Abstract
We used a real-world database (GuardantINFORMTM) to analyze the treatment choices for patients with mCRC who underwent next-generation sequencing of circulating tumor DNA (ctDNA) using a commercially available test (Guardant360®) after first- or second-line therapy. From 18,875 patients with claims for CRC, 1064 had confirmed metastatic disease and sufficient histories for analysis (median age 59 years, 44.8% female, 44.5% left-sided). ctDNA was detectable for 997/1064 (93.7%) patients. Clinically actionable molecular profiles were present for 507/1064 (47.7%) patients, including those who had not received targeted therapy in the previous line (410/926, 44.3%). Second- or third-line targeted therapies were administered to 338/1064 patients (31.8%) and were considered matched for 193/338 (57.1%) patients. Therapies administered after testing were informed by the ctDNA results in 56.7% of patients overall (603/1064). Time to treatment discontinuation was most favorable for patients with a clinically actionable ctDNA profile who received matched therapy. This analysis demonstrates the real-world clinical value of plasma-based comprehensive genomic profiling for selecting appropriate molecular-targeted therapies in mCRC patients with disease progression after first- or second-line therapy.
1. Introduction
Advancements in molecular-targeted therapies and biomarker detection have improved in recent years [1,2]. Such technologies have been successfully applied to people living with metastatic colorectal cancer (mCRC). In particular, the treatment of mCRC has progressed significantly over the past 20 years, with the addition of new chemotherapy to fluoropyrimidine backbones along with the introduction of agents that target vascular endothelial growth factor (VEGF) and molecular aberrations of the cancer cells themselves. Such advancements in molecular-targeted therapies and biomarker detection have resulted in improvements in tumor response rates and overall survival [3,4]. The presence of specific biomarkers in these tumors can predict the response to targeted therapies. For example, tumors with wild-type KRAS and NRAS (which account for 30%–50% of cases) [5,6,7,8,9,10,11,12] are more likely to respond to EGFR-targeted therapies than tumors with mutations in either of these genes [13,14]. Other examples of targeted therapies for mCRC include immune checkpoint inhibitors (ICIs) for microsatellite-instability-high (MSI-H) tumors [15], tyrosine kinase inhibitors for BRAF V600E [16], and ERBB2-targeted therapies for ERBB2 amplification [17,18]. Therefore, US treatment guidelines recommend testing for mutations in KRAS, NRAS, and BRAF; ERBB2 amplification; NTRK1/2/3 rearrangements; and MSI-high status in patients with advanced CRC [19].
Resistance mutations may emerge after treatment with targeted therapies. For example, EGFR-targeted therapy may lead to alterations such as KRAS, NRAS, BRAF, and EGFR extracellular domain mutations or ERBB2 amplification [20,21,22]. Some of these can be successfully treated with additional targeted therapies, such as trastuzumab-based therapies or trastuzumab deruxtecan (DS-8201) for ERBB2 amplification [18,23,24,25,26,27]. However, for tumors lacking these mechanisms of resistance, repeated treatment with EGFR-targeted therapy is feasible and effective [20].
Therefore, genomic assessment of mCRC after progression on initial treatment can guide therapy selection. While tissue biopsies have traditionally been used for such assessment, access to tumor tissue for biomarker testing may be challenging. Furthermore, archived tissue samples may not represent the current state of the tumor and it may be difficult to obtain a fresh tissue biopsy.
Recent advances in next-generation sequencing (NGS) of circulating tumor DNA (ctDNA) have made liquid biopsy a viable testing modality for sequencing mCRC after disease progression. Comprehensive genomic profiling of ctDNA can detect mutations, gene rearrangements, and amplification in genes that may be targets for subsequent therapy or provide information on resistance mechanisms to help clinicians avoid ineffective therapies [28,29].
In this study, we sought to evaluate how clinicians apply ctDNA testing results when making treatment decisions for patients with mCRC that had progressed on prior therapy. Using a real-world database (GuardantINFORMTM, Guardant Health, Redwood City, CA, USA), we analyzed the treatment choices for patients with mCRC who underwent NGS-based ctDNA testing (Guardant360®, Guardant Health) after completing first- or second-line therapy. The goal was to evaluate the real-world application of ctDNA testing and its ability to identify clinically relevant genomic alterations and how such findings influence the subsequent treatment decisions and clinical outcomes for people living with mCRC.
2. Methods
2.1. Ethics
The GuardantINFORM database is fully compliant and deidentified in accordance with the US patient confidentiality requirements set forth in Sections 164.514 (a)–(b)1ii of the Health Insurance Portability and Accountability Act (HIPAA) regarding the determination and documentation of statistically deidentified data. Institutional Review Board approval to conduct this study was not required because this study used deidentified patient records and did not involve the collection, use, or transmission of individually identifiable data.
