Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Specimens
2.2. RNA Isolation and Droplet Digital PCR (ddPCR) Analysis of miRNAs
2.3. DNA Isolation and ddPCR Analysis of Bacterial Abundances
2.4. Statistical Analysis
3. Results
3.1. The Demographic and Clinical Variables of Cases and Controls
3.2. The Diagnostic Performance of Plasma miRNA and Sputum Bacterial Biomarkers in Distinguishing between Lung Cancer Patients and Cancer-Free Controls
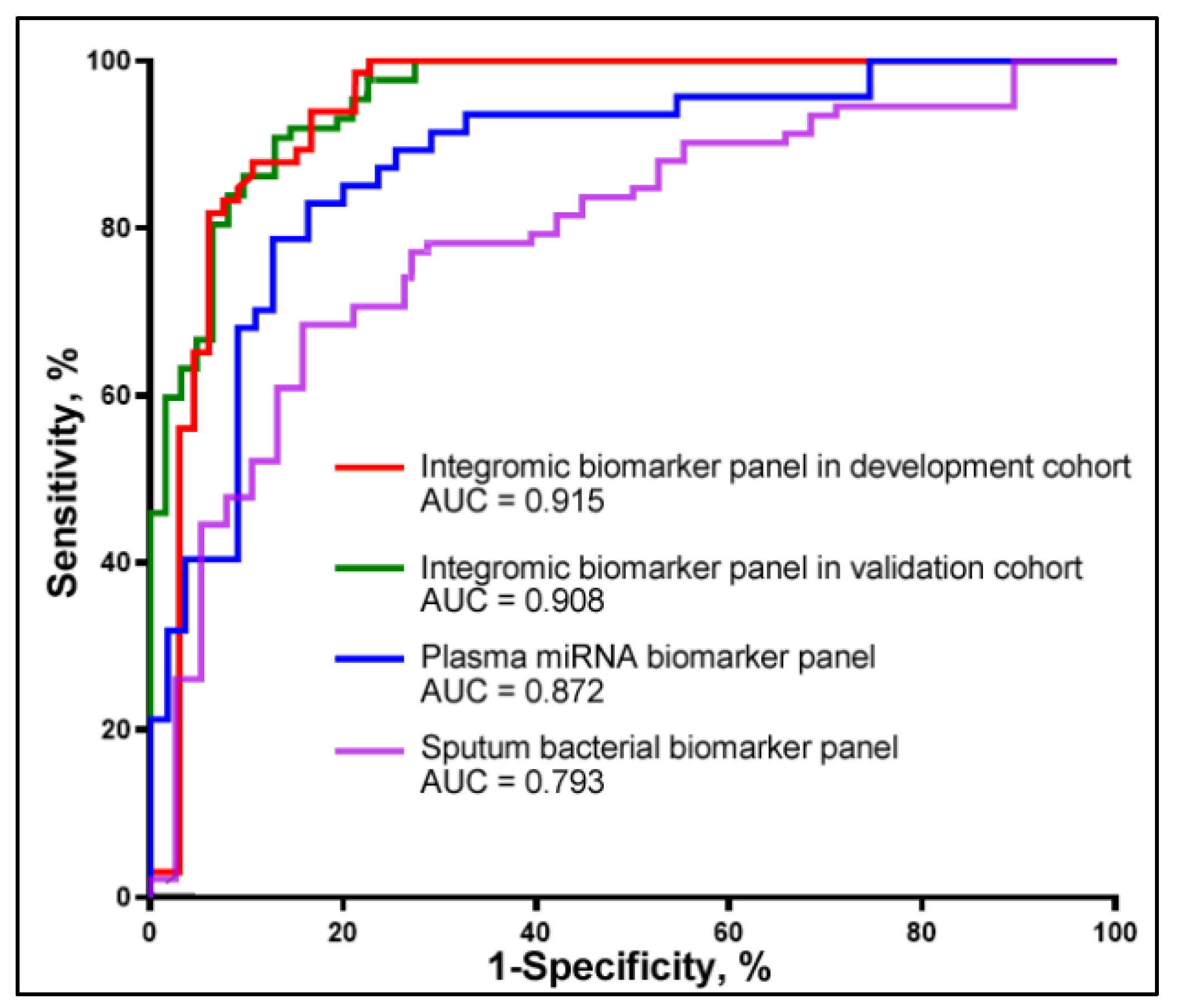
3.3. Combined Use of Plasma miRNA and Sputum Bacterial Biomarkers for Early Detection of NSCLC
3.4. Validation of the Integromic Biomarker Panel in a Different Cohort for the Diagnosis of NSCLC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [PubMed]
- Wilson, D.O. Lung cancer screening with low-dose CT (LDCT) is ready for prime time in the USA. Evid. Based Med. 2014, 19, 150. [Google Scholar] [CrossRef]
- Marcus, P.M. Lung cancer screening with low dose computed tomography (LDCT): Looking back and moving forward. Ann. Transl. Med. 2015, 3, S41. [Google Scholar]
- Oudkerk, M.; Liu, S.; Heuvelmans, M.A.; Walter, J.E.; Field, J.K. Lung cancer LDCT screening and mortality reduction - evidence, pitfalls and future perspectives. Nat. Rev. Clin. Oncol. 2021, 18, 135–151. [Google Scholar] [CrossRef]
- Pelosi, G.; Sonzogni, A.; Veronesi, G.; De Camilli, E.; Maisonneuve, P.; Spaggiari, L.; Manzotti, M.; Masullo, M.; Taliento, G.; Fumagalli, C.; et al. Pathologic and molecular features of screening low-dose computed tomography (LDCT)-detected lung cancer: A baseline and 2-year repeat study. Lung Cancer 2008, 62, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, H.; Shen, Y.; Li, W.; Chen, Y.; Wang, H. Low-dose computed tomography (LDCT) versus other cancer screenings in early diagnosis of lung cancer: A meta-analysis. Medicine 2018, 97, e11233. [Google Scholar] [CrossRef]
- Hashemi, M.; Khosroshahi, E.M.; Chegini, M.K.; Abedi, M.; Matinahmadi, A.; Hosnarody, Y.S.D.; Rezaei, M.; Saghari, Y.; Fattah, E.; Abdi, S.; et al. miRNAs and exosomal miRNAs in lung cancer: New emerging players in tumor progression and therapy response. Pathol. Res. Pract. 2023, 251, 154906. [Google Scholar] [CrossRef]
- Hajipour, S.; Hosseini, S.M.; Irani, S.; Tavallaie, M. Identification of novel potential drugs and miRNAs biomarkers in lung cancer based on gene co-expression network analysis. Genomics Inform. 2023, 21, e38. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Burdennyy, A.M.; Loginov, V.I.; Dmitriev, A.A.; Pronina, I.V.; Morozov, S.G. Various LncRNA Mechanisms in Gene Regulation Involving miRNAs or RNA-Binding Proteins in Non-Small-Cell Lung Cancer: Main Signaling Pathways and Networks. Int. J. Mol. Sci. 2023, 24, 13617. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, J.; Chen, Y.; Ma, S.; Bai, W.; Peng, Y.; Shi, G. Identification of serum MiRNAs as candidate biomarkers for non-small cell lung cancer diagnosis. BMC Pulm. Med. 2022, 22, 479. [Google Scholar] [CrossRef]
