DPYD Exon 4 Deletion Associated with Fluoropyrimidine Toxicity and Importance of Copy Number Variation
Abstract
1. Introduction
2. Methods
2.1. Patient Cohort
2.2. Detection of DPYD Exon 4 Deletion
2.3. Literature Review for DPYD CNV
3. Results
3.1. Study Population
3.2. Fluoropyrimidine-Associated Toxicity
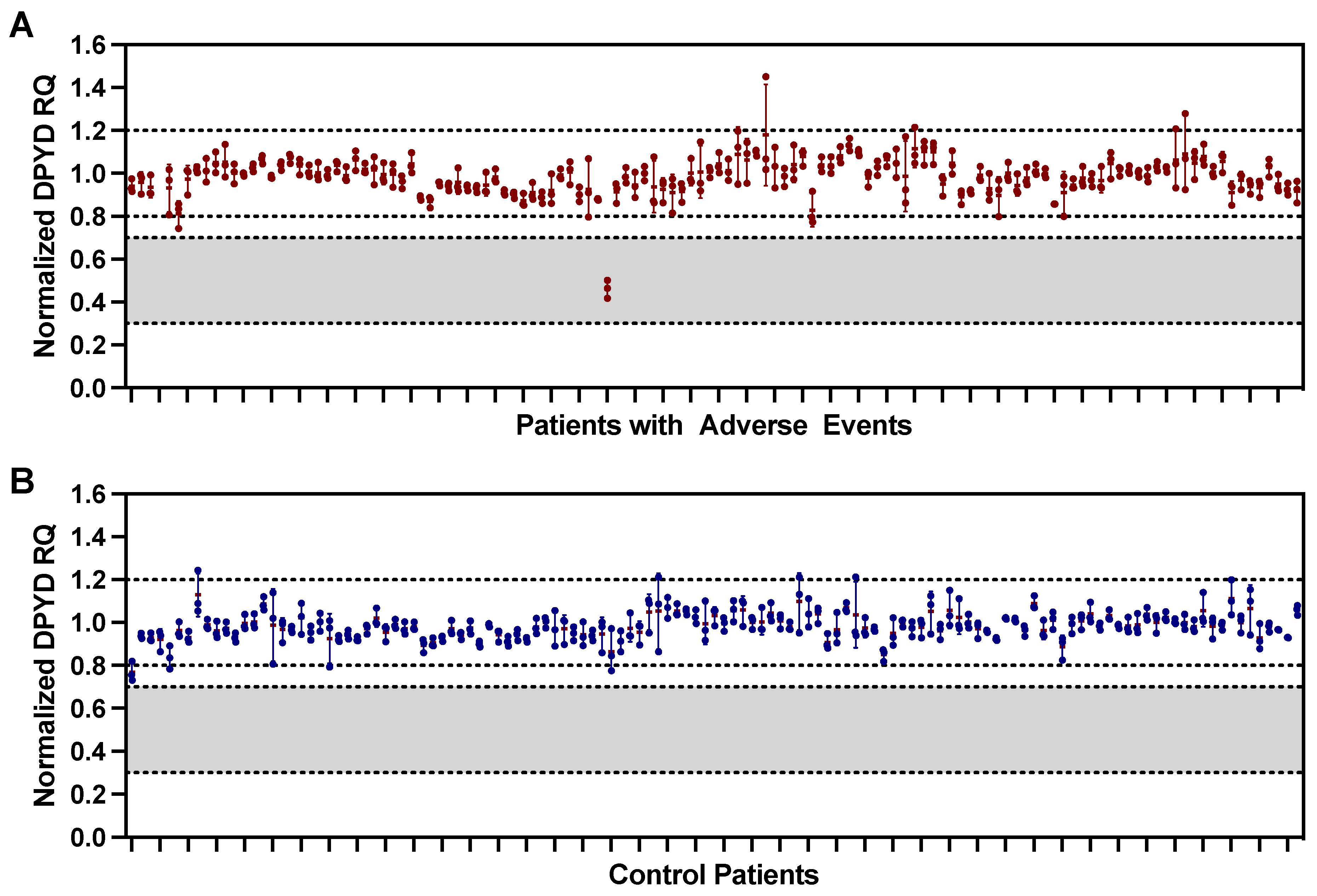
3.3. Exon 4 Deletion
3.4. Literature Review of DPYD Copy Number Variation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wigle, T.J.; Tsvetkova, E.V.; Welch, S.A.; Kim, R.B. Fluorouracil-Based Chemotherapy: Mini Review and Case Report. Pharmaceutics 2019, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Mikhail, S.E.; Sun, J.F.; Marshall, J.L. Safety of capecitabine: A review. Expert Opin. Drug Saf. 2010, 9, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Heggie, G.D.; Sommadossi, J.P.; Cross, D.S.; Huster, W.J.; Diasio, R.B. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987, 47, 2203–2206. [Google Scholar] [PubMed]
- Diasio, R.B.; Beavers, T.L.; Carpenter, J.T. Familial deficiency of dihydropyrimidine dehydrogenase. Biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. J. Clin. Investig. 1988, 81, 47–51. [Google Scholar] [CrossRef]
- Loriot, M.A.; Ciccolini, J.; Thomas, F.; Barin-Le-Guellec, C.; Royer, B.; Milano, G.; Picard, N.; Becquemont, L.; Verstuyft, C.; Narjoz, C.; et al. Dihydropyrimidine déhydrogenase (DPD) deficiency screening and securing of fluoropyrimidine-based chemotherapies: Update and recommendations of the French GPCO-Unicancer and RNPGx networks. Bull. Cancer 2018, 105, 397–407. [Google Scholar] [CrossRef]
- Lunenburg, C.A.T.C.; van der Wouden, C.H.; Nijenhuis, M.; Crommentuijn-van Rhenen, M.H.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2020, 28, 508–517. [Google Scholar] [CrossRef]
- Paulsen, N.H.; Vojdeman, F.; Andersen, S.E.; Bergmann, T.K.; Ewertz, M.; Plomgaard, P.; Hansen, M.R.; Esbech, P.S.; Pfeiffer, P.; Qvortrup, C.; et al. DPYD genotyping and dihydropyrimidine dehydrogenase (DPD) phenotyping in clinical oncology. A clinically focused minireview. Basic Clin. Pharmacol. Toxicol. 2022, 131, 325–346. [Google Scholar] [CrossRef]
- Johnson, M.R.; Wang, K.; Tillmanns, S.; Albin, N.; Diasio, R.B. Structural organization of the human dihydropyrimidine dehydrogenase gene. Cancer Res. 1997, 57, 1660–1663. [Google Scholar]
- Wei, X.; Elizondo, G.; Sapone, A.; McLeod, H.L.; Raunio, H.; Fernandez-Salguero, P.; Gonzalez, F.J. Characterization of the human dihydropyrimidine dehydrogenase gene. Genomics 1998, 51, 391–400. [Google Scholar] [CrossRef]
- Meulendijks, D.; Henricks, L.M.; Sonke, G.S.; Deenen, M.J.; Froehlich, T.K.; Amstutz, U.; Largiadèr, C.R.; Jennings, B.A.; Marinaki, A.M.; Sanderson, J.D.; et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: A systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015, 16, 1639–1650. [Google Scholar] [CrossRef]
