signals in MSI colorectal carcinoma cells displaying BRAFV600E but not KRAS mutations. J. Pathol. 2008, 214, 320–327. [Google Scholar] [CrossRef]
- Kundu, S.; Ali, M.A.; Handin, N.; Conway, L.P.; Rendo, V.; Artursson, P.; He, L.; Globisch, D.; Sjöblom, T. Common and mutation specific phenotypes of KRAS and BRAF mutations in colorectal cancer cells revealed by integrative -omics analysis. J. Exp. Clin. Cancer Res. 2021, 40, 225. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Liu, C.; Velazquez, R.; Wang, H.; Dunkl, L.M.; Kazic-Legueux, M.; Haberkorn, A.; Billy, E.; Manchado, E.; Brachmann, S.M.; et al. SHP2 Inhibition Overcomes RTK-Mediated Pathway Reactivation in KRAS-Mutant Tumors Treated with MEK Inhibitors. Mol. Cancer Ther. 2019, 18, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Kano, Y.; Gebregiworgis, T.; Marshall, C.B.; Radulovich, N.; Poon, B.P.K.; St-Germain, J.; Cook, J.D.; Valencia-Sama, I.; Grant, B.M.M.; Herrera, S.G.; et al. Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation. Nat. Commun. 2019, 10, 224. [Google Scholar] [CrossRef]
- Gebregiworgis, T.; Kano, Y.; St-Germain, J.; Radulovich, N.; Udaskin, M.L.; Mentes, A.; Huang, R.; Poon, B.P.K.; He, W.; Valencia-Sama, I.; et al. The Q61H mutation decouples KRAS from upstream regulation and renders cancer cells resistant to SHP2 inhibitors. Nat. Commun. 2021, 12, 6274. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.F.; Branon, T.C.; Udeshi, N.D.; Myers, S.A.; Carr, S.A.; Ting, A.Y. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 2020, 15, 3971–3999. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, B.; Zou, M.; Zhou, H.; Ding, Y.; Ren, W.; Fang, L.; Zhang, J. Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics. Biology 2025, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Masson, G.R.; Burke, J.E.; Ahn, N.G.; Anand, G.S.; Borchers, C.; Brier, S.; Bou-Assaf, G.M.; Engen, J.R.; Englander, S.W.; Faber, J.; et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 2019, 16, 595–602. [Google Scholar] [CrossRef]
- Hernández-Porras, I.; Schuhmacher, A.J.; Garcia-Medina, R.; Jiménez, B.; Cañamero, M.; de Martino, A.; Guerra, C. K-RasV14I--induced Noonan syndrome predisposes to tumour development in mice. J. Pathol. 2016, 239, 206–217. [Google Scholar] [CrossRef]
- Bera, A.K.; Lu, J.; Wales, T.E.; Gondi, S.; Gurbani, D.; Nelson, A.; Engen, J.R.; Westover, K.D. Structural basis of the atypical activation mechanism of KRASV14I. J. Biol. Chem. 2019, 294, 13964–13972. [Google Scholar] [CrossRef]
- Rennella, E.; Henry, C.; Dickson, C.J.; Georgescauld, F.; Wales, T.E.; Erdmann, D.; Cotesta, S.; Maira, M.; Sedrani, R.; Brachmann, S.M.; et al. Dynamic conformational equilibria in the active states of KRAS and NRAS. RSC Chem. Biol. 2025, 6, 106–118. [Google Scholar] [CrossRef]
- Dharmaiah, S.; Tran, T.H.; Messing, S.; Agamasu, C.; Gillette, W.K.; Yan, W.; Waybright, T.; Alexander, P.; Esposito, D.; Nissley, D.V.; et al. Structures of N-terminally processed KRAS provide insight into the role of N-acetylation. Sci. Rep. 2019, 9, 10512. [Google Scholar] [CrossRef]
- Lu, J.; Harrison, R.A.; Li, L.; Zeng, M.; Gondi, S.; Scott, D.; Gray, N.S.; Engen, J.R.; Westover, K.D. KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry. Structure 2017, 25, 1442–1448.e1443. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef]