2.2. Data Source and Extraction
We used secondary data from the GuardantINFORM database, which comprises anonymized genomic data for patients with advanced stage solid tumors in the US who underwent testing with a Guardant360 assay. This assay is certified according to Clinical Laboratory Improvement Amendments and accredited by the College of American Pathologists and has been approved by the US Food and Drug Administration (FDA) and New York State Department of Health for clinical testing of patients with advanced stage (stage III–IV) solid tumors. The assay utilizes hybrid capture technology and NGS to identify genomic alterations in 74 genes [30,31]. The panel covers all coding exons in KRAS, NRAS, and BRAF, and detects ERBB2 amplification, NTRK1 fusion, and MSI-high status.
In addition to the genetic data, the GuardantINFORM database includes structured commercial payer claims data collected from inpatient and outpatient facilities in academic and community settings. Claims data are provided through a commercial agreement with a data aggregator (the ‘parent dataset’), which collates high-quality industry-standard sources of anonymized patient level data and a further 150 fully privileged payer complete datasets that include a combination of open and closed claims. By percentage, the prescription claim coverage of the full parent dataset by payer type is: Commercial 78%, Medicare 0%, Medicare Advantage 16%, and Medicaid 6%. However, it does not capture clinical features that are not coded as claims, such as the results of tumor biomarker testing from other sources and the clinical response to cancer treatment. Deaths are sourced from third-party providers and aggregated with administrative claims data; for at least half of the CDC-reported deaths in the country, the parent dataset has an encounter within ≤1 month of the date of death. GuardantINFORM is refreshed every quarter. At the time of this study, GuardantINFORM contained records for more than 190,000 patients across 60 cancer types, representing approximately 7000 oncologists from US-based academic and community cancer practices.
2.3. Study Population
For this study, we included all CRC patients who underwent testing between 1 June 2014 and 30 September 2021, with a Guardant360 assay performed within 90 days prior to initiating second or third-line systemic therapy, had a 6-month clean window of claims information (≥2 medical or pharmacy claims) without treatment prior to the first observed therapy date, and had claims data indicating the presence of metastatic disease. The clean window was implemented to ensure that the first observed systemic therapy date corresponds to first-line treatment. If patients underwent ctDNA testing more than once after initial treatment, data from the test conducted immediately after the earliest line of targeted therapy were analyzed; otherwise, the earlier test was used regardless of prior therapy.
2.4. Lines of Therapy
The patients’ treatment regimens were reconstructed over the study period based on the claims data. All systemic cancer therapies recommended for mCRC in the NCCN guidelines [19] were included. The same regimens used within 90 days from the end of one to the start of the next administration date were combined and considered the same line of therapy. Any treatment modifications or additions to targeted therapies more than 60 days after treatment initiation were considered a new line of therapy, whereas changes to chemotherapies more than 15 days after treatment initiation were considered a new line of therapy. Changes in treatments involving fluorouracil or bevacizumab were not considered as new therapy. Discontinuing a medicine without adding a new medicine to a regimen was not considered a new line of therapy.
2.5. Genomic Profiling
Clinically informative genomic alterations were those classified as Therapeutic Levels 1, 2 or R1 by OncoKB [32]; namely, KRAS and NRAS point mutations, BRAF V600E, ERBB2 amplification, NTRK1 fusion, and MSI-high status. The Guardant360 assay used at the time of data collection did not include NTRK2 or NTRK3 fusions, nor did it assess tumor mutation burden status.
For determination of actionable genomic profiles and molecularly informed therapy, only the results of Guardant360 tests were considered because the GuardantINFORM database did not include biomarker results for other testing modalities. Samples with detectable ctDNA but neither KRAS nor NRAS mutations were classified as having an actionable molecular profile, and patients with such profiles were categorized as appropriate candidates for EGFR-targeted therapy except in the case of concurrent BRAF V600E or primary tumor location in the right colon [33]. Immune checkpoint inhibitors were considered molecularly informed only if MSI-high were detected. BRAF-directed therapies were considered informed for BRAF V600E, ERBB2-targeted therapies were considered informed for ERBB2 amplification, and NTRK inhibitors were considered informed for NTRK1 fusions. Chemotherapy and/or VEGF-targeted therapy were classified as molecularly informed in the absence of ERBB2 amplification, BRAF V600E, NTRK1 fusion, and MSI-high or the presence of a mutation in KRAS or NRAS without other actionable biomarkers. Non-targeted therapy was also considered informed for patients with right-sided tumors and ctDNA profiles lacking RAS mutations or other actionable alterations. Throughout this manuscript, “targeted therapy” refers to systemic anti-cancer agents that are targeted primarily against aberrant proteins expressed by cancer cells or to immune checkpoint inhibitors; VEGF-targeted therapy is not included in this definition.
2.6. Patient Outcomes
As a surrogate for progression-free survival, we evaluated the time to treatment discontinuation (TTD) for each line of therapy, which was defined as the time from the first day therapy to the estimated last day of therapy, the last claim activity date, or death, whichever occurred first. Patients with a last claim activity date prior to the end of therapy or <90 days after the end of therapy were censored due to lack of follow-up information. We also evaluated overall survival, which was defined as the time from the ctDNA report date prior to the start of second- or third-line therapy (for second- or third-line therapy patients, respectively) until death. Patients were censored at last claim activity date.