- Ma, J.; Mannoor, K.; Gao, L.; Tan, A.; Guarnera, M.A.; Zhan, M.; Shetty, A.; Stass, S.A.; Xing, L.; Jiang, F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol. Oncol. 2014, 8, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Todd, N.W.; Qiu, Q.; Fan, T.; Zhao, R.Y.; Rodgers, W.H.; Fang, H.B.; Katz, R.L.; Stass, S.A.; Jiang, F. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin. Cancer Res. 2007, 13, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Glyn, T.; Purcell, R. Circulating Bacterial DNA: A New Paradigm for Cancer Diagnostics. Front. Med. 2022, 9, 831096. [Google Scholar] [CrossRef]
- Li, N.; Zhou, H.; Holden, V.K.; Deepak, J.; Dhilipkannah, P.; Todd, N.W.; Stass, S.A.; Jiang, F. Streptococcus pneumoniae promotes lung cancer development and progression. iScience 2023, 26, 105923. [Google Scholar] [CrossRef]
- Mao, Q.; Jiang, F.; Yin, R.; Wang, J.; Xia, W.; Dong, G.; Ma, W.; Yang, Y.; Xu, L.; Hu, J. Interplay between the lung microbiome and lung cancer. Cancer Lett 2018, 415, 40–48. [Google Scholar] [CrossRef]
- Zhou, H.; Liao, J.; Leng, Q.; Chinthalapally, M.; Dhilipkannah, P.; Jiang, F. Circulating Bacterial DNA as Plasma Biomarkers for Lung Cancer Early Detection. Microorganisms 2023, 11, 582. [Google Scholar] [CrossRef]
- Leng, Q.; Holden, V.K.; Deepak, J.; Todd, N.W.; Jiang, F. Microbiota Biomarkers for Lung Cancer. Diagnostics 2021, 11, 407. [Google Scholar] [CrossRef]
- Shi, H.; Bi, H.; Sun, X.; Dong, H.; Jiang, Y.; Mu, H.; Li, W.; Liu, G.; Gao, R.; Su, J. Tubeimoside-1 inhibits the proliferation and metastasis by promoting miR-126-5p expression in non-small cell lung cancer cells. Oncol. Lett. 2018, 16, 3126–3134. [Google Scholar] [CrossRef]
- Huang, B.; Wu, G.; Peng, C.; Peng, X.; Huang, M.; Ding, J.; Zhang, H.; Wu, X. miR-126 regulates the proliferation, migration, invasion, and apoptosis of non-small lung cancer cells via AKT2/HK2 axis. IUBMB Life 2023, 75, 186–195. [Google Scholar] [CrossRef]