- Osaka, N.; Hirota, Y.; Ito, D.; Ikeda, Y.; Kamata, R.; Fujii, Y.; Chirasani, V.R.; Campbell, S.L.; Takeuchi, K.; Senda, T.; et al. Divergent Mechanisms Activating RAS and Small GTPases Through Post-translational Modification. Front. Mol. Biosci. 2021, 8, 707439. [Google Scholar] [CrossRef]
- Gillette, W.K.; Esposito, D.; Abreu Blanco, M.; Alexander, P.; Bindu, L.; Bittner, C.; Chertov, O.; Frank, P.H.; Grose, C.; Jones, J.E.; et al. Farnesylated and methylated KRAS4b: High yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 2015, 5, 15916. [Google Scholar] [CrossRef]
- Wang, W.-h.; Yuan, T.; Qian, M.-j.; Yan, F.-j.; Yang, L.; He, Q.-j.; Yang, B.; Lu, J.-j.; Zhu, H. Post-translational modification of KRAS: Potential targets for cancer therapy. Acta Pharmacol. Sin. 2021, 42, 1201–1211. [Google Scholar] [CrossRef]
- Ahearn, I.M.; Haigis, K.; Bar-Sagi, D.; Philips, M.R. Regulating the regulator: Post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2012, 13, 39–51. [Google Scholar] [CrossRef]
- Inouye, K.; Mizutani, S.; Koide, H.; Kaziro, Y. Formation of the Ras Dimer Is Essential for Raf-1 Activation. J. Biol. Chem. 2000, 275, 3737–3740. [Google Scholar] [CrossRef]
- Ambrogio, C.; Köhler, J.; Zhou, Z.-W.; Wang, H.; Paranal, R.; Li, J.; Capelletti, M.; Caffarra, C.; Li, S.; Lv, Q.; et al. KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell 2018, 172, 857–868.e815. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.D.; Scott, E.; Chang, J.-Y.; Bahramimoghaddam, H.; Lynn, M.; Lantz, C.; Russell, D.H.; Laganowsky, A. Capturing RAS oligomerization on a membrane. Proc. Natl. Acad. Sci. USA 2024, 121, e2405986121. [Google Scholar] [CrossRef] [PubMed]
- Ahearn, I.; Zhou, M.; Philips, M.R. Posttranslational Modifications of RAS Proteins. Cold Spring Harb. Perspect. Med. 2018, 8, a031484. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-Y.; Fan, S.; Zheng, H.; Guo, W.; Zheng, Z.; Sun, Y.; Zhong, C.; Zeng, J.; Li, S.; Zhang, M.; et al. Methylation of KRAS by SETD7 promotes KRAS degradation in non-small cell lung cancer. Cell Rep. 2023, 42, 113003. [Google Scholar] [CrossRef]
- Bivona, T.G.; Quatela, S.E.; Bodemann, B.O.; Ahearn, I.M.; Soskis, M.J.; Mor, A.; Miura, J.; Wiener, H.H.; Wright, L.; Saba, S.G.; et al. PKC Regulates a Farnesyl-Electrostatic Switch on K-Ras that Promotes its Association with Bcl-XL on Mitochondria and Induces Apoptosis. Mol. Cell 2006, 21, 481–493. [Google Scholar] [CrossRef]
- Yang, M.H.; Laurent, G.; Bause, A.S.; Spang, R.; German, N.; Haigis, M.C.; Haigis, K.M. HDAC6 and SIRT2 Regulate the Acetylation State and Oncogenic Activity of Mutant K-RAS. Mol. Cancer Res. 2013, 11, 1072–1077. [Google Scholar] [CrossRef]
- Song, H.Y.; Biancucci, M.; Kang, H.-J.; O’Callaghan, C.; Park, S.-H.; Principe, D.R.; Jiang, H.; Yan, Y.; Satchell, K.F.; Raparia, K.; et al. SIRT2 deletion enhances KRAS-induced tumorigenesis in vivo by regulating K147 acetylation status. Oncotarget 2016, 7, 80336. [Google Scholar] [CrossRef]
- Knyphausen, P.; Lang, F.; Baldus, L.; Extra, A.; Lammers, M. Insights into K-Ras 4B regulation by post-translational lysine acetylation. Biol. Chem. 2016, 397, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Jo, J.Y.; Choi, M.; Kim, K.-H.; Bae, Y.-K.; Kim, S.S. Oncogenic KRAS mutation confers chemoresistance by upregulating SIRT1 in non-small cell lung cancer. Exp. Mol. Med. 2023, 55, 2220–2237. [Google Scholar] [CrossRef]