- Rosmarin, D.; Palles, C.; Church, D.; Domingo, E.; Jones, A.; Johnstone, E.; Wang, H.; Love, S.; Julier, P.; Scudder, C.; et al. Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: Investigation in the QUASAR2 study, systematic review, and meta-analysis. J. Clin. Oncol. 2014, 32, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Terrazzino, S.; Cargnin, S.; Del Re, M.; Danesi, R.; Canonico, P.L.; Genazzani, A.A. DPYD IVS14+1G>A and 2846A>T genotyping for the prediction of severe fluoropyrimidine-related toxicity: A meta-analysis. Pharmacogenomics 2013, 14, 1255–1272. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.M.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Henricks, L.M.; Lunenburg, C.A.T.C.; de Man, F.M.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.J.; Baars, A.; Dezentjé, V.O.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: A prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467. [Google Scholar] [CrossRef]
- Wigle, T.J.; Povitz, B.L.; Medwid, S.; Teft, W.A.; Legan, R.M.; Lenehan, J.; Nevison, S.; Panuganty, V.; Keller, D.; Mailloux, J.; et al. Impact of pretreatment dihydropyrimidine dehydrogenase genotype-guided fluoropyrimidine dosing on chemotherapy associated adverse events. Clin. Transl. Sci. 2021, 14, 1338–1348. [Google Scholar] [CrossRef]
- Schwab, M.; Zanger, U.M.; Marx, C.; Schaeffeler, E.; Klein, K.; Dippon, J.; Kerb, R.; Blievernicht, J.; Fischer, J.; Hofmann, U.; et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: A prospective clinical trial by the German 5-FU Toxicity Study Group. J. Clin. Oncol. 2008, 26, 2131–2138. [Google Scholar] [CrossRef]
- Saarenheimo, J.; Wahid, N.; Eigeliene, N.; Ravi, R.; Salomons, G.S.; Ojeda, M.F.; Vijzelaar, R.; Jekunen, A.; van Kuilenburg, A.B.P. Preemptive screening of DPYD as part of clinical practice: High prevalence of a novel exon 4 deletion in the Finnish population. Cancer Chemother. Pharmacol. 2021, 87, 657–663. [Google Scholar] [CrossRef]
- van Kuilenburg, A.B.; Meijer, J.; Mul, A.N.; Hennekam, R.C.; Hoovers, J.M.; de Die-Smulders, C.E.; Weber, P.; Mori, A.C.; Bierau, J.; Fowler, B.; et al. Analysis of severely affected patients with dihydropyrimidine dehydrogenase deficiency reveals large intragenic rearrangements of DPYD and a de novo interstitial deletion del(1)(p13.3p21.3). Hum. Genet. 2009, 125, 581–590. [Google Scholar] [CrossRef]
- Carter, M.T.; Nikkel, S.M.; Fernandez, B.A.; Marshall, C.R.; Noor, A.; Lionel, A.C.; Prasad, A.; Pinto, D.; Joseph-George, A.M.; Noakes, C.; et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin. Genet. 2011, 80, 435–443. [Google Scholar] [CrossRef]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef]