2.7. Statistical Analysis
All patients in the study population were included in the baseline descriptive statistics. Means, medians, and standard deviations were calculated for continuous variables whereas counts and proportions were calculated for categorical variables. Proportions were compared using the two proportion z-test or Fisher’s Exact test for small samples.
The TTD for anti-cancer therapies administered after ctDNA testing was evaluated using the Kaplan–Meier method for the following four subgroups of patients: (1) patients with a clinically actionable genomic profile who received the matched therapy, (2) patients with a clinically actionable genomic profile who did not receive a matched therapy, (3) patients without a clinically actionable genomic profile, and (4) patients in whom ctDNA was not detected. Log-rank tests were used to compare TTD across these subgroups. In addition, a Cox proportional hazards model was performed with these four subgroups to generate pairwise hazard ratios (HRs) with 95% confidence intervals. A p-value of <0.05 was considered statistically significant.
Subgroup analyses were performed for patients who received EGFR-targeted therapy or immune checkpoint inhibitors, and the TTD was compared between patients whose ctDNA profiles supported the choice of the targeted treatment (matched) and those whose profiles did not support such treatment (unmatched).
All analyses were conducted using SAS software package 9.4 (SAS Institute, Cary, NC, USA).
3. Results
3.1. Patients
A total of 18,875 patients with primary colon or rectal cancer were identified in the GuardantINFORM database. Of these, a Guardant360 test was performed within 90 days of starting second- or third-line therapy in 1646 patients, 1569 of whom had at least two medical or pharmacy claims in the 6 months prior to first-line therapy. A total of 1064 patients with metastatic disease that could be confirmed by a relevant claim were analyzed (Figure 1). Subsequent treatment information was available for all 1064 patients.
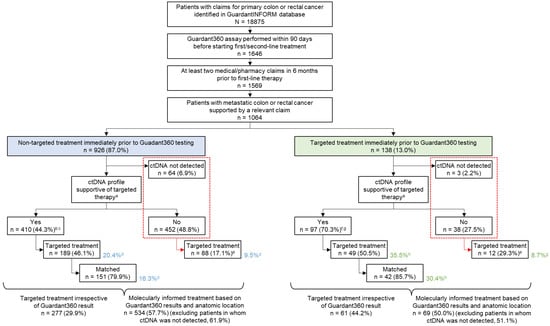
Figure 1.
Disposition of patients. Percentages were calculated using the value in the preceding box as the denominator. a Presence of OncoKB level 1–2 or lack of R1; b 47.6% of samples with ctDNA present; c Excludes 48 patients with right-sided mCRC and RAS wild-type only; d Percentage calculated using non-targeted treatment (blue shaded box) as the denominator; e Percentage calculated using ‘No’ and ‘ctDNA not detected’ (outlined with red dashed box) as the denominator; f 71.9% of samples with ctDNA present; g Excludes 9 patients with right-sided mCRC and RAS wild-type only; h Percentage calculated using targeted treatment (green shaded box) as the denominator; Matched, patients who received a treatment matched to the molecular alterations revealed by the ctDNA assay; targeted, patients who received a molecularly targeted treatment.
Of 1064 patients, 642 patients underwent ctDNA testing prior to second-line treatment (second-line cohort) and 422 patients underwent testing prior to third-line treatment (third-line cohort). Overall, 62 of 642 patients in the second-line cohort received first-line targeted therapy and 76 of 422 patients in the third-line cohort received second-line targeted therapy before ctDNA testing. Patients who received prior targeted therapies were assumed to be more likely to have tumors with relevant genomic biomarkers; therefore, these two independent groups were combined (n = 138, 13% of all patients in the cohort; green box in Figure 1) to increase confidence in subsequent analyses, and results were compared to those from patients who had not received immediate prior targeted therapy (n = 926; blue box in Figure 1).
The median age for patients included in this analysis was 59 years and 44.8% were female. The anatomic location of the primary tumor as described in the claims was left-sided for 44.5%, right-sided for 18.9%, mixed for 25.8%, and unspecified for 10.8%. The most common treatments received immediately prior to ctDNA testing were chemotherapy alone (365/1064, 34.3%) or combination regimens including VEGF-targeted treatment, but without other targeted agents (561/1064). Targeted therapy alone or in combination with other medicines was administered to 138 patients, most of whom received EGFR-targeted treatment (125/138, 90.6%) (Table 1).

Table 1.
Treatments immediately prior to ctDNA testing.