- Yang, M.H.; Nickerson, S.; Kim, E.T.; Liot, C.; Laurent, G.; Spang, R.; Philips, M.R.; Shan, Y.; Shaw, D.E.; Bar-Sagi, D.; et al. Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. USA 2012, 109, 10843–10848. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.T.; Carracedo, A.; Locasale, J.W.; Anastasiou, D.; Takeuchi, K.; Kahoud, E.R.; Haviv, S.; Asara, J.M.; Pandolfi, P.P.; Cantley, L.C. Ubiquitination of K-Ras Enhances Activation and Facilitates Binding to Select Downstream Effectors. Sci. Signal. 2011, 4, ra13. [Google Scholar] [CrossRef] [PubMed]
- Magits, W.; Steklov, M.; Jang, H.; Sewduth, R.N.; Florentin, A.; Lechat, B.; Sheryazdanova, A.; Zhang, M.; Simicek, M.; Prag, G.; et al. K128 ubiquitination constrains RAS activity by expanding its binding interface with GAP proteins. EMBO J. 2024, 43, 2862–2877. [Google Scholar] [CrossRef]
- Baker, R.; Wilkerson, E.M.; Sumita, K.; Isom, D.G.; Sasaki, A.T.; Dohlman, H.G.; Campbell, S.L. Differences in the Regulation of K-Ras and H-Ras Isoforms by Monoubiquitination. J. Biol. Chem. 2013, 288, 36856–36862. [Google Scholar] [CrossRef]
- Ntai, I.; Fornelli, L.; DeHart, C.J.; Hutton, J.E.; Doubleday, P.F.; LeDuc, R.D.; van Nispen, A.J.; Fellers, R.T.; Whiteley, G.; Boja, E.S.; et al. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. USA 2018, 115, 4140–4145. [Google Scholar] [CrossRef]
- Simão, S.; Agostinho, R.R.; Martínez-Ruiz, A.; Araújo, I.M. Regulation of Ras Signaling by S-Nitrosylation. Antioxidants 2023, 12, 1562. [Google Scholar] [CrossRef]
- Batista, W.L.; Ogata, F.T.; Curcio, M.F.; Miguel, R.B.; Arai, R.J.; Matsuo, A.L.; Moraes, M.S.; Stern, A.; Monteiro, H.P. S-Nitrosoglutathione and Endothelial Nitric Oxide Synthase-Derived Nitric Oxide Regulate Compartmentalized Ras S-Nitrosylation and Stimulate Cell Proliferation. Antioxid. Redox Signal. 2012, 18, 221–238. [Google Scholar] [CrossRef]
- Kramer-Drauberg, M.; Petrini, E.; Mira, A.; Patrucco, E.; Scardaci, R.; Savinelli, I.; Wang, H.; Qiao, K.; Carrà, G.; Nokin, M.-J.; et al. Oncogenic mutant KRAS inhibition through oxidation at cysteine 118. Mol. Oncol. 2025, 19, 311–328. [Google Scholar] [CrossRef]
- Salamh, S.; Sayyed-Ahmad, A. Investigating the effects of cysteine-118 oxidation on G12D KRas structure and dynamics: Insights from MD simulations. J. Biomol. Struct. Dyn. 2024, 42, 6968–6981. [Google Scholar] [CrossRef]
- Kramer-Drauberg, M.; Ambrogio, C. Discoveries in the redox regulation of KRAS. Int. J. Biochem. Cell Biol. 2021, 131, 105901. [Google Scholar] [CrossRef] [PubMed]
- Isermann, T.; Sers, C.; Der, C.J.; Papke, B. KRAS inhibitors: Resistance drivers and combinatorial strategies. Trends Cancer 2025, 11, 91–116. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.; Moffat, J.; Schaefer, G.; Chan, J.; Wang, X.; Orr, C.; Cheng, J.; Hunsaker, T.; Shao, L.; Wang, S.J. Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLoS ONE 2017, 12, e0185862. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Goel, H.; Solanki, R.; Rawat, L.; Tabasum, S.; Tanwar, P.; Pal, S.; Sabarwal, A. Recent developments in receptor tyrosine kinase inhibitors: A promising mainstay in targeted cancer therapy. Med. Drug Discov. 2024, 23, 100195. [Google Scholar] [CrossRef]