- Willemsen, M.H.; Vallès, A.; Kirkels, L.A.; Mastebroek, M.; Olde Loohuis, N.; Kos, A.; Wissink-Lindhout, W.M.; de Brouwer, A.P.; Nillesen, W.M.; Pfundt, R.; et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J. Med. Genet. 2011, 48, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Brečević, L.; Rinčić, M.; Krsnik, Ž.; Sedmak, G.; Hamid, A.B.; Kosyakova, N.; Galić, I.; Liehr, T.; Borovečki, F. Association of new deletion/duplication region at chromosome 1p21 with intellectual disability, severe speech deficit and autism spectrum disorder-like behavior: An all-in approach to solving the. Transl. Neurosci. 2015, 6, 59–86. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, C.S.; Moller Dos Santos, M.F.; Alonso, L.G.; Koiffmann, C.P. Two New Cases of 1p21.3 Deletions and an Unbalanced Translocation t(8;12) among Individuals with Syndromic Obesity. Mol. Syndromol. 2015, 6, 63–70. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, C.S.; Varela, M.C.; de Castro, C.I.E.; Otto, P.A.; Perez, A.B.A.; Lourenço, C.M.; Kim, C.A.; Bertola, D.R.; Kok, F.; Garcia-Alonso, L.; et al. Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Mol. Cytogenet. 2018, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Tabata, H.; Sone, K.; Kobayashi, T.; Yanagisawa, T.; Tamura, T.; Shimizu, N.; Kanbe, Y.; Tashiro, M.; Ono, S.; Kuroume, T. Short arm deletion of chromosome 1: Del(1)(p13.3 p22.3) in a female infant with an extreme tetralogy of Fallot. Clin. Genet. 1991, 39, 132–135. [Google Scholar] [CrossRef]
- Dockery, H.; Van der Westhuyzen, J. Monosomy of 1p13.3-22.3 in twins. Clin. Genet. 1991, 39, 223–227. [Google Scholar] [CrossRef]
- Mattia, F.R.; Wardinsky, T.D.; Tuttle, D.J.; Grix, A.; Smith, K.A.; Walling, P. Interstitial deletion of the short arm of chromosome 1 (46XY, del(1)(p13p22.3)). Am. J. Med. Genet. 1992, 44, 551–554. [Google Scholar] [CrossRef]
- Santos, M.; Niemi, M.; Hiratsuka, M.; Kumondai, M.; Ingelman-Sundberg, M.; Lauschke, V.M.; Rodríguez-Antona, C. Novel copy-number variations in pharmacogenes contribute to interindividual differences in drug pharmacokinetics. Genet. Med. 2018, 20, 622–629. [Google Scholar] [CrossRef]
- van Kuilenburg, A.B.; Meijer, J.; Maurer, D.; Dobritzsch, D.; Meinsma, R.; Los, M.; Knegt, L.C.; Zoetekouw, L.; Jansen, R.L.; Dezentjé, V.; et al. Severe fluoropyrimidine toxicity due to novel and rare DPYD missense mutations, deletion and genomic amplification affecting DPD activity and mRNA splicing. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 721–730. [Google Scholar] [CrossRef]
- He, Y.; Hoskins, J.M.; McLeod, H.L. Copy number variants in pharmacogenetic genes. Trends Mol. Med. 2011, 17, 244–251. [Google Scholar] [CrossRef]
