ITGAV Promotes the Progression of Head and Neck Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of ITGAV Gene Expression in HNSCC and Its Clinical Significance
2.2. Cell Culture
2.3. Quantitative Polymerase Chain Reaction (qPCR)
2.4. Western Blotting
2.5. siRNA Knockdown of ITGAV in UM1 and UMSCC5 Cells
2.6. Transwell Migration Assay
2.7. Transwell Invasion Assay
2.8. Colony Formation Assay
2.9. CCK-8 Assay
2.10. Chromatin Immunoprecipitation (ChIP)
2.11. Statistical Analysis
3. Results
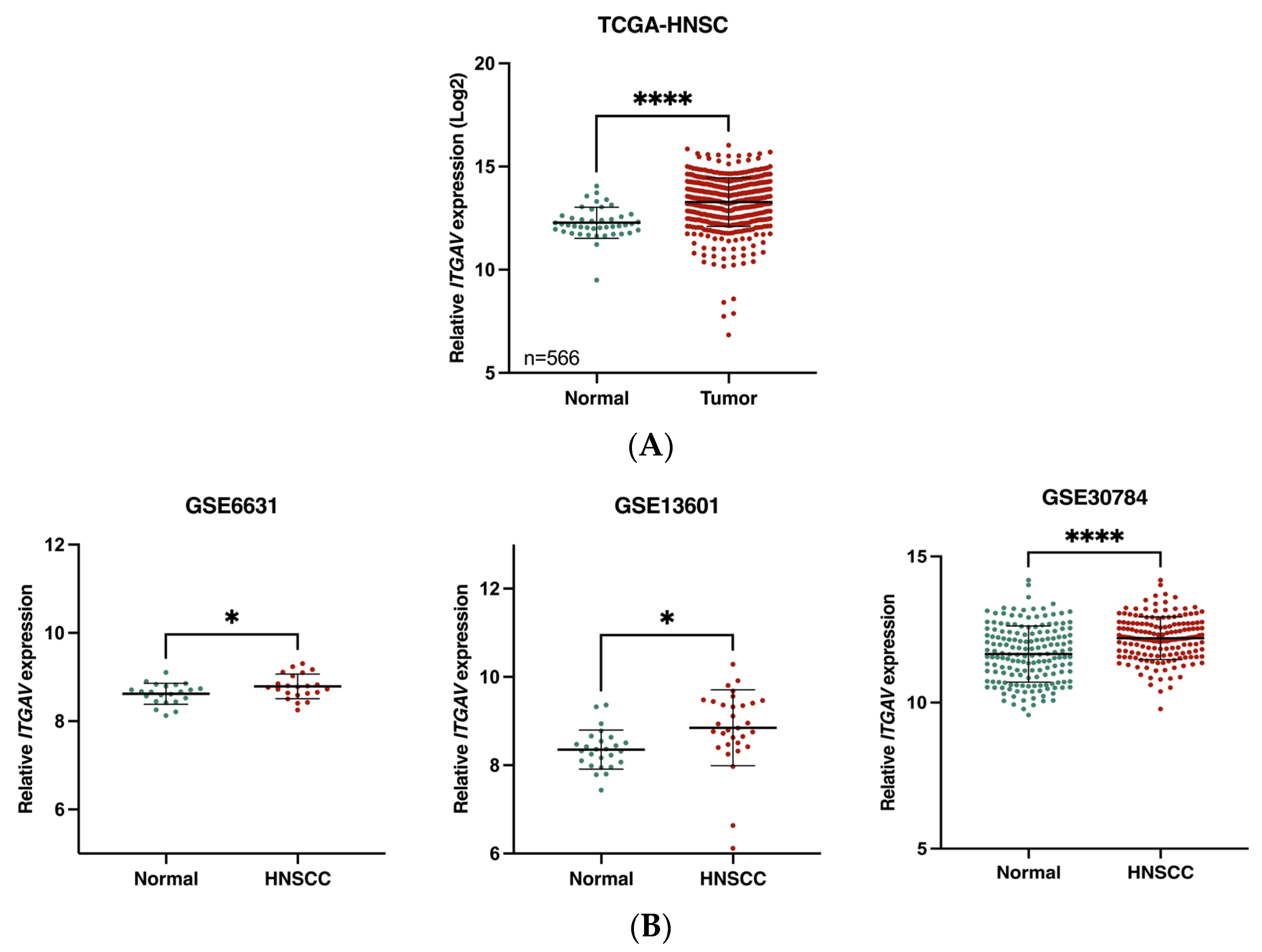
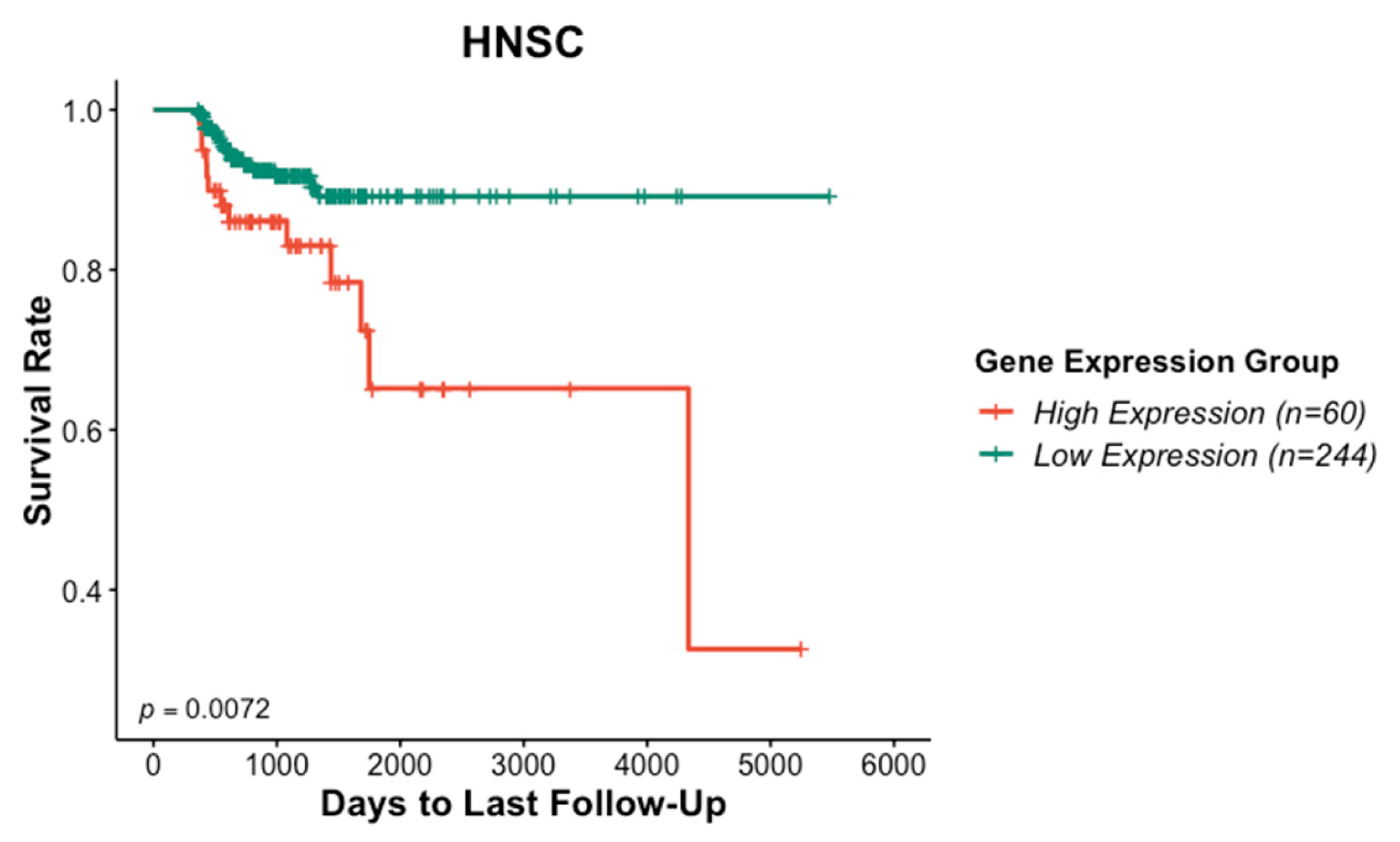
3.1. Association of ITGAV Overexpression with Poor Prognosis in HNSCC
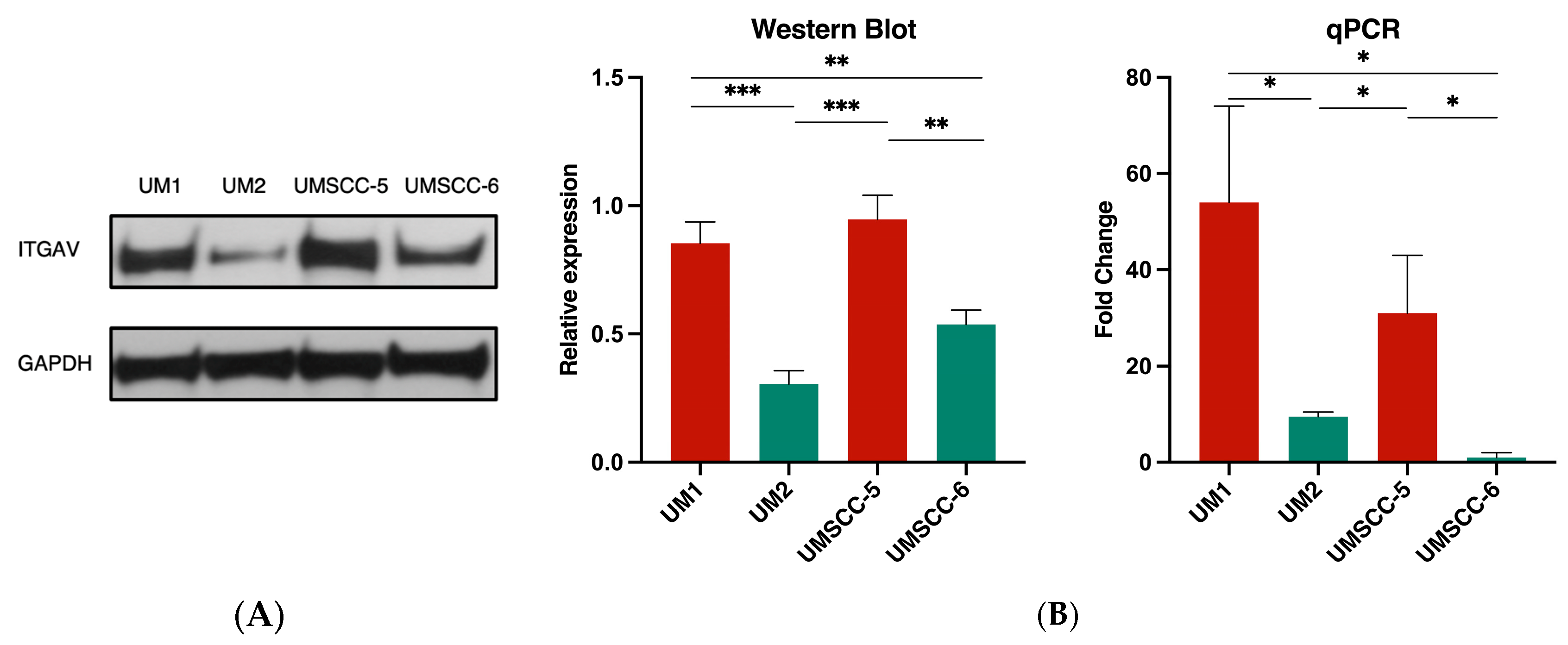
3.2. ITGAV Expression in High- and Low-Invasive HNSCC Cell Lines
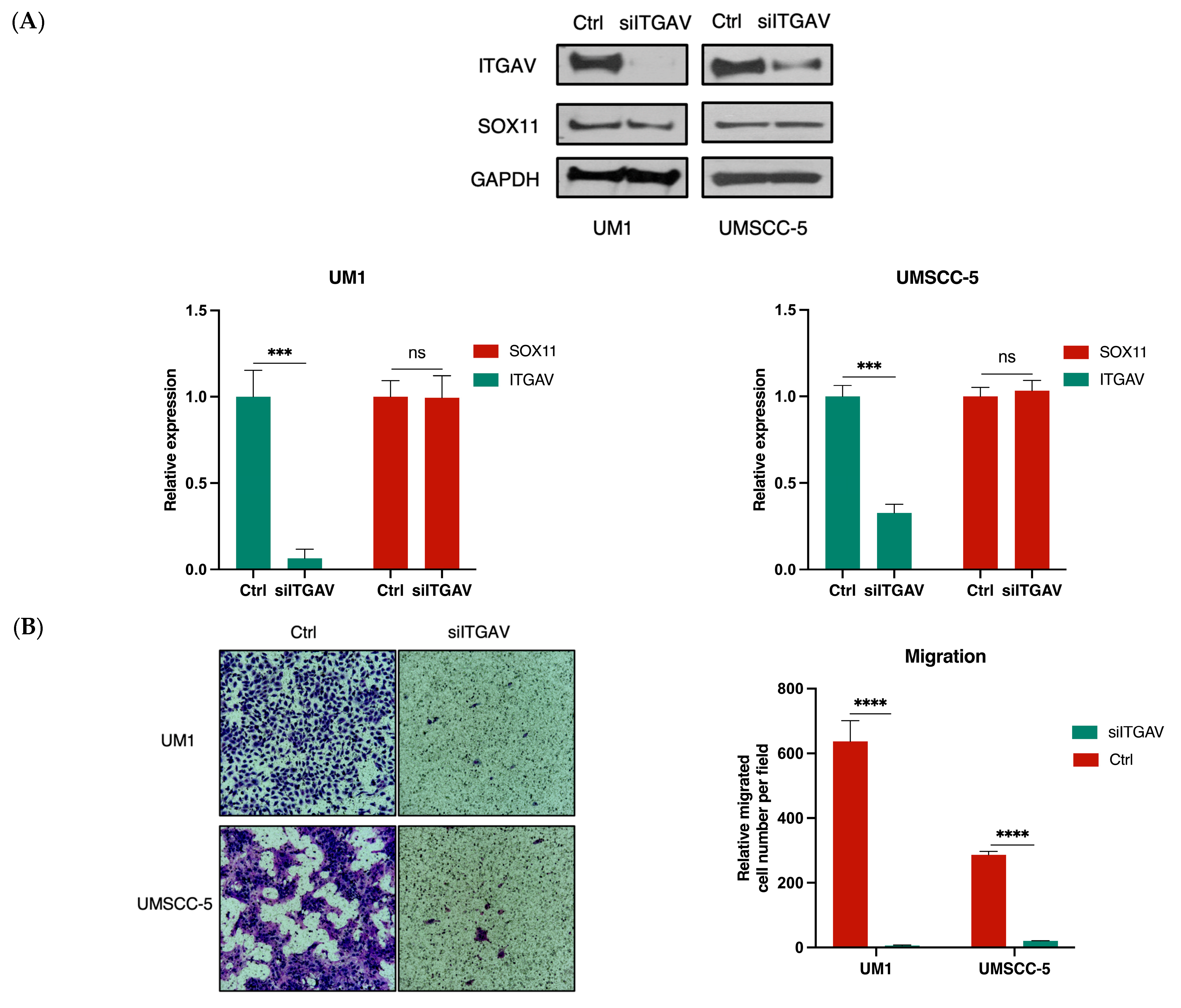
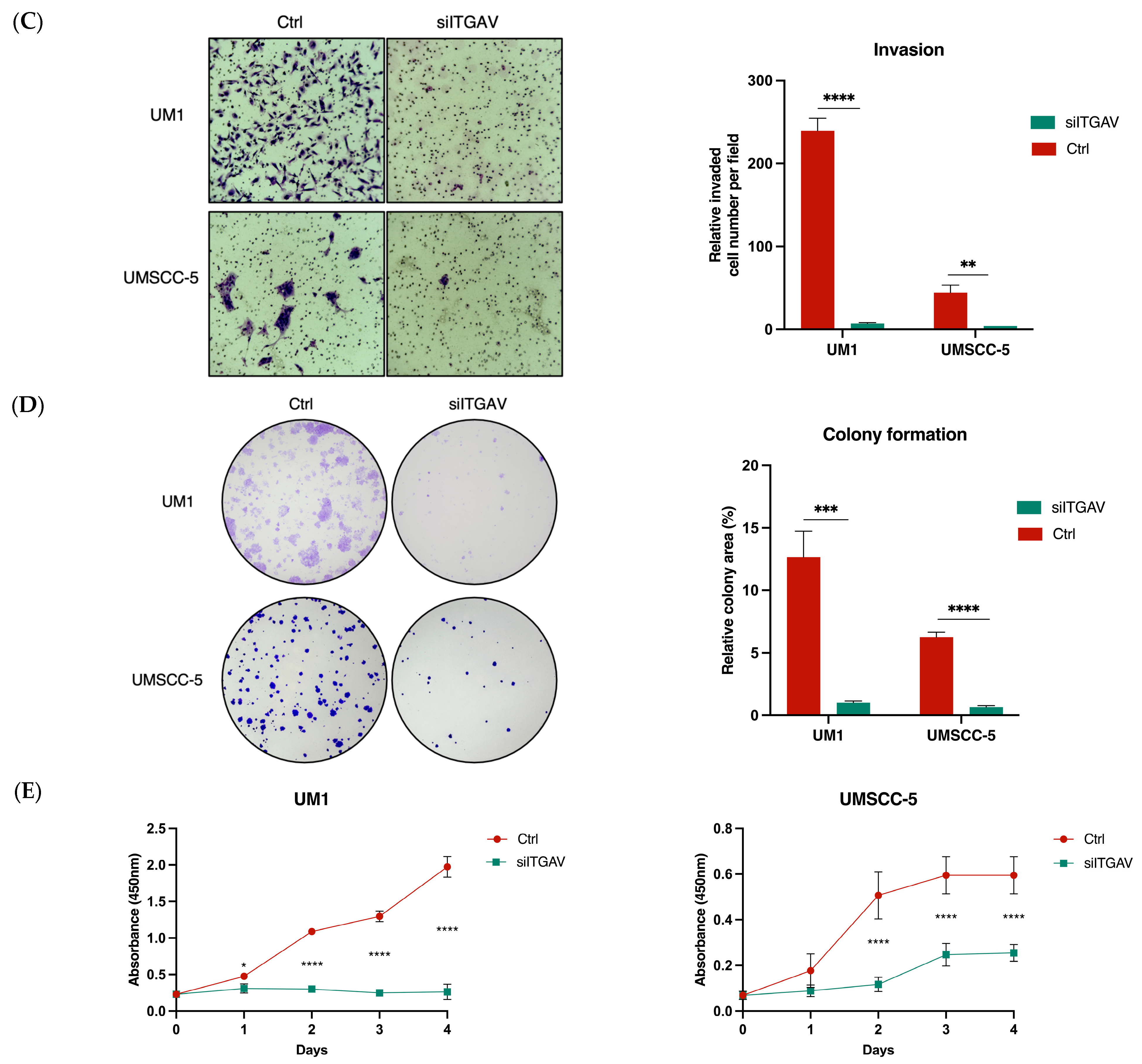
3.3. Knockdown of ITGAV in UM1 and UMSCC-5 Cells Inhibits Cell Migration, Invasion, Viability, and Proliferation
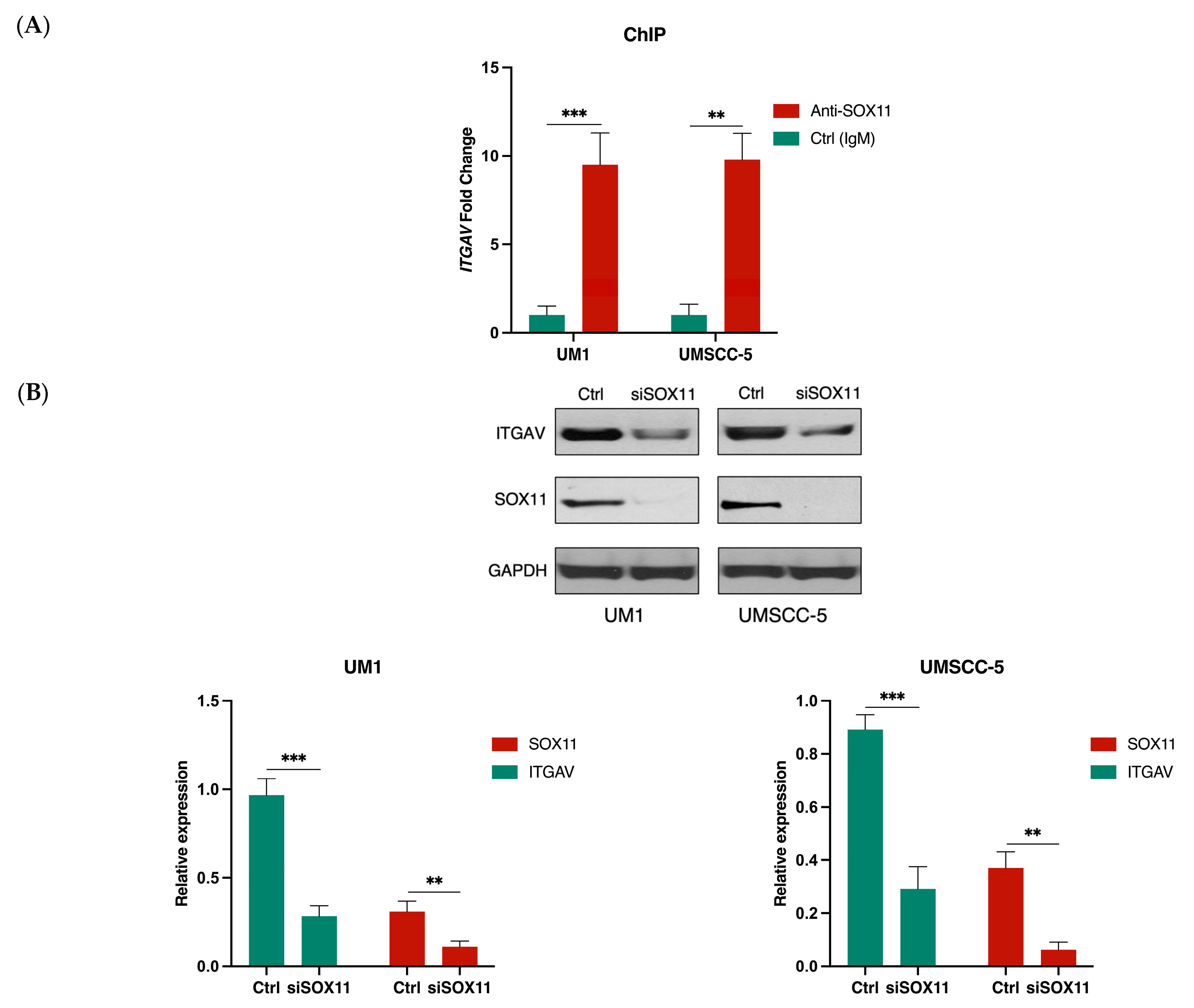