- Hayes, T.K.; Neel, N.F.; Hu, C.; Gautam, P.; Chenard, M.; Long, B.; Aziz, M.; Kassner, M.; Bryant, K.L.; Pierobon, M.; et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 2016, 29, 75–89. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e517. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Fell, J.B.; Fischer, J.P.; Baer, B.R.; Ballard, J.; Blake, J.F.; Bouhana, K.; Brandhuber, B.J.; Briere, D.M.; Burgess, L.E.; Burkard, M.R. Discovery of tetrahydropyridopyrimidines as irreversible covalent inhibitors of KRAS-G12C with in vivo activity. ACS Med. Chem. Lett. 2018, 9, 1230–1234. [Google Scholar] [CrossRef]
- Fell, J.B.; Fischer, J.P.; Baer, B.R.; Blake, J.F.; Bouhana, K.; Briere, D.M.; Brown, K.D.; Burgess, L.E.; Burns, A.C.; Burkard, M.R.; et al. Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer. J. Med. Chem. 2020, 63, 6679–6693. [Google Scholar] [CrossRef]
- Riedl, J.M.; Fece de la Cruz, F.; Lin, J.J.; Parseghian, C.; Kim, J.E.; Matsubara, H.; Barnes, H.; Caughey, B.; Norden, B.L.; Morales-Giron, A.A.; et al. Genomic landscape of clinically acquired resistance alterations in patients treated with KRASG12C inhibitors. Ann. Oncol. 2025, 36, 682–692. [Google Scholar] [CrossRef]
- Purkey, H. Abstract ND11: Discovery of GDC-6036, a clinical stage treatment for KRAS G12C-positive cancers. Cancer Res. 2022, 82, ND11. [Google Scholar] [CrossRef]
- Brazel, D.; Nagasaka, M. Divarasib in the Evolving Landscape of KRAS G12C Inhibitors for NSCLC. Target. Oncol. 2024, 19, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Sacher, A.; LoRusso, P.; Patel, M.R.; Miller, W.H.; Garralda, E.; Forster, M.D.; Santoro, A.; Falcon, A.; Kim, T.W.; Paz-Ares, L.; et al. Single-Agent Divarasib (GDC-6036) in Solid Tumors with a KRAS G12C Mutation. N. Engl. J. Med. 2023, 389, 710–721. [Google Scholar] [CrossRef]
- Skoulidis, F.; Solomon, B.J.; Frost, N.S.W.; Heist, R.; Zhou, C.; Zarak Crnkovic, M.; Antic, V.; Prizant, H.; Mayo, M.C.; Meyenberg, C. Krascendo 2: A phase III study of divarasib and pembrolizumab vs pembrolizumab and chemotherapy in patients with previously untreated, advanced or metastatic, KRAS G12C-mutated non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2025, 43, TPS8656. [Google Scholar] [CrossRef]
- Drosten, M.; Barbacid, M. KRAS inhibitors: Going noncovalent. Mol. Oncol. 2022, 16, 3911–3915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guiley, K.Z.; Shokat, K.M. Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S). Nat. Chem. Biol. 2022, 18, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, G.A.; Baker, N.M.; Miermont, A.M.; Thurman, R.D.; Pierobon, M.; Tran, T.H.; Anderson, A.O.; Waters, A.M.; Diehl, J.N.; Papke, B. Atypical KRASG12R mutant is impaired in PI3K signaling and macropinocytosis in pancreatic cancer. Cancer Discov. 2020, 10, 104–123. [Google Scholar] [CrossRef]
- Zhang, Z.; Shokat, K.M. Covalent inhibitors of K-Ras G12S, G12R, and G12D. In RAS Drug Discovery; Gill, A.L., Shokat, K.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2025; pp. 379–402. [Google Scholar]
- Zhang, Z.; Morstein, J.; Ecker, A.K.; Guiley, K.Z.; Shokat, K.M. Chemoselective Covalent Modification of K-Ras(G12R) with a Small Molecule Electrophile. J. Am. Chem. Soc. 2022, 144, 15916–15921. [Google Scholar] [CrossRef]