3.2. Overview of Alterations Detected
ctDNA was detected in plasma from 997 of 1064 patients tested (93.7%). Clinically actionable molecular profiles were detected in 507 samples (47.7% of all patients tested; 50.9% of samples with detectable ctDNA). Actionable profiles were more frequently detected in patients who had previously received targeted therapy (70.3% (97/138) of all samples tested; 71.9% (97/135) of samples with detectable ctDNA) than in patients who had received non-targeted therapy (44.3% (410/926); 47.6% (410/862) of samples with detectable ctDNA) (p < 0.001 for comparisons of all samples and samples with ctDNA detected), suggesting the persistence of actionable profiles after treatment (Figure 1).
Considering only the samples with ctDNA present, mutations in RAS genes (KRAS or NRAS) were found in 45.6% (455/997). The frequency of KRAS mutations was lower (21.5% vs. 45.8%, p < 0.001) but the frequencies of BRAF V600E (12.6% vs. 6.5%, p = 0.019) and ERBB2 amplification (5.9% vs. 2.3%, p = 0.043) were higher among patients who had previously received targeted therapy than in patients who had received non-targeted therapy (Table 2). There was a non-significant trend for more NRAS mutations after targeted treatment (8.9% vs. 4.6%, p = 0.063). KRAS G12C was detected in 41 samples (4.1%). The assay also detected rare alterations such as MET exon 14 skipping and fusions involving FGFR3, RET, ALK, and ROS1 for which targeted therapies exist in other diseases but did not meet the definition for actionability in this analysis.

Table 2.
Frequencies of clinically informative alterations detected by ctDNA.
3.3. Evaluation of Molecularly Informed Therapy
Irrespective of ctDNA test results, targeted therapy was administered to 338 of 1064 patients (31.8%) in the subsequent treatment line immediately after ctDNA testing. However, patients with samples harboring clinically actionable molecular alterations according to ctDNA were more likely to receive targeted therapy than those who did not have actionable profiles (46.9% (238/507) vs. 18.0% (100/557; including patients with nondetectable ctDNA), p < 0.001). Targeted therapy for actionable profiles was administered to 189 of 926 patients (20.4%) without targeted treatment prior to ctDNA testing and to 49 of 138 patients (35.5%) with an immediate past history of targeted treatment (Figure 1). Among patients with samples containing clinically actionable molecular profiles, the rates of targeted treatment were similar between those who had received recent targeted therapy (49/97, 50.5%) and those who had not (189/410, 46.1%, p = 0.502).
3.4. Types of Treatments Administered after ctDNA Testing
The types of treatments administered after ctDNA testing are summarized in Table 3.

Table 3.
Systemic treatments after ctDNA testing.
Non-targeted therapies (chemotherapy and/or VEGF targeting agents) were administered to approximately two-thirds of patients (726/1064, 68.2%) and were considered molecularly informed in 56.2% (408/726). Among patients whose treatment was not molecularly informed, 50 samples lacked ctDNA and 269 samples had molecular profiles associated with eligibility for targeted therapy: 27 with BRAF V600E, 6 with ERBB2 amplification, 1 with MSI-high, 1 with MSI-high and concurrent BRAF V600E, and 234 without KRAS or NRAS mutations. Of these 234 patients without KRAS or NRAS mutations, 195 had no previous exposure to EGFR-targeting agents.
EGFR-targeted therapy was administered to 233 patients (21.9%), of whom 156 (67.0%) received treatment and were considered to be molecularly informed by ctDNA testing and primary tumor site (Table 3). Patients whose treatment was classified as not molecularly informed included 14 with no ctDNA detected and 23 with RAS mutations identified in ctDNA. Eighteen patients without RAS mutations had right-sided colon cancer. Other patients had actionable mutations in ctDNA, including 21 with BRAF V600E and 1 with MSI-high.
Immune checkpoint inhibitors either alone or in combination with other agents were administered to 78 patients (7.3%), 16 (20.5%) of whom had evidence of MSI-high status in ctDNA. Among the remaining 62 patients, 3 had undetectable ctDNA, and 20 had alternative genomic targets: 4 with BRAF V600E, 2 with ERBB2 amplification, and 14 without RAS mutations including 11 without prior EGFR antibody treatment. Of the remaining 39 patients treated with immunotherapy, 18 had not received all possible standard of care treatment including fluoropyrimidine, oxaliplatin, irinotecan, and VEGF-targeted therapy. For 21 patients who had exhausted standard of care therapies, information on tumor mutation burden was not available.
Among 16 patients treated with BRAF-targeted agents and 21 treated with ERBB2-targeted agents, ctDNA was unable to detect the relevant alteration in 3 and 2 patients, respectively.