3.4. SOX11 Regulates ITGAV Expression in HNSCC Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Marur, S.; Forastiere, A.A. Head and neck cancer: Changing epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2008, 83, 489–501. [Google Scholar] [CrossRef]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Viana, L.D.; Affonso, R.J., Jr.; Silva, S.R.; Denadai, M.V.; Matos, D.; Salinas de Souza, C.; Waisberg, J. Relationship between the expression of the extracellular matrix genes SPARC, SPP1, FN1, ITGA5 and ITGAV and clinicopathological parameters of tumor progression and colorectal cancer dissemination. Oncology 2013, 84, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Versatility, modulation, and signaling in cell adhesion. Cell 1992, 69, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Jin, H.; Varner, J. Integrins: Roles in cancer development and as treatment targets. Br. J. Cancer 2004, 90, 561–565. [Google Scholar] [CrossRef]
- Hynes, R.O. A reevaluation of integrins as regulators of angiogenesis. Nat. Med. 2002, 8, 918–921. [Google Scholar] [CrossRef]
- Schwartz, M.A.; Ginsberg, M.H. Networks and crosstalk: Integrin signalling spreads. Nat. Cell Biol. 2002, 4, E65–E68. [Google Scholar] [CrossRef]
- Hood, J.D.; Cheresh, D.A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2002, 2, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.H.; Carman, C.V.; Springer, T.A. Structural basis of integrin regulation and signaling. Ann. Rev. Immunol. 2007, 25, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.D.; Byron, A.; Humphries, M.J. Integrin ligands at a glance. J. Cell Sci. 2006, 119 Pt 19, 3901–3903. [Google Scholar] [CrossRef] [PubMed]
- Weis, S.M.; Cheresh, D.A. αV integrins in angiogenesis and cancer. Cold Spring Harb. Perspect. Med. 2011, 1, a006478. [Google Scholar] [CrossRef] [PubMed]
- Aoudjit, F.; Vuori, K. Integrin signaling in cancer cell survival and chemoresistance. Chemother. Res. Pract. 2012, 2012, 283181. [Google Scholar] [CrossRef] [PubMed]