- Kim, D.; Herdeis, L.; Rudolph, D.; Zhao, Y.; Böttcher, J.; Vides, A.; Ayala-Santos, C.I.; Pourfarjam, Y.; Cuevas-Navarro, A.; Xue, J.Y.; et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature 2023, 619, 160–166. [Google Scholar] [CrossRef]
- Tedeschi, A.; Schischlik, F.; Rocchetti, F.; Popow, J.; Ebner, F.; Gerlach, D.; Geyer, A.; Santoro, V.; Boghossian, A.S.; Rees, M.G. Pan-KRAS Inhibitors BI-2493 and BI-2865 Display Potent Antitumor Activity in Tumors with KRAS Wild-type Allele Amplification. Mol. Cancer Ther. 2025, 24, 550–562. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Mares, A.; Smith, I.E.D.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef]
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C.M. Targeted Degradation of Oncogenic KRASG12C by VHL-Recruiting PROTACs. ACS Cent. Sci. 2020, 6, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Popow, J.; Farnaby, W.; Gollner, A.; Kofink, C.; Fischer, G.; Wurm, M.; Zollman, D.; Wijaya, A.; Mischerikow, N.; Hasenoehrl, C.; et al. Targeting cancer with small-molecule pan-KRAS degraders. Science 2024, 385, 1338–1347. [Google Scholar] [CrossRef]
- Ash, L.J.; Busia-Bourdain, O.; Okpattah, D.; Kamel, A.; Liberchuk, A.; Wolfe, A.L. KRAS: Biology, Inhibition, and Mechanisms of Inhibitor Resistance. Curr. Oncol. 2024, 31, 2024–2046. [Google Scholar] [CrossRef] [PubMed]
- Yunchang, L.; Lanlin, H.; Xinhao, P.; Huasheng, X.; Bo, T.; Chuan, X. Resistance to immune checkpoint inhibitors in KRAS-mutant non-small cell lung cancer. Cancer Drug Resist. 2022, 5, 129–146. [Google Scholar] [CrossRef]
- Oya, Y.; Mitsudomi, T. Is adagrasib just another sotorasib?—Or, should we differentiate their usage according to patients’ clinical presentation? Transl. Lung Cancer Res. 2023, 12, 940–943. [Google Scholar] [CrossRef]
- Jänne, P.A.; Riely, G.J.; Gadgeel, S.M.; Heist, R.S.; Ou, S.-H.I.; Pacheco, J.M.; Johnson, M.L.; Sabari, J.K.; Leventakos, K.; Yau, E.; et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring KRASG12C Mutation. N. Engl. J. Med. 2022, 387, 120–131. [Google Scholar] [CrossRef]
- Negrao, M.V.; Araujo, H.A.; Lamberti, G.; Cooper, A.J.; Akhave, N.S.; Zhou, T.; Delasos, L.; Hicks, J.K.; Aldea, M.; Minuti, G.; et al. Comutations and KRASG12C Inhibitor Efficacy in Advanced NSCLC. Cancer Discov. 2023, 13, 1556–1571. [Google Scholar] [CrossRef]
- Awad, M.M.; Liu, S.; Rybkin, I.I.; Arbour, K.C.; Dilly, J.; Zhu, V.W.; Johnson, M.L.; Heist, R.S.; Patil, T.; Riely, G.J.; et al. Acquired Resistance to KRASG12C Inhibition in Cancer. N. Engl. J. Med. 2021, 384, 2382–2393. [Google Scholar] [CrossRef]
- Dilly, J.; Hoffman, M.T.; Abbassi, L.; Li, Z.; Paradiso, F.; Parent, B.D.; Hennessey, C.J.; Jordan, A.C.; Morgado, M.; Dasgupta, S.; et al. Mechanisms of Resistance to Oncogenic KRAS Inhibition in Pancreatic Cancer. Cancer Discov. 2024, 14, 2135–2161. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Lu, X.; Cao, J.; Luo, Q.; Ding, Y.; Peng, F.; Pataer, A.; Lu, D.; Han, D.; Malmberg, E.; et al. Modulation of the proteostasis network promotes tumor resistance to oncogenic KRAS inhibitors. Science 2023, 381, eabn4180. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H. ER stress and diseases. FEBS J. 2007, 274, 630–658. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Osgood, T.; Ma, S.; Estrada, J.; Arias, V.; Yamawaki, T.; Hoang, U.; Rex, K.; DeVoss, J.; Canon, J.; et al. KRAS inhibition triggers coordinated neoplastic and immune remodeling in the tumor microenvironment. J. Immunol. 2024, 212, 0875_7666. [Google Scholar] [CrossRef]