- Hormozian, F.; Schmitt, J.G.; Sagulenko, E.; Schwab, M.; Savelyeva, L. FRA1E common fragile site breaks map within a 370kilobase pair region and disrupt the dihydropyrimidine dehydrogenase gene (DPYD). Cancer Lett. 2007, 246, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Van Kuilenburg, A.B.; Vreken, P.; Abeling, N.G.; Bakker, H.D.; Meinsma, R.; Van Lenthe, H.; De Abreu, R.A.; Smeitink, J.A.; Kayserili, H.; Apak, M.Y.; et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum. Genet. 1999, 104, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bakkeren, J.A.; De Abreu, R.A.; Sengers, R.C.; Gabreëls, F.J.; Maas, J.M.; Renier, W.O. Elevated urine, blood and cerebrospinal fluid levels of uracil and thymine in a child with dihydrothymine dehydrogenase deficiency. Clin. Chim. Acta 1984, 140, 247–256. [Google Scholar] [CrossRef] [PubMed]
- van Gennip, A.H.; Busch, S.; Elzinga, L.; Stroomer, A.E.; van Cruchten, A.; Scholten, E.G.; Abeling, N.G. Application of simple chromatographic methods for the diagnosis of defects in pyrimidine degradation. Clin. Chem. 1993, 39, 380–385. [Google Scholar] [CrossRef] [PubMed]
- van Gennip, A.H.; Abeling, N.G.; Stroomer, A.E.; van Lenthe, H.; Bakker, H.D. Clinical and biochemical findings in six patients with pyrimidine degradation defects. J. Inherit. Metab. Dis. 1994, 17, 130–132. [Google Scholar] [CrossRef]
- van Gennip, A.H.; Abeling, N.G.; Vreken, P.; van Kuilenburg, A.B. Inborn errors of pyrimidine degradation: Clinical, biochemical and molecular aspects. J. Inherit. Metab. Dis. 1997, 20, 203–213. [Google Scholar] [CrossRef]
- Ticha, I.; Kleiblova, P.; Fidlerova, J.; Novotny, J.; Pohlreich, P.; Kleibl, Z. Lack of large intragenic rearrangements in dihydropyrimidine dehydrogenase (DPYD) gene in fluoropyrimidine-treated patients with high-grade toxicity. Cancer Chemother. Pharmacol. 2009, 64, 615–618. [Google Scholar] [CrossRef]
- Paré, L.; Paez, D.; Salazar, J.; Del Rio, E.; Tizzano, E.; Marcuello, E.; Baiget, M. Absence of large intragenic rearrangements in the DPYD gene in a large cohort of colorectal cancer patients treated with 5-FU-based chemotherapy. Br. J. Clin. Pharmacol. 2010, 70, 268–272. [Google Scholar] [CrossRef]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002, 30, e57. [Google Scholar] [CrossRef]
| Characteristic | Case (N = 125) | Control (N = 125) |
|---|---|---|
| Sex, N (%) | ||
| Female | 63 (50) | 63 (50) |
| Male | 62 (50) | 62 (50) |
| Age in years, mean (SD) | 65.0 (10.4) | 65.6 (9.9) |
| Ethnicity, N (%) a | ||
| Caucasian | 124 (99) | 114 (91) |
| African | 0 (0) | 2 (2) |
| Asian | 1 (1) | 1 (1) |
| Unknown | 0 (0) | 8 (6) |
| Treatment characteristics | ||
| BSA (m2), mean (SD) b | 1.9 (0.2) | 1.9 (0.3) |
| Initial Dose Intensity, mean (SD) c | 91 (14) | 87 (14) |
| Average Dose Intensity, mean (SD) | 81 (15) | 84 (13) |
| Treatment Cycles, median (IQR) d | 6 (3–7) | 6 (3–8) |
| Regimen, N (%) | ||
| Capecitabine with radiation | 14 (11) | 13 (10) |
| Capecitabine monotherapy | 35 (28) | 35 (28) |