- Danen, E.H.; Berge, P.T.; Muijen, G.V. Emergence of alpha 5 beta 1 fibronectin- and alpha v beta 3 vitronectin-receptor expression in melanocytic tumour progression. Histopathology 1994, 24, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Hosotani, R.; Kawaguchi, M.; Masui, T.; Koshiba, T.; Ida, J.; Fujimoto, K.; Wada, M.; Doi, R. Expression of integrin alphaVbeta3 in pancreatic carcinoma: Relation to MMP-2 activation and lymph node metastasis. Pancreas 2002, 25, e30–e35. [Google Scholar] [CrossRef]
- Bauer, K.; Mierke, C.; Behrens, J. Expression profiling reveals genes associated with transendothelial migration of tumor cells: A functional role for alphavbeta3 integrin. Int. J. Cancer 2007, 121, 1910–1918. [Google Scholar] [CrossRef]
- Brooks, P.C.; Clark, R.A.; Cheresh, D.A. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science 1994, 264, 569–571. [Google Scholar] [CrossRef]
- Lu, X.; Lu, D.; Scully, M.; Kakkar, V. The role of integrins in cancer and the development of anti-integrin therapeutic agents for cancer therapy. Perspect. Med. Chem. 2008, 2, 57–73. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Pytela, R.; Pierschbacher, M.D.; Ruoslahti, E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell 1985, 40, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Penzo-Méndez, A.I. Critical roles for SoxC transcription factors in development and cancer. Int. J. Biochem. Cell Biol. 2010, 42, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Dy, P.; Penzo-Mendez, A.; Wang, H.; Pedraza, C.E.; Macklin, W.B.; Lefebvre, V. The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008, 36, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.M.; Oliemuller, E.; Howard, B.A. Regulatory roles for SOX11 in development, stem cells and cancer. Semin. Cancer Biol. 2020, 67, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ji, E.H.; Zhao, X.; Cui, L.; Misuno, K.; Guo, M.; Huang, Z.; Chen, X.; Hu, S. Sox11 promotes head and neck cancer progression via the regulation of SDCCAG8. J. Exp. Clin. Cancer Res. 2019, 38, 138. [Google Scholar] [CrossRef]