3.5. Patient Outcomes
The median TTD for patients treated in the second- and third-line settings after ctDNA testing were similar (4.2 vs. 3.8 months, p = 0.1764; Supplementary Figure S1). Therefore, we included patients from both groups in the subsequent analyses to increase the sample size and to allow for a more robust analysis. As shown in Figure 2, we assessed TTD in the following four groups: (1) patients with a clinically actionable molecular profile according to ctDNA who received matched therapy, (2) patients with a clinically actionable molecular profile who did not receive matched therapy, (3) patients without a clinically actionable genomic profile, and (4) ctDNA not detected. TTD was significantly different among the four groups (log-rank p = 0.0073). The median TTD was longest among patients with an actionable profile who received matched therapy (5.3 months) and similar for the other groups (3.9 months for actionable profiles without matched therapy and 3.5 months each for non-actionable profiles or those without ctDNA detected). Among patients with actionable ctDNA profiles, there was a non-significant trend for longer TTD for those who received matched versus unmatched therapy (hazard ratio 1.18, 95% confidence interval, 0.96–1.46; p = 0.116). An analysis of overall survival, separated by line of therapy due to clear differences in outcomes favoring longer duration in the earlier line of treatment, revealed no clear patterns among the four groups (Supplementary Figure S2).
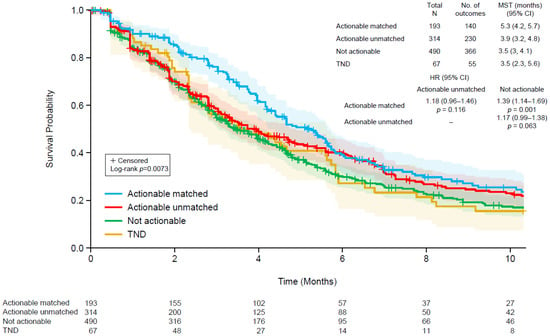
Figure 2.
Kaplan–Meier plots of time to discontinuation of therapy following ctDNA testing using a Guardant360 assay in four groups: (1) patients with a clinically actionable genomic profile who received matched therapy (blue), (2) patients with a clinically actionable genomic profile who did not receive a matched therapy (red), (3) patients without a clinically actionable genomic profile (green), and (4) patients in whom ctDNA was not detected (brown). The shaded regions indicate 95% confidence intervals. CI, confidence interval; HR, hazard ratio; MST, median survival time.
We also analyzed TTD for the 233 patients who received EGFR-targeted therapy (Figure 3A) and the 78 patients treated with immune checkpoint inhibitors (Figure 3B) because these were the most common types of targeted therapy used after ctDNA testing. We classified patients as those whose ctDNA profiles supported the choice of the targeted treatment (matched) and those whose profiles did not support such treatment (unmatched). Among patients who received EGFR-targeted therapy, the median TTD was significantly longer in those with matched treatment (5.5 months) than in those with unmatched treatment (3.2 months; log-rank p = 0.0040; Figure 3A). Among patients who received immunotherapy, the median TTD tended to be longer in those with matched treatment (6.0 months) than in those with unmatched treatment (4.2 months; log-rank p = 0.2024; Figure 3B).
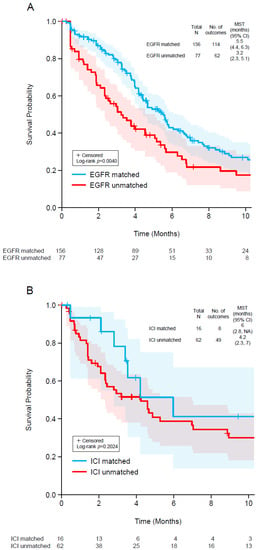
Figure 3.
Kaplan–Meier plots of time to discontinuation of EGFR-targeted therapy (A) and immune checkpoint inhibitor therapy (B) in two groups: (1) patients with a clinically actionable genomic profile who received matched targeted therapy (blue) and (2) patients whose genomic profile did not support the therapy used. The shaded regions indicate 95% confidence intervals. CI, confidence interval; EGFR, epidermal growth factor receptor; ICI, immune checkpoint inhibitor; MST, median survival time.
4. Discussion
For patients with mCRC and disease progression, we analyzed a real-world medical claims database combined with ctDNA test results to assess the types of therapeutic decisions made based on ctDNA profiling and the clinical outcomes associated with those decisions.
The assay used in this analysis detected ctDNA in 93.7% of the samples, a similar result to that previously reported for plasma samples from a large cohort of mCRC patients tested in routine clinical practice [34]. Clinically actionable ctDNA profiles were detected in approximately half (507/1064, 47.7%) of all patients tested, not only among those who had received targeted therapy immediately prior to testing (97/138, 70.3%) but also in those with disease progression after non-targeted therapy (410/926, 44.3%).
