High Expression of Fas-Associated Factor 1 Indicates a Poor Prognosis in Non-Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Preparation
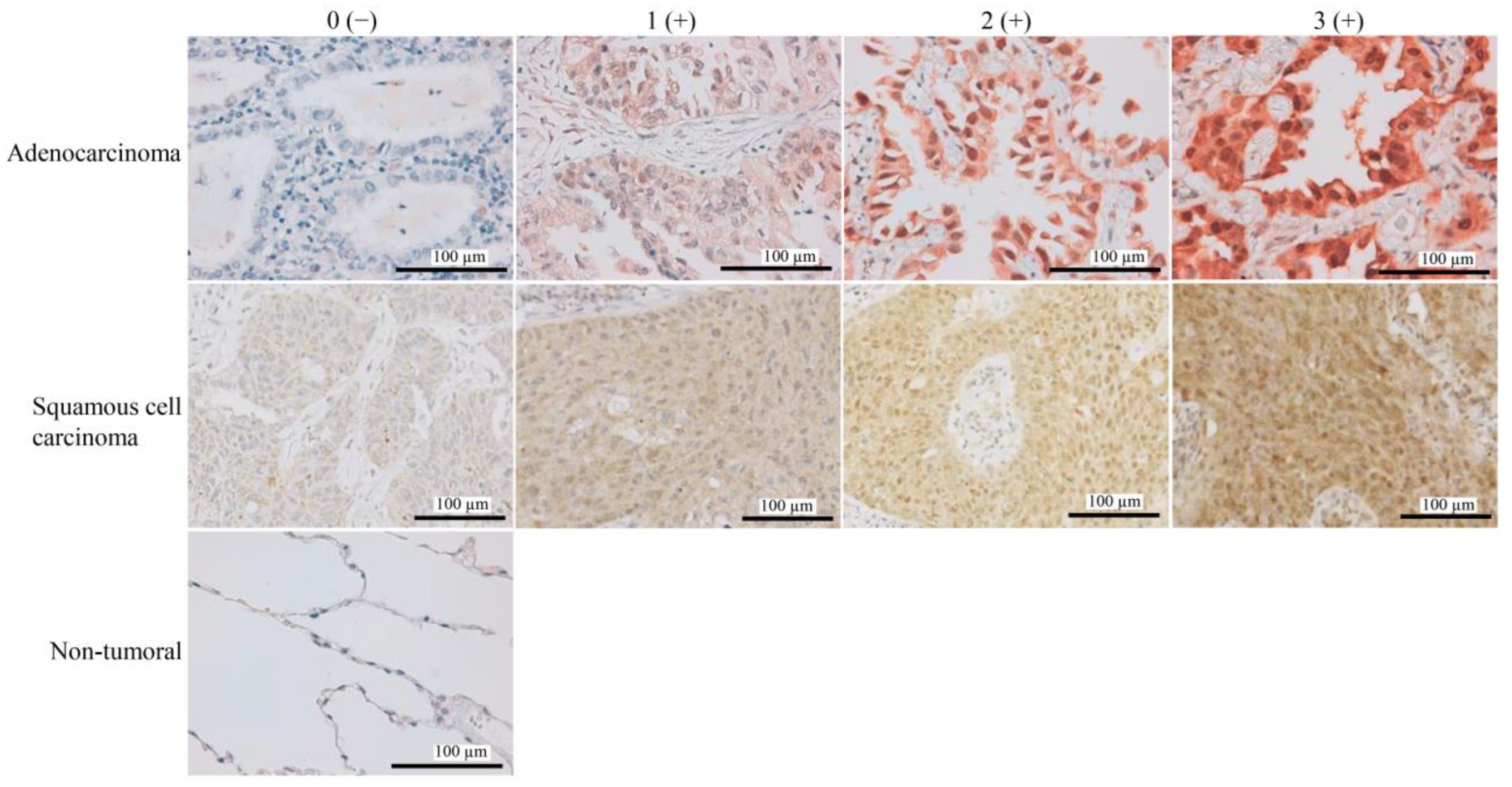
2.2. Immunohistochemistry (IHC) Analysis
2.3. Survival Analysis Using Kaplan–Meier Plotter
2.4. TUNEL Assay
2.5. Cell Culture
2.6. Small Interfering RNA (siRNA) Transfection
2.7. Plasmid Construction and Transfection
2.8. Western Blot Analysis
2.9. RNA Isolation and Quantitative Real-Time PCR (qPCR)
2.10. Immunofluorescence Analysis
2.11. Cell Viability Assay
2.12. Apoptosis Analysis
2.13. Statistical Analysis
3. Results
3.1. High FAF1 Expression Is Observed in NSCLC, but Not in Non-Tumor Lung Tissues
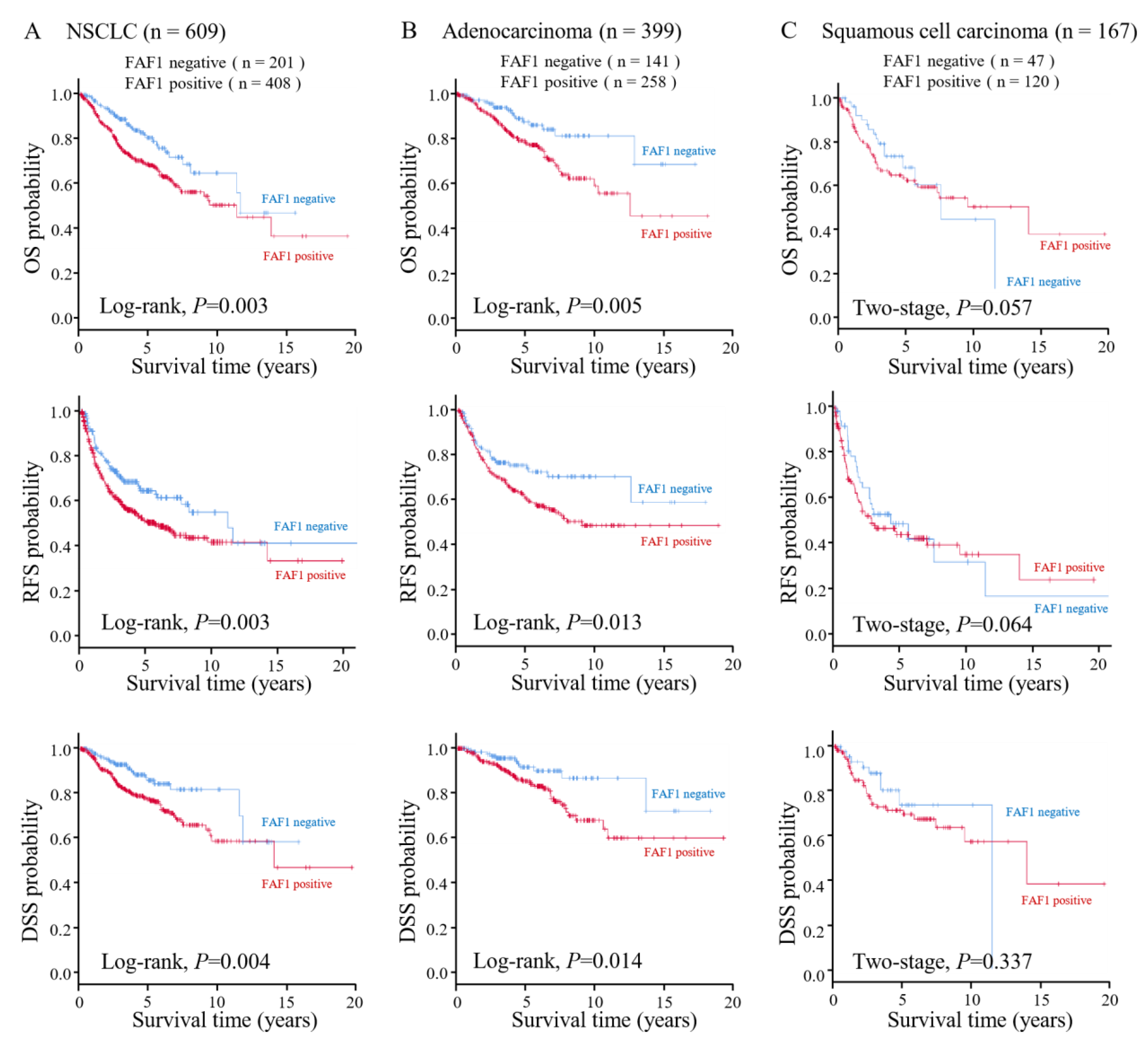
3.2. High FAF1 Expression Indicates Poor Prognosis in NSCLC
3.3. The Relationship between FAF1 Expression and Apoptosis in NSCLC
3.4. Diverse Expression of FAF1 in Lung Cancer Cell Lines
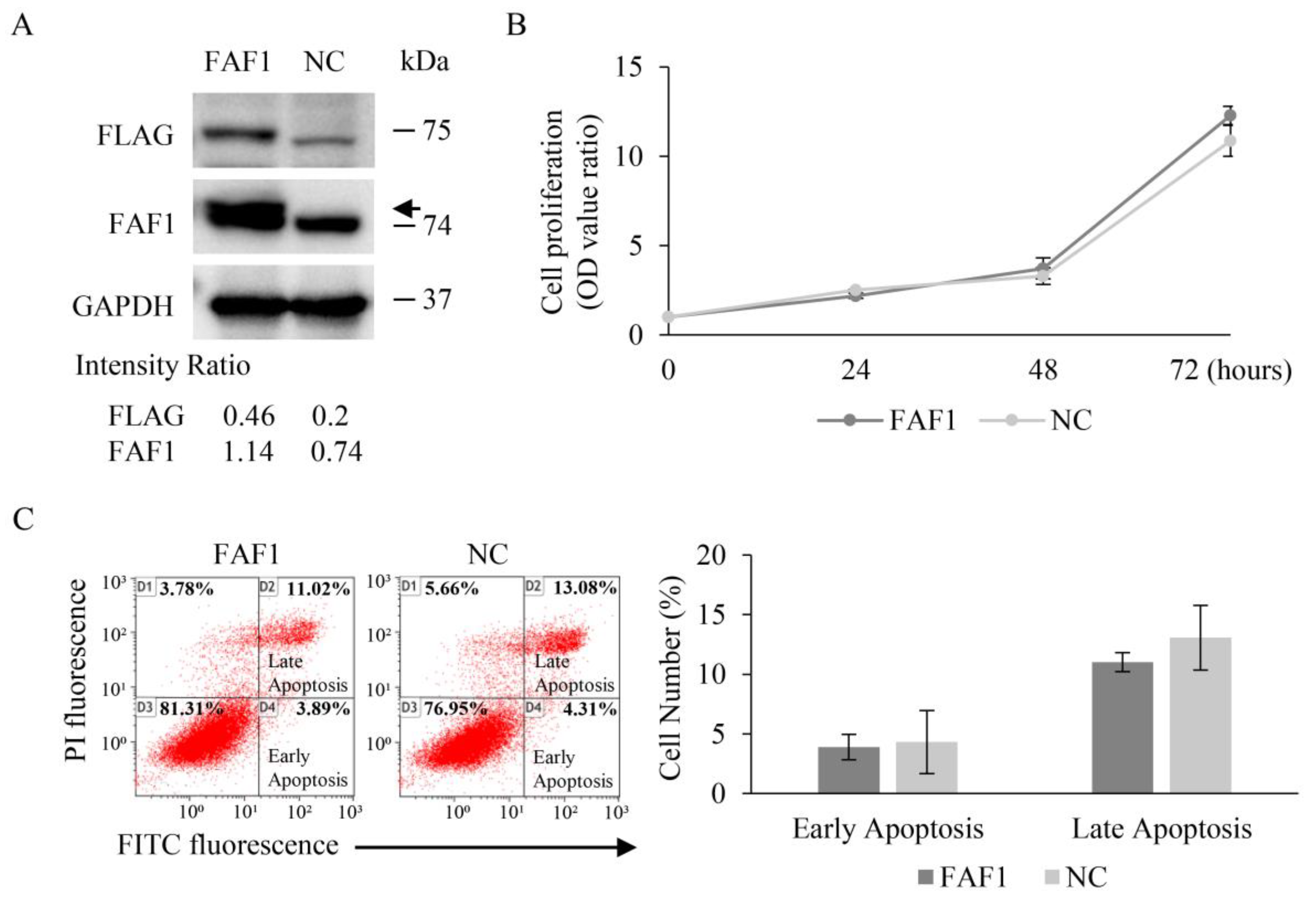
3.5. FAF1 Downregulation Inhibits the Growth of the Lung Cancer Cell Line A549
3.6. FAF1 Overexpression Has No Effect on the Growth of the Lung Cancer Cell Line H460
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Ashley, E.A. Towards Precision Medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef]
- Roos, A.; Byron, S.A. Genomics-Enabled Precision Medicine for Cancer. Cancer Treat. Res. 2019, 178, 137–169. [Google Scholar] [CrossRef]
- Chu, K.; Niu, X.; Williams, L.T. A Fas-Associated Protein Factor, FAF1, Potentiates Fas-Mediated Apoptosis. Proc. Natl. Acad. Sci. USA 1995, 92, 11894–11898. [Google Scholar] [CrossRef]
- Ryu, S.-W.; Chae, S.-K.; Lee, K.-J.; Kim, E. Identification and Characterization of Human Fas Associated Factor 1, hFAF1. Biochem. Biophys. Res. Commun. 1999, 262, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.-W.; Lee, S.-J.; Park, M.-Y.; Jun, J.; Jung, Y.-K.; Kim, E. Fas-Associated Factor 1, FAF1, Is a Member of Fas Death-Inducing Signaling Complex. J. Biol. Chem. 2003, 278, 24003–24010. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.; Li, Y.; Drabsch, Y.; Zhang, J.; van Dam, H.; Ten Dijke, P. Fas-Associated Factor 1 Is a Scaffold Protein That Promotes β-Transducin Repeat-Containing Protein (β-TrCP)-Mediated β-Catenin Ubiquitination and Degradation. J. Biol. Chem. 2012, 287, 30701–30710. [Google Scholar] [CrossRef]