- Chin, D.; Boyle, G.M.; Porceddu, S.; Theile, D.R.; Parsons, P.G.; Coman, W.B. Head and neck cancer: Past, present and future. Expert. Rev. Anticancer. Ther. 2006, 6, 1111–1118. [Google Scholar] [CrossRef]
- Gourin, C.G.; McAfee, W.J.; Neyman, K.M.; Howington, J.W.; Podolsky, R.H.; Terris, D.J. Effect of comorbidity on quality of life and treatment selection in patients with squamous cell carcinoma of the head and neck. Laryngoscope 2005, 115, 1371–1375. [Google Scholar] [CrossRef]
- Gourin, C.G.; Boyce, B.J.; Vaught, C.C.; Burkhead, L.M.; Podolsky, R.H. Effect of comorbidity on post-treatment quality of life scores in patients with head and neck squamous cell carcinoma. Laryngoscope 2009, 119, 907–914. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Felding-Habermann, B.; Mueller, B.; Romerdahl, C.A.; Cheresh, D.A. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J. Clin. Investig. 1992, 89, 2018–2022. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Ren, Y.; Geng, J.; He, X.; Ban, C.; Wang, S.; Jiang, D.; Luo, S.; Chen, Q.; Liu, M. Idiopathic Pulmonary Fibrosis Registry China study (PORTRAY): Protocol for a prospective, multicentre registry study. BMJ Open 2020, 10, e036809. [Google Scholar] [CrossRef] [PubMed]
- Abeysekera, K.W.; Fernandes, G.S.; Hammerton, G.; Portal, A.J.; Gordon, F.H.; Heron, J.; Hickman, M. Prevalence of steatosis and fibrosis in young adults in the UK: A population-based study. Lancet Gastroenterol. Hepatol. 2020, 5, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X. Diversity and Complexity in DNA Recognition by Transcription Factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Srivastava, Y.; Jauch, R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017, 63, 2–12. [Google Scholar] [CrossRef]
- Jauch, R.; Ng, C.K.; Narasimhan, K.; Kolatkar, P.R. The crystal structure of the Sox4 HMG domain–DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem. J. 2012, 443, 39–47. [Google Scholar] [CrossRef]
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ITGAV Promoter 1 | TGCCCTGCGAATCCTTTCTT | CGTGTTTCTGCTGCTTAGCC |