The frequencies of informative genomic alterations detected in this study were generally consistent with those described in previous studies [35,36,37,38,39]. KRAS mutations were the most common (424 of 997 samples with ctDNA detected, 42.5%), followed by BRAF V600E (7.3%), NRAS (5.2%), and ERBB2 amplification (2.8%). MSI-high assessment was added to the ctDNA assay in 2018, but the database includes patients tested from 2014; therefore, the observed proportion of samples with MSI-high status (2.0%) most likely underestimates the true prevalence, which is expected to be up to 5% [40]. Among patients with detectable ctDNA, the proportion of patients with KRAS mutations was lower in those who had previously received targeted therapy (21.5%) than in those who had not (45.8%), presumably due to prior selection for RAS wild-type profiles in consideration of EGFR-targeted regimens, which were the most commonly used targeted therapies prior to ctDNA testing. However, there was a trend for more NRAS mutations among patients exposed to prior targeted treatment (8.9%) vs. patients not exposed to prior targeted therapy (4.6%) (p = 0.063). As expected, prior targeted therapy, which was overwhelmingly EGFR-targeted, increased the proportion of cancers with BRAF V600E (12.6% vs. 6.5%, p = 0.019) and ERBB2 amplification (5.9% vs. 2.3%, p = 0.043).
Targeted therapies were more likely to be administered to patients with actionable ctDNA profiles (238/507, 46.9%) than to those without actionable profiles (100/557, 18.0%), but only slightly more than one-third of patients with actionable profiles received appropriately matched therapy (38.1%, 193/507). Excluding patients in whom ctDNA could not be detected, molecularly informed therapy, targeted or not, was administered to 60.5% of patients (603/997). However, because the claims database used in this study does not include results of other biomarker tests, we cannot exclude the possibility that some treatment decisions were molecularly informed by other assays.
In one recent study of 78 patients with mCRC, alterations that could be targeted by available therapies were detected using ctDNA in 59 (75.6%), and 45 satisfied OncoKB criteria in a manner similar to that employed in our study [36]. In another study of 94 patients who underwent ctDNA testing, non-synonymous mutations were detected in 74 patients (79%), of whom 69 (73%) had potentially actionable alterations [37]. In a study that used cell-free DNA (cfDNA) samples from 128 patients with mCRC, somatic alterations were detected in 100 (78%), of which 50% had potentially actionable alterations [41]. However, even under controlled study circumstances, targeted therapy may not be administered to all potentially eligible patients. For example, in the MOSCATO 01 trial in which NGS was used to guide treatment decisions in a cohort of 948 patients with advanced cancers who underwent tumor biopsy, a molecular portrait was developed for 843. An actionable alteration was found in 411 patients, but only 199 (48.4%) received a targeted therapy matched to the alteration [42].
Our findings support the clinical utility of matched therapy for mCRC patients with actionable molecular profiles detected by ctDNA. Median TTD, a surrogate for progression-free survival, was longest for patients with actionable profiles who received matched targeted treatment (5.3 months) than in those with actionable profiles who received unmatched treatment (3.9 months). Patients without actionable ctDNA profiles or those in whom ctDNA was not detected had the shortest median TTD (3.5 months each). These results are consistent with those of other studies demonstrating the importance of matching treatment to tissue-based molecular profiles [36,37,42]. The survival benefit of molecularly matched therapy based on tissue-based testing was previously reported [43,44,45,46]. However, the utility of ctDNA genotyping for molecularly matched therapy in real-world clinical practice, especially for mCRC patients following prior targeted therapies, has not been fully addressed. The clinical outcomes of 35 mCRC patients from a single institution suggested that matched therapy based on ctDNA results achieved a higher tumor response rate than unmatched therapy, with a trend for longer progression-free survival [37]. Our study, which included patients from academic and community-based settings, confirmed the usefulness of ctDNA genotyping for identification of patients who would benefit from targeted therapies.
One important role of ctDNA testing prior to later-line therapies is identification of resistance mutations. For example, testing identified patients who are less likely to respond to EGFR-targeted therapy. We observed that TTD following EGFR-targeted therapy was significantly prolonged when treatment was matched to the ctDNA profile. Furthermore, even though the absence of detectable ctDNA in plasma samples of mCRC patients is associated with a better prognosis [47], the TTD was longer in patients with a clinically actionable genomic profile who received matched therapy than in ctDNA-negative patients. This underscores the efficacy of matched targeted therapy based on ctDNA genotyping.
Although ctDNA testing identified patients who are less likely to benefit from EGFR-targeted treatments, this class of therapy appeared to be under-utilized in this cohort. A small proportion of patients received EGFR-targeted therapies immediately prior to ctDNA testing (125/1064, 11.7%), although it is expected that about 40% should be eligible [9,12]. After ctDNA testing, 248 patients with molecular and clinical profiles supporting EGFR-targeted therapies received other types of therapies instead. For most of these, there was no prior history of EGFR-targeted treatment. Although chemotherapy with or without anti-VEGF therapy can be effective in this setting, clinicians and patients should be aware of alternative treatment options. It is also important to consider that ctDNA can identify patients who should not receive EGFR-targeted therapy. The median TTD was shorter for patients treated with EGFR-targeted agents with unfavorable ctDNA results (77/233 treated; 3.2 months) compared with patients whose treatment was matched to the ctDNA profile (5.5 months).