- Lee, J.-J.; Kim, Y.M.; Jeong, J.; Bae, D.S.; Lee, K.-J. Ubiquitin-Associated (UBA) Domain in Human Fas Associated Factor 1 Inhibits Tumor Formation by Promoting Hsp70 Degradation. PLoS ONE 2012, 7, e40361. [Google Scholar] [CrossRef] [PubMed]
- Menges, C.W.; Altomare, D.A.; Testa, J.R. FAS-Associated Factor 1 (FAF1): Diverse Functions and Implications for Oncogenesis. Cell Cycle 2009, 8, 2528–2534. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-Y.; Ryu, S.-W.; Kim, K.D.; Lim, J.-S.; Lee, Z.-W.; Kim, E. Fas-Associated Factor-1 Mediates Chemotherapeutic-Induced Apoptosis via Death Effector Filament Formation. Int. J. Cancer 2005, 115, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.-S.; Sul, J.-W.; Choi, B.-J.; Lee, S.-J.; Suh, J.-H.; Kim, N.-S.; Kim, W.H.; Lim, D.-S.; Lee, C.-W.; Kim, E. Negative Feedback Regulation of Aurora-A via Phosphorylation of Fas-Associated Factor-1. J. Biol. Chem. 2008, 283, 32344–32351. [Google Scholar] [CrossRef]
- De Zio, D.; Ferraro, E.; D’Amelio, M.; Simoni, V.; Bordi, M.; Soroldoni, D.; Berghella, L.; Meyer, B.I.; Cecconi, F. Faf1 Is Expressed during Neurodevelopment and Is Involved in Apaf1-Dependent Caspase-3 Activation in Proneural Cells. Cell. Mol. Life Sci. 2008, 65, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.A.; Menges, C.W.; Pei, J.; Zhang, L.; Skele-Stump, K.L.; Carbone, M.; Kane, A.B.; Testa, J.R. Activated TNF-α/NF-κB Signaling via down-Regulation of Fas-Associated Factor 1 in Asbestos-Induced Mesotheliomas from Arf Knockout Mice. Proc. Natl. Acad. Sci. USA 2009, 106, 3420–3425. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.-C.; Lee, C.-W.; Lee, Y.-S.; Lian, J.-H.; Tsai, C.-L.; Liu, Y.-P.; Wu, C.-H.; Tsai, C.-N. Prediction of Early-Stage Hepatocellular Carcinoma Using OncoScan Chromosomal Copy Number Aberration Data. World J. Gastroenterol. 2017, 23, 7818–7829. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, A.; Baudis, M.; Petersen, I.; Arreola, H.; Piña, P.; Vázquez-Ortiz, G.; Hernández, D.; González, J.; Lazos, M.; López, R.; et al. Microarray Comparative Genomic Hybridization Detection of Chromosomal Imbalances in Uterine Cervix Carcinoma. BMC Cancer 2005, 5, 77. [Google Scholar] [CrossRef]