| Capecitabine with oxaliplatin | 26 (21) | 26 (21) |
| FOLFOX e | 31 (25) | 31 (25) |
| FOLFIRI/FOLFIRINOX e | 13 (10) | 14 (11) |
| 5-FU with radiation | 6 (5) | 6 (5) |
| Category | No. |
|---|---|
| No. of Patients | 125 |
| No. of Adverse Events | 157 |
| Gastrointestinal | |
| Diarrhea | 47 |
| Colitis | 11 |
| Mucositis a | 6 |
| Nausea/Vomiting b | 4 |
| Myelosuppression | |
| Neutropenia | 31 |
| Febrile Neutropenia | 11 |
| Anemia | 2 |
| HFS c | 25 |
| Other d | 15 |
| Death | 5 |
| Gene Changes | Size | Effect on DPYD | Other Genes Affected | Phenotype | Ref. |
|---|---|---|---|---|---|
| 1p21.3 deletion | 10 kb | Deletion of exon 6 | None | Autism, language delay | [19] |
| c.1340–3473_c.1525 + 10,154del1,3812 | ~13.8 kb | Deletion of exon 12 | None | Seizures, aggressive attitude, developmental delay, muscular hypotonia, microcephaly, autistic-like behavior | [18] |
| c.1741_2058del | ~122 kb | Deletion of exon 14–16 | None | Amniotic infections, Respiratory insufficiency, developmental delay, facial and skeletal abnormalities, dysostosis multiplex | [18] |
| 1p21.3 deletion | 1.1 Mb | Whole DPYD deletion | MIR137 | Severe language delay, aggressive behavior, autism, seizure | [19,20] |
| 1p21.3 deletion | 1.41 Mb | Whole DPYD deletion | LOC729987, MIR137 | Mild intellectual disability, features of autism, tendency to overeat, remarkably shy and friendly, speech deficits, ocular problems | [21] |
| 1p21.3 deletion | 1.5 Mb | Whole DPYD deletion | PTBP2 | Severe language delay, fine motor skill delay, autism, dysmorphic features | [19] |
| 1p21.3 deletion | 1.75 Mb | Whole DPYD deletion | LOC729987, SNX7, LPPR5, MIR137 | Mild to moderate intellectual disability, features of autism, tendency to overeat, remarkably shy and friendly, ocular problem, facial structure abnormalities | [21] |
| 1p21.3 deletion | 2.45 Mb | Whole DPYD deletion | LOC729987, PTBP2, MIR137, LOC101928241 | Mild intellectual disability, remarkable shy and friendly, aggressive outbursts | [21] |
| 1p21.3p21.2 duplication | 3.56 Mb | Whole DPYD duplication | LOC101928241, PTBP2, MIR137 LOC729987, SNX7, LPPR5, LPPR4 | Intellectual disability, pervasive developmental disorder, febrile convulsions, psychomotor restlessness, hyperactivity, facial and skeletal abnormalities, clinodactyly | [22] |
| 1p21.3 deletion | 3.68 Mb | Whole DPYD duplication | LOC101928241, PTBP2, MIR137, SNX7, LPPR5, LOC729987, LPPR4, PALMD, FRRS1, MIR548 | Intellectual disability | [22] |
| 1p22.1p21.3 deletion | 4.58 Mb | Whole DPYD deletion | LOC101928241, PTBP2, MIR137, LOC729987 | Intellectual disability and obesity | [23] |
| 1p21.3p21 deletion | 5.43 Mb | Whole DPYD deletion | PTBP2, MIR137, SNX7, LPPR5, LOC729987, LPPR4, LOC100129620 | Intellectual disability, autistic spectrum disorder | [23] |