Most patients treated with immune checkpoint inhibitors did not have MSI-high detected by ctDNA (62/78, 79.5%). There was a trend toward longer median TTD among patients treated with immunotherapy and MSI-high samples (6 months) compared to those without MSI-high (4.2 months). However, this was not statistically significant due to the small number of patients with MSI-high and treated with immune checkpoint inhibitors. The ctDNA assay used in this study introduced MSI-high assessment several years into the observation period and did not include all potential genomic information that could inform the suitability of immunotherapy. For example, POLD1 and POLE are not in the panel. Mutations in these genes, which occur in <1% of cases, are associated with response to immune checkpoint inhibitors [40,48]. Furthermore, the assay used in this study did not assess for the presence of high tumor mutation burden, which can be detected in approximately 3% of patients with mCRC without MSI-high [40]. The US FDA approved pembrolizumab for the treatment of solid tumors with a high tumor mutation burden (defined as ≥10 mutations/megabase), as determined by an FDA-approved test, in patients with disease progression after prior treatment and who have no satisfactory alternative treatment options [49]. However, standard treatment was not exhausted by over one-quarter of patients without MSI-high (18/62) who received an immune checkpoint inhibitor. Furthermore, even among mCRC patients with a high tumor mutation burden in the absence of MSI-high, the effectiveness of immunotherapy remains controversial [39,50].
There are some limitations of this study. In particular, the database lacked complete clinical information, such as performance status of the patients, physician-assessed tumor response, and results of concurrent biomarker testing that may have informed clinical decisions. Furthermore, because all treatments were identified using healthcare claims data, the database did not evaluate participation in clinical trials. Despite these factors, the genomic landscape observed in this study is generally consistent with that reported in other studies [51,52].
This study was designed to determine the proportion of patients for whom the potential use of targeted therapy would be appropriate following ctDNA testing. Therefore, our definition of molecularly informed therapy was intentionally narrow. Many of the treatment decisions made in this real-world setting may be valid, considering additional clinical information that was not available when these patients were treated. Tumor molecular information is necessary to make informed treatment decisions for patients with mCRC, but it should not be the only consideration.
In summary, this analysis of a real-world database demonstrates the clinical value of plasma-based comprehensive genomic profiling in mCRC patients with disease progression after one or two lines of prior therapy. Analysis of ctDNA identifies patients who are candidates for new or further targeted therapy, regardless of prior treatment history. Most importantly, in standard clinical practice, patients who received targeted treatments that matched their ctDNA test results had more favorable treatment outcomes than patients who received unmatched therapies. The modest improvement in TTD with the use of matched therapy for actionable alterations underscores the need for continued improvement in the identification of predictive biomarkers and the development of therapeutic agents to target such biomarkers. Future analyses of real-world clinical practice databases will help to further characterize the evolving role of ctDNA testing and its potential to influence the clinical outcomes of people living with advanced CRC.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/curroncol29050277/s1, Figure S1: Kaplan–Meier plots of time to treatment discontinuation following second-line (blue) or third-line (red) therapy after Guardant360 testing. The shaded regions indicate 95% confidence intervals. CI, confidence interval; MST, median survival time, Figure S2: Kaplan–Meier plots of overall survival following second- (A) or third-(B) line therapy in patients who underwent Guardant360 testing: (1) patients with a clinically actionable genomic profile who received matched therapy (blue); (2) patients with a clinically actionable genomic profile who did not receive matched therapy (red); (3) patients without a clinically actionable genomic profile (green); and (4) patients in whom ctDNA was not detected (brown). The shaded regions indicate 95% confidence intervals.
Author Contributions
Conceptualization, Y.N., S.O., N.Z., T.Y.; methodology, Y.N., S.O., N.Z., J.L., T.Y.; software, JL; formal analysis, J.L., N.Z., S.O.; resources, S.O., N.Z.; writing—original draft preparation, S.O.; writing—review and editing, Y.N., S.O., N.Z., J.L., T.Y.; funding acquisition, S.O. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Guardant Health.
Institutional Review Board Statement
Ethics approval and participant consent were not necessary because this study used a deidentified database in accordance with US patient confidentiality requirements set forth in Sections 164.514 (a)–(b)1ii of the Health Insurance Portability and Accountability Act (HIPAA) regarding the determination and documentation of statistically deidentified data.
Informed Consent Statement
Ethics approval and participant consent were not necessary because this study used a deidentified database in accordance with US patient confidentiality requirements set forth in Sections 164.514 (a)–(b)1ii of the Health Insurance Portability and Accountability Act (HIPAA) regarding the determination and documentation of statistically deidentified data.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available and cannot be shared due to the use of a third-party healthcare claims database. Researchers interested in replicating our study or pursuing new research topics should contact Guardant Health (https://guardanthealth.com/products/biopharma-solutions/real-world-evidence/, accessed on 18 March 2022) directly.
Acknowledgments
The authors thank Nicholas D. Smith (EMC K.K.) for medical writing support, which was funded by Guardant Health.