- Zheng, W.; Zhou, Y.; Lu, J.; Xu, H.; Lei, L.; Chen, C.; Zhao, J.; Xu, L. The prognostic value of miR-126 expression in non-small-cell lung cancer: A meta-analysis. Cancer Cell Int. 2017, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Jung, S.B.; Kim, J.S.; Roh, M.S.; Lee, J.H.; Lee, E.H.; Lee, H.W. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows. Arch. 2014, 465, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, H.; Long, L.; Hui, L.; Chen, H.; Wang, X.; Shen, H.; Xu, W. miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta. Biochim. Biophys Sin. 2012, 44, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Peng, X.C.; Zheng, X.L.; Wang, J.; Qin, Y.W. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 2009, 66, 169–175. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Cheng, J.; Yu, X. Effects of miR-126 on the STAT3 signaling pathway and the regulation of malignant behavior in lung cancer cells. Oncol. Lett. 2018, 15, 8412–8416. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Zhang, J.X.; Yang, J.J.; Wei, Y.B.; Peng, J.F.; Fu, C.J.; Huang, M.H.; Wang, R.; Wang, P.Y.; Sun, G.B.; et al. MiR-205-5p promotes lung cancer progression and is valuable for the diagnosis of lung cancer. Thorac. Cancer 2022, 13, 832–843. [Google Scholar] [CrossRef]
- Tsay, J.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, H.; Zhang, J.; Shen, L.; Yang, J.; Wang, Y.; Ma, J.; Zhuan, B. miR-210-3p Promotes Lung Cancer Development and Progression by Modulating USF1 and PCGF3. Onco. Targets Ther. 2021, 14, 3687–3700. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, X. Association of Exosomal miR-210 with Signaling Pathways Implicated in Lung Cancer. Genes 2021, 12, 1248. [Google Scholar] [CrossRef]
- Eilertsen, M.; Andersen, S.; Al-Saad, S.; Richardsen, E.; Stenvold, H.; Hald, S.M.; Al-Shibli, K.; Donnem, T.; Busund, L.T.; Bremnes, R.M. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer 2014, 83, 272–278. [Google Scholar] [CrossRef]
- Fan, J.; Xu, G.; Chang, Z.; Zhu, L.; Yao, J. miR-210 transferred by lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin. Sci. 2020, 134, 807–825. [Google Scholar] [CrossRef] [PubMed]
- He, R.Q.; Cen, W.L.; Cen, J.M.; Cen, W.N.; Li, J.Y.; Li, M.W.; Gan, T.Q.; Hu, X.H.; Chen, G. Clinical Significance of miR-210 and its Prospective Signaling Pathways in Non-Small Cell Lung Cancer: Evidence from Gene Expression Omnibus and the Cancer Genome Atlas Data Mining with 2763 Samples and Validation via Real-Time Quantitative PCR. Cell Physiol. Biochem. 2018, 46, 925–952. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Peng, Q.; Zhu, J.; Shen, Y.; Lin, K.; Shen, Y.; Zhu, Y. Identification of miR-210 and combination biomarkers as useful agents in early screening non-small cell lung cancer. Gene 2020, 729, 144225. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I.; et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018, 19, 123. [Google Scholar] [CrossRef]