| 1p22.1p21.2 deletion | 5.9 Mb | Whole DPYD deletion | F3, LOC101928241, PTBP2, MIR137, SNX7, LPPR5, LOC729987, LPPR4, LOC100129620 | Neonatal hypotonia, psychomotor and speech delay, intellectual disability, obesity, hyperphagia, macrocephaly, ocular problems, clinodactyly | [23,24] |
| 1p21.3p13.3 deletion | 9.9 Mb | Partial DPYD deletion | MIR137, SNX7, LPPR5, LOC729987, LPPR4, LOC100129620 | Delayed speech | [23] |
| 1p21.3p13.3 deletion | 11.19 Mb | Whole DPYD deletion | LOC101928241, PTBP2, MIR137, SNX7, LPPR5, LOC729987, LPPR4, LOC100129620 | Intellectual disability and obesity | [23] |
| 1p21.3p13.3 deletion | 12 Mb | Whole DPYD deletion | LOC101928241, PTBP2, MIR137, SNX7, LPPR5, LOC729987, LPPR4, LOC100129620, VCAM1, COL11A1, AMY2B, AMY2A, AMY1A | Obesity, hyperphagia, psychomotor delay, speech delay, intellectual disability, macrocephaly precocious puberty | [23,24] |
| 1p21.3p13.3 deletion | ~14 Mb | Whole DPYD deletion | Multiple genes, including WNT2B and NTNG1 | Intellectual disability, epilepsy, psychomotor and speech impairment, hypotonic and hypermobile, toe abnormalities, coloboma | [25] |
| 1p21.3p13.3 deletion | 14 Mb | Whole DPYD deletion | 57 genes, including, MIR137, PTBP2, SNX7 | Hypertonia and irritability at birth, hypotonia, areflexia, intellectual disability, facial and skeletal abnormalities, macrocephaly, epiphyseal dysplasia | [18] |
| 1p22.3p13.3 deletion | Not reported | Whole DPYD deletion | Multiple genes | Intellectual disability, language delay, hypotonia, facial abnormalities, digitalized thumbs | [27] |
| 1p22.3p13.3 deletion | Not reported | Whole DPYD deletion | Multiple genes | Intellectual disability, hearing loss, digitalized thumbs, toe abnormalities, facial abnormalities | [26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wigle, T.J.; Medwid, S.; Ross, C.; Schwarz, U.I.; Kim, R.B. DPYD Exon 4 Deletion Associated with Fluoropyrimidine Toxicity and Importance of Copy Number Variation. Curr. Oncol. 2023, 30, 663-672. https://doi.org/10.3390/curroncol30010051
Wigle TJ, Medwid S, Ross C, Schwarz UI, Kim RB. DPYD Exon 4 Deletion Associated with Fluoropyrimidine Toxicity and Importance of Copy Number Variation. Current Oncology. 2023; 30(1):663-672. https://doi.org/10.3390/curroncol30010051
Chicago/Turabian StyleWigle, Theodore J., Samantha Medwid, Cameron Ross, Ute I. Schwarz, and Richard B. Kim. 2023. "DPYD Exon 4 Deletion Associated with Fluoropyrimidine Toxicity and Importance of Copy Number Variation" Current Oncology 30, no. 1: 663-672. https://doi.org/10.3390/curroncol30010051
APA StyleWigle, T. J., Medwid, S., Ross, C., Schwarz, U. I., & Kim, R. B. (2023). DPYD Exon 4 Deletion Associated with Fluoropyrimidine Toxicity and Importance of Copy Number Variation. Current Oncology, 30(1), 663-672. https://doi.org/10.3390/curroncol30010051