Conflicts of Interest
Y.N.: Research grants from Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Genomedia Inc., Guardant Health Inc., Daiichi Sankyo Co., Ltd., Roche Diagnostics K.K., and Seagen Inc. S.O.: Employment: Guardant Health AMEA, Inc; Stock ownership: Guardant Health, Inc and AstraZeneca. N.Z.: Employment: Guardant Health, Inc. J.L.: Employment: Guardant Health, Inc. T.Y.: Research grants to the institution from Ono Pharmaceutical Co., Ltd., Sanofi K.K., Daiichi Sankyo Co., Ltd., PAREXEL International Inc., Pfizer Japan Inc., Taiho Pharmaceutical Co., Ltd., MSD K.K., Amgen K.K., Genomedia Inc., Sysmex Corporation, Chugai Pharmaceutical Co., Ltd., and Nippon Boehringer Ingelheim Co., Ltd.; Payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing or educational events from Taiho Pharmaceutical Co., Ltd. Chugai Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Merck Biopharma Co., Ltd., Bayer Yakuhin, Ltd., Ono Pharmaceutical Co., Ltd., and MSD K.K.
References
- Alexovič, M.; Urban, P.L.; Tabani, H.; Sabo, J. Recent advances in robotic protein sample preparation for clinical analysis and other biomedical applications. Clin. Chim. Acta 2020, 507, 104–116. [Google Scholar] [CrossRef]
- Ou, F.S.; Michiels, S.; Shyr, Y.; Adjei, A.A.; Oberg, A.L. Biomarker discovery and validation: Statistical considerations. J. Thorac. Oncol. 2021, 16, 537–545. [Google Scholar] [CrossRef]
- Rogers, J.E.; Dasari, A. Pharmacotherapy for unresectable metastatic colorectal cancer. Expert Opin. Pharm. 2022, 23, 211–220. [Google Scholar] [CrossRef]
- Yu, I.S.; Kopetz, S. The emergence of targetable pathways in colorectal cancer. Clin. Adv. Hematol. Oncol. 2021, 19, 774–783. [Google Scholar]
- De Roock, W.; De Vriendt, V.; Normanno, N.; Ciardiello, F.; Tejpar, S. KRAS, BRAF, PIK3CA, and PTEN mutations: Implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011, 12, 594–603. [Google Scholar] [CrossRef]
- Bokemeyer, C.; Köhne, C.H.; Ciardiello, F.; Lenz, H.J.; Heinemann, V.; Klinkhardt, U.; Beier, F.; Duecker, K.; van Krieken, J.H.; Tejpar, S. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. Eur. J. Cancer 2015, 51, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.Y.; Oliner, K.S.; Siena, S.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013, 369, 1023–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douillard, J.Y.; Siena, S.; Cassidy, J.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Final results from PRIME: Randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann. Oncol. 2014, 25, 1346–1355. [Google Scholar] [CrossRef]
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef] [Green Version]
- Van Cutsem, E.; Köhne, C.H.; Hitre, E.; Zaluski, J.; Chang Chien, C.R.; Makhson, A.; D’Haens, G.; Pintér, T.; Lim, R.; Bodoky, G.; et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 1408–1417. [Google Scholar] [CrossRef] [Green Version]
- Van Cutsem, E.; Lenz, H.J.; Köhne, C.H.; Heinemann, V.; Tejpar, S.; Melezínek, I.; Beier, F.; Stroh, C.; Rougier, P.; van Krieken, J.H.; et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J. Clin. Oncol. 2015, 33, 692–700. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zheng, J.; Yang, Y.; Lu, J.; Gao, J.; Lu, T.; Sun, J.; Jiang, H.; Zhu, Y.; Zheng, Y.; et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: Analysis of 1110 cases. Sci. Rep. 2015, 5, 18678. [Google Scholar] [CrossRef]
- Lièvre, A.; Laurent-Puig, P. Toward an individualizing therapy for colorectal cancer: The example of the anti-EGFR monoclonal antibodies. Per. Med. 2009, 6, 145–157. [Google Scholar] [CrossRef]
- Stein, A.; Bokemeyer, C. How to select the optimal treatment for first line metastatic colorectal cancer. World J. Gastroenterol. 2014, 20, 899–907. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [Green Version]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Okamoto, W.; Kato, T.; Esaki, T.; Kato, K.; Komatsu, Y.; Yuki, S.; Masuishi, T.; Nishina, T.; Ebi, H.; et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: A phase 2 trial. Nat. Med. 2021, 27, 1899–1903. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology. Colon Cancer Version 3.2021—10 September 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. (accessed on 17 February 2022).
- Parseghian, C.M.; Napolitano, S.; Loree, J.M.; Kopetz, S. Mechanisms of Innate and Acquired Resistance to Anti-EGFR Therapy: A Review of Current Knowledge with a Focus on Rechallenge Therapies. Clin. Cancer Res. 2019, 25, 6899–6908. [Google Scholar] [CrossRef]
- Piawah, S.; Venook