- Beà, S.; Salaverria, I.; Armengol, L.; Pinyol, M.; Fernández, V.; Hartmann, E.M.; Jares, P.; Amador, V.; Hernández, L.; Navarro, A.; et al. Uniparental Disomies, Homozygous Deletions, Amplifications, and Target Genes in Mantle Cell Lymphoma Revealed by Integrative High-Resolution Whole-Genome Profiling. Blood 2009, 113, 3059–3069. [Google Scholar] [CrossRef]
- Bjørling-Poulsen, M.; Seitz, G.; Guerra, B.; Issinger, O.-G. The Pro-Apoptotic FAS-Associated Factor 1 Is Specifically Reduced in Human Gastric Carcinomas. Int. J. Oncol. 2003, 23, 1015–1023. [Google Scholar] [CrossRef]
- Liu, A.; Ge, L.; Ye, X.; Luo, X.; Luo, Y. Reduced FAF1 Expression and Helicobacter Infection: Correlations with Clinicopathological Features in Gastric Cancer. Gastroenterol. Res. Pract. 2012, 2012, 153219. [Google Scholar] [CrossRef]
- Wei, G.; Xu, Y.; Peng, T.; Yan, J.; Wang, Z.; Sun, Z. Sanguinarine Exhibits Antitumor Activity via Up-Regulation of Fas-Associated Factor 1 in Non-Small Cell Lung Cancer. J. Biochem. Mol. Toxicol. 2017, 31, e21914. [Google Scholar] [CrossRef]
- Weber, G.F. Molecular Analysis of a Recurrent Sarcoma Identifies a Mutation in FAF1. Sarcoma 2015, 2015, 839182. [Google Scholar] [CrossRef]
- Willett, C.G.; Chang, D.T.; Czito, B.G.; Meyer, J.; Wo, J. The Cancer genome Atlas Network Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Bonjoch, L.; Franch-Expósito, S.; Garre, P.; Belhadj, S.; Muñoz, J.; Arnau-Collell, C.; Díaz-Gay, M.; Gratacós-Mulleras, A.; Raimondi, G.; Esteban-Jurado, C.; et al. Germline Mutations in FAF1 Are Associated With Hereditary Colorectal Cancer. Gastroenterology 2020, 159, 227–240.e7. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Lee, H.-C.; Kim, J.-H.; Hewawaduge, C.Y.; Chathuranga, K.; Chathuranga, W.A.G.; Ekanayaka, P.; Wijerathne, H.M.S.M.; Kim, C.-J.; Kim, E.; et al. Fas-Associated Factor 1 Mediates NADPH Oxidase-Induced Reactive Oxygen Species Production and Proinflammatory Responses in Macrophages against Listeria Infection. PLoS Pathog. 