| ITGAV Promoter 2 | GCCTTATTTCACCGGTGTGC | AAGGATTCGCAGGGCAAAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Barrett, J.G.; Peng, J.; Li, S.; Messadi, D.; Hu, S. ITGAV Promotes the Progression of Head and Neck Squamous Cell Carcinoma. Curr. Oncol. 2024, 31, 1311-1322. https://doi.org/10.3390/curroncol31030099
Xu L, Barrett JG, Peng J, Li S, Messadi D, Hu S. ITGAV Promotes the Progression of Head and Neck Squamous Cell Carcinoma. Current Oncology. 2024; 31(3):1311-1322. https://doi.org/10.3390/curroncol31030099
Chicago/Turabian StyleXu, Lingyi, Jeremy G Barrett, Jiayi Peng, Suk Li, Diana Messadi, and Shen Hu. 2024. "ITGAV Promotes the Progression of Head and Neck Squamous Cell Carcinoma" Current Oncology 31, no. 3: 1311-1322. https://doi.org/10.3390/curroncol31030099
APA StyleXu, L., Barrett, J. G., Peng, J., Li, S., Messadi, D., & Hu, S. (2024). ITGAV Promotes the Progression of Head and Neck Squamous Cell Carcinoma. Current Oncology, 31(3), 1311-1322. https://doi.org/10.3390/curroncol31030099