| Category | Key Alterations | References |
|---|---|---|
| Global (Phospho)Proteomic Signatures | Distinct proteomic/phosphoproteomic subtypes | [30] |
| ↑ Feedback regulators (DUSPs, SPRYs) | ||
| ↑ MAPK and PI3K phosphorylation | [24] | |
| ↑ Mutation-specific interactome changes | [31] | |
| Metabolic Reprogramming | ↑ Glycolysis (HK2, PKM2, LDHA) and glucose transporter level | [32] |
| ↑ Glutamine metabolism (GLS) | ||
| ↑ Altered lipid synthesis (FASN, CPT1A) | [3,21] | |
| ↑ Redox/antioxidant proteins (NRF2 targets) | [33] | |
| Cell Cycle and Proliferation | ↑ Cyclins (D1, E2) and CDKs (CDK4/6, and CDK2) | [34] |
| ↓ Inhibitors p21 (CDKN1A), p27 (CDKN1B) | [35] | |
| ↑ DNA replication proteins (MCMs, PCNA, DNA polymerases, replication origin licensing factors) | [36] | |
| ↑ Mitotic regulators (Aurora, PLK1) and checkpoint kinases (Chk1/2) | [37] | |
| Stress Response and Proteostasis | ↑ Chaperones (HSP70, HSP90, HSP27) | [38] |
| ↑ Proteasome activity | [39] | |
| ↑ Autophagy (LC3, ATG) | [40] | |
| ↑ UPR (BiP, CHOP) | [41] | |
| Tumor Microenvironment | ↑ Cytokines (IL-6, TGF-β) | [42] |
| ↑ ECM remodeling (MMPs, integrins) | [4] | |
| ↑ Exosome-mediated signaling | [43] | |
| ↑ Stromal interactions alter proteome | [44] |
| PTM on KRAS | Residues | Implication in Signaling | References |
|---|---|---|---|
| Farnesylation | C185 | Anchoring to PM and activation | [68] |
| methylation | K182 andK184 | Downregulation of KRAS | [69] |
| Phosphorylation | Y32, Y64 | Downregulation of KRAS | [50] |
| S181 | Dissociation from PM | [70] | |
| Acetylation | K104 and K147 | Inconclusive | [73,74,75] |
| Ubiquitination | K104 | No effect | [78] |
| K128 | Increased signaling | [77] | |
| K147 | Increased signaling | [76] | |
| Nitrosylation | C118 | Increased signaling if the mutant KRAS allele is depleted | [79] |
| KRAS Mutant | Tumor Region | Primary Resistance | Acquired Resistance | References |
|---|---|---|---|---|
| G12C | NSCLC, CRC, and appendiceal cancer |
|
| [113,114] |
| G12C | PDAC |
|
| [115] |
| G12D | PDAC |
|
| [115] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karki, R.; Chen, R.; Pan, S. Proteomic Perspectives on KRAS-Driven Cancers and Emerging Therapeutic Approaches. Curr. Oncol. 2025, 32, 614. https://doi.org/10.3390/curroncol32110614
Karki R, Chen R, Pan S. Proteomic Perspectives on KRAS-Driven Cancers and Emerging Therapeutic Approaches. Current Oncology. 2025; 32(11):614. https://doi.org/10.3390/curroncol32110614
Chicago/Turabian StyleKarki, Ramesh, Ru Chen, and Sheng Pan. 2025. "Proteomic Perspectives on KRAS-Driven Cancers and Emerging Therapeutic Approaches" Current Oncology 32, no. 11: 614. https://doi.org/10.3390/curroncol32110614
APA StyleKarki, R., Chen, R., & Pan, S. (2025). Proteomic Perspectives on KRAS-Driven Cancers and Emerging Therapeutic Approaches. Current Oncology, 32(11), 614. https://doi.org/10.3390/curroncol32110614