2019, 15, e1008004. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Lee, J.-J.; Kim, H.-J.; Lee, J.Y.; Chang, J.; Lee, K.-J. Fas-Associated Factor 1 Negatively Regulates the Antiviral Immune Response by Inhibiting Translocation of Interferon Regulatory Factor 3 to the Nucleus. Mol. Cell Biol. 2016, 36, 1136–1151. [Google Scholar] [CrossRef]
- Mi, D.; Cai, C.; Zhou, B.; Liu, X.; Ma, P.; Shen, S.; Lu, W.; Huang, W. Long Non-coding RNA FAF1 Promotes Intervertebral Disc Degeneration by Targeting the Erk Signaling Pathway. Mol. Med. Rep. 2018, 17, 3158–3163. [Google Scholar] [CrossRef]
- Rezvani, K. UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer. Int. J. Mol. Sci. 2016, 17, 1724. [Google Scholar] [CrossRef]
- Eba, J.; Nakamura, K. Overview of the Ethical Guidelines for Medical and Biological Research Involving Human Subjects in Japan. Jpn. J. Clin. Oncol. 2022, 52, 539–544. [Google Scholar] [CrossRef]
- Sugimura, H.; Mori, H.; Nagura, K.; Kiyose, S.; Hong, T.; Isozaki, M.; Igarashi, H.; Shinmura, K.; Hasegawa, A.; Kitayama, Y.; et al. Fluorescence in Situ Hybridization Analysis with a Tissue Microarray: ‘FISH and Chips’ Analysis of Pathology Archives. Pathol. Int. 2010, 60, 543–550. [Google Scholar] [CrossRef]
- Inoue, Y.; Yoshimura, K.; Kurabe, N.; Kahyo, T.; Kawase, A.; Tanahashi, M.; Ogawa, H.; Inui, N.; Funai, K.; Shinmura, K.; et al. Prognostic Impact of CD73 and A2A Adenosine Receptor Expression in Non-Small-Cell Lung Cancer. Oncotarget 2017, 8, 8738–8751. [Google Scholar] [CrossRef]
- Sauter, J.L.; Dacic, S.; Galateau-Salle, F.; Attanoos, R.L.; Butnor, K.J.; Churg, A.; Husain, A.N.; Kadota, K.; Khoor, A.; Nicholson, A.G.; et al. The 2021 World Health Organization Classification of Tumors of the Pleura: Advances since the 2015 Classification. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Igarashi, H.; Sugimura, H.; Maruyama, K.; Kitayama, Y.; Ohta, I.; Suzuki, M.; Tanaka, M.; Dobashi, Y.; Kino, I. Alteration of Immunoreactivity by Hydrated Autoclaving, Microwave Treatment, and Simple Heating of Paraffin-Embedded Tissue Sections. APMIS 1994, 102, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Suzuki, Y.; Inoue, Y.; Tsuchiya, K.; Karayama, M.; Iwashita, Y.; Kahyo, T.; Kawase, A.; Tanahashi, M.; Ogawa, H.; et al. CD200 and CD200R1 Are Differentially Expressed and Have Differential Prognostic Roles in Non-Small Cell Lung Cancer. Oncoimmunology 2020, 9, 1746554. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Protein Extraction from Mammalian Tissues. In Circadian Rhythms: Methods and Protocols; Rosato, E., Ed.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2007; pp. 385–389. ISBN 978-1-59745-257-1. [Google Scholar]
- Tsuchiya, K.; Yoshimura, K.; Inoue, Y.; Iwashita, Y.; Yamada, H.; Kawase, A.; Watanabe, T.; Tanahashi, M.; Ogawa, H.; Funai, K.; et al. YTHDF1 and YTHDF2 Are Associated with Better Patient Survival and an Inflamed Tumor-Immune Microenvironment in Non–Small-Cell Lung Cancer. OncoImmunology 2021, 10, 1962656. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.-J.; Han, L.-H.; Cong, R.-S.; Liang, J. Caspase Family Proteases and Apoptosis. ABBS 2005, 37, 719–727. [Google Scholar] [CrossRef]
- Chang, H.Y.; Yang, X. Proteases for Cell Suicide: Functions and Regulation of Caspases. Microbiol. Mol. Biol. Rev. 2000, 64, 821–846. [Google Scholar] [CrossRef]
- Ye, M.-F.; Lin, D.; Li, W.-J.; Xu, H.-P.; Zhang, J. MiR-26a-5p Serves as an Oncogenic MicroRNA in Non-Small Cell Lung Cancer by Targeting FAF1. Cancer Manag. Res. 2020, 12, 7131–7142. [Google Scholar] [CrossRef]
- Buccitelli, C.; Selbach, M. mRNAs, Proteins and the Emerging Principles of Gene Expression Control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Lehmann, R. Translational Control during Developmental Transitions. Cold Spring Harb. Perspect. Biol. 2019, 11, a032987. [Google Scholar] [CrossRef]
- Abbott, A. Cell Culture: Biology’s New Dimension. Nature 2003, 424, 870–873. [Google Scholar] [CrossRef]
- Song, E.J.; Yim, S.-H.; Kim, E.; Kim, N.-S.; Lee, K.-J. Human Fas-Associated Factor 1, Interacting with Ubiquitinated Proteins and Valosin-Containing Protein, Is Involved in the Ubiquitin-Proteasome Pathway. Mol. Cell. Biol. 2005, 25, 2511–2524. [Google Scholar] [CrossRef]