- Yan, X.; Yang, M.; Liu, J.; Gao, R.; Hu, J.; Li, J.; Zhang, L.; Shi, Y.; Guo, H.; Cheng, J.; et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015, 5, 3111–3122. [Google Scholar]
- Gomes, S.; Cavadas, B.; Ferreira, J.C.; Marques, P.I.; Monteiro, C.; Sucena, M.; Sousa, C.; Vaz Rodrigues, L.; Teixeira, G.; Pinto, P.; et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019, 9, 12838. [Google Scholar] [CrossRef]

| miRNAs or Bacteria | Mean (SD) in Cancer-Free Smokers | Mean (SD) NSCLC | p | AUC (95% CI) | Sensitivity % | Specificity % |
|---|---|---|---|---|---|---|
| miR-126-3p | 0.269 (0.419) | 0.608 (1.393) | 0.0003 | 0.702 (0 0.606 to 0.798) | 68.97 | 66.63 |
| miR-205-5p | 0.0729 (0.183) | 0.2276 (1.013) | 0.0001 | 0.812 (0.735 to 0.889) | 69.26 | 82.26 |
| miR-210-3p | 3.700 (2.697) | 7.802 (13.689) | <0.001 | 0.806 (0.726 to 0.885) | 77.59 | 78.69 |
| Acidovorax | 1.106 (0.537) | 1.572 (0.786) | 0.0010 | 0.680 (0.581 to 0.779) | 62.07 | 79.03 |
| Capnocytophaga | 3.070 (0.957) | 3.667 (0.737) | 0.0009 | 0.656 (0.558 to 0.756) | 63.79 | 61.29 |
| Streptococus | 1.312 (0.662) | 2.111 (1.065) | <0.0001 | 0.729 (0.639 to 0.820) | 70.86 | 54.29 |
| Veillonella | 1.010 (0.436) | 1.778 (0.737) | 0.0004 | 0.678 (0.579 to 0.776) | 68.97 | 58.06 |
| Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| Three-plasma miRNAs biomarker panel | 81.82% | 85.48% | 84.21% | 72.0% | 83.87% | 80.46% | 79.31% | 85.48% | 82.50% |
| Four-sputum bacterial biomarker panel | 66.67% | 72.58% | 70.52 | 76.0% | 85.48% | 82.76% | 68.97% | 79.03% | 74.17% |
| The integromic biomarker panel | 84.85% | 90.32% | 88.42% | 84.0% | 90.32% | 88.51% | 84.48% | 90.32% | 87.50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhilipkannah, P.; Sachdeva, A.; Holden, V.K.; Jiang, F. Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA. Curr. Oncol. 2024, 31, 5949-5959. https://doi.org/10.3390/curroncol31100444
Dhilipkannah P, Sachdeva A, Holden VK, Jiang F. Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA. Current Oncology. 2024; 31(10):5949-5959. https://doi.org/10.3390/curroncol31100444
Chicago/Turabian StyleDhilipkannah, Pushpa, Ashutosh Sachdeva, Van K. Holden, and Feng Jiang. 2024. "Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA" Current Oncology 31, no. 10: 5949-5959. https://doi.org/10.3390/curroncol31100444
APA StyleDhilipkannah, P., Sachdeva, A., Holden, V. K., & Jiang, F. (2024). Integrative Biomarker Panel for Improved Lung Cancer Diagnosis Using Plasma microRNAs and Sputum Bacterial DNA. Current Oncology, 31(10), 5949-5959. https://doi.org/10.3390/curroncol31100444