- Crinò, L.; Weder, W.; van Meerbeeck, J.; Felip, E. Early Stage and Locally Advanced (Non-Metastatic) Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2010, 21, v103–v115. [Google Scholar] [CrossRef] [PubMed]
- Caballero-López, M.J.; Nieto-Díaz, M.; Yunta, M.; Reigada, D.; Muñoz-Galdeano, T.; del Águila, Á.; Navarro-Ruíz, R.; Pita-Thomas, W.; Lindholm, D.; Maza, R.M. XIAP Interacts with and Regulates the Activity of FAF1. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2017, 1864, 1335–1348. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zeng, X.; Jiang, G.; Liao, X.; Liu, C.; Li, J.; Jin, H.; Zhu, J.; Sun, H.; Wu, X.-R.; et al. XIAP BIR Domain Suppresses miR-200a Expression and Subsequently Promotes EGFR Protein Translation and Anchorage-Independent Growth of Bladder Cancer Cell. J. Hematol. Oncol. 2017, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, A.H.; Babiker, A.Y.; Anwar, S. Hesperidin, a Bioflavonoid in Cancer Therapy: A Review for a Mechanism of Action through the Modulation of Cell Signaling Pathways. Molecules 2023, 28, 5152. [Google Scholar] [CrossRef]


| All Cases | FAF1 in Nuclei | Ad | FAF1 in Nuclei | Sq | FAF1 in Nuclei | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor | Total n = 609 | Negative n = 201 (33%) | Positive n = 408 (67%) | p-Value | Total n = 399 | Negative n = 141 (35.3%) | Positive n = 258 (64.7%) | p-Value | Total n = 167 | Negative n = 47 (28.1%) | Positive n = 120 (71.9%) | p-Value |
| Mean Age (range) | 67 (23~88) | 66 (23~86) | 70 (39~88) | |||||||||
| <Mean Age | 248 (40.7) | 78 (31.4) | 170 (68.6) | 0.499 | 179 (44.9) | 62 (34.6) | 117 (65.4) | 0.791 | 83 (49.7) | 21 (25.3) | 62 (74.7) | 0.417 |
| ≥Mean Age | 361 (59.3) | 123 (34.1) | 238 (65.9) | 220 (55.1) | 79 (35.9) | 141 (64.1) | 84 (50.3) | 26 (31) | 58 (69) | |||
| Gender | ||||||||||||
| Female | 194 (31.9) | 69 (35.6) | 125 (64.4) | 0.358 | 178 (44.6) | 65 (36.5) | 113 (63.5) | 0.659 | 156 (93.4) | 44 (28.2) | 112 (71.8) | 0.947 |
| Male | 415 (68.1) | 132 (31.8) | 283 (68.2) | 221 (55.4) | 76 (34.4) | 145 (65.6) | 11 (6.6) | 3 (27.3) | 8 (72.7) | |||
| Smoking History | ||||||||||||
| Never | 181 (29.7) | 63 (34.8) | 118 (65.2) | 0.656 | 174 (43.6) | 62 (35.6) | 112 (64.4) | 0.928 | 5 (3.0) | 1 (20) | 4 (80) | 0.666 |
| Ever | 416 (68.3) | 137 (32.9) | 279 (67.1) | 219 (54.9) | 79 (36.1) | 140 (63.9) | 156 (93.4) | 45 (28.8) | 111 (71.2) | |||
| Unknown | 12 (2.0) | 1 | 11 | 6 (1.5) | 0 | 6 | 6 (3.6) | 1 | 5 | |||
| Pathological Stage | ||||||||||||
| I | 377 (61.9) | 148 (39.3) | 229 (60.7) | <0.001 | 276 (69.2) | 111 (40.2) | 165 (59.8) | 0.002 | 84 (50.3) | 30 (35.7) | 54 (64.3) | 0.029 |
| II, III | 232 (38.1) | 53 (22.8) | 179 (77.2) | 123 (30.8) | 30 (24.4) | 93 (75.6) | 83 (49.7) | 17 (20.5) | 66 (79.5) | |||
| Primary Tumor | ||||||||||||
| T1 | 246 (40.4) | 95 (38.6) | 151 (61.4) | 0.015 | 199 (49.9) | 75 (37.7) | 124 (62.3) | 0.327 | 38 (22.8) | 15 (39.5) | 23 (60.5) | 0.077 |
| T2,3,4 | 363 (59.6) | 106 (29.2) | 257 (70.8) | 200 (50.1) | 66 (33.0) | 134 (67.0) | 129 (77.2) | 32 (24.8) | 97 (75.2) | |||
| Lymph Nodes Met | ||||||||||||
| N0 | 452 (74.2) | 168 (37.2) | 284 (62.8) | <0.001 | 312 (78.2) | 120 (38.5) | 192 (61.5) | 0.014 | 117 (70.1) | 38 (32.5) | 79 (67.5) | 0.057 |
| N1,2,3 | 157 (25.8) | 33 (21.0) | 124 (79.0) | 87 (21.8) | 21 (24.1) | 66 (75.9) | 50 (29.9) | 9 (18.0) | 41 (82.0) | |||
| Pathological Histology | ||||||||||||
| Ad | 399 (65.5) | 141 (35.3) | 258 (64.7) | 0.233 | ||||||||
| Sq | 167 (27.4) | 47 (28.1) | 120 (71.9) | |||||||||
| Others | 43 (7.1) | 13 (30.2) | 30 (69.8) | |||||||||
| EGFR Mutation | ||||||||||||
| Negative | 484 (79.5) | 170 (35.1) | 314 (64.9) | 0.029 | 286 (71.7) | 113 (39.5) | 173 (60.5) | 0.006 | 159 (95.2) | 44 (27.7) | 115 (72.3) | 0.546 |
| Positive | 125 (20.5) | 31 (24.8) | 94 (75.2) | 113 (28.3) | 28 (24.8) | 85 (75.2) | 8 (4.8) | 3 (37.5) | 5 (62.5) | |||
| All Cases | Ad | Sq | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Factor | HR | 95.0% CI | p-Value | HR | 95.0% CI | p-Value | HR | 95.0% CI | p-Value |
| Gender (female, male) | 0.791 | 0.466–1.344 | 0.387 | 1.253 | 0.655–2.398 | 0.495 | 0.792 | 0.210–2.987 | 0.731 |
| Smoking History (never, ever) | 0.679 | 0.366–1.259 | 0.219 | 0.400 | 0.194–0.824 | 0.013 | 2.338 | 0.485–11.275 | 0.290 |
| Pathological Stage (I, II, III) | 1.026 | 1.004–1.049 | 0.020 | 1.049 | 1.018–1.081 | 0.002 | 1.004 | 0.967–1.042 | 0.841 |
| Primary Tumor (T1, T234) | 1.002 | 1.001–1.004 | 0.008 | 1.003 | 1.001–1.006 | 0.006 | 1.001 | 0.998–1.004 | 0.446 |
| Regional Lymph Nodes (N0, N123) | 1.311 | 0.826–2.081 | 0.251 | 0.912 | 0.487–1.707 | 0.772 | 1.960 | 0.870–4.413 | 0.104 |
| Pathological Histology (Ad, Sq, others) | 1.423 | 1.113–1.820 | 0.005 | ||||||
| EGFR Mutation (negative, positive) | 0.940 | 0.589–1.502 | 0.797 | 0.963 | 0.547–1.695 | 0.895 | 0.530 | 0.115–2.451 | 0.417 |
| FAF1 in Nuclei (negative, positive) | 1.313 | 0.901–1.912 | 0.156 | 1.709 | 0.984–2.968 | 0.057 | 1.002 | 0.545–1.840 | 0.996 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, D.; Yamada, H.; Yoshimura, K.; Ohta, T.; Tsuchiya, K.; Inoue, Y.; Funai, K.; Suda, T.; Iwashita, Y.; Watanabe, T.; et al. High Expression of Fas-Associated Factor 1 Indicates a Poor Prognosis in Non-Small-Cell Lung Cancer. Curr. Oncol. 2023, 30, 9484-9500. https://doi.org/10.3390/curroncol30110687
Hu D, Yamada H, Yoshimura K, Ohta T, Tsuchiya K, Inoue Y, Funai K, Suda T, Iwashita Y, Watanabe T, et al. High Expression of Fas-Associated Factor 1 Indicates a Poor Prognosis in Non-Small-Cell Lung Cancer. Current Oncology. 2023; 30(11):9484-9500. https://doi.org/10.3390/curroncol30110687
Chicago/Turabian StyleHu, De, Hidetaka Yamada, Katsuhiro Yoshimura, Tsutomu Ohta, Kazuo Tsuchiya, Yusuke Inoue, Kazuhito Funai, Takafumi Suda, Yuji Iwashita, Takuya Watanabe, and et al. 2023. "High Expression of Fas-Associated Factor 1 Indicates a Poor Prognosis in Non-Small-Cell Lung Cancer" Current Oncology 30, no. 11: 9484-9500. https://doi.org/10.3390/curroncol30110687
APA StyleHu, D., Yamada, H., Yoshimura, K., Ohta, T., Tsuchiya, K., Inoue, Y., Funai, K., Suda, T., Iwashita, Y., Watanabe, T., Ogawa, H., Kurono, N., Shinmura, K., & Sugimura, H. (2023). High Expression of Fas-Associated Factor 1 Indicates a Poor Prognosis in Non-Small-Cell Lung Cancer. Current Oncology, 30(11), 9484-9500. https://doi.org/10.3390/curroncol30110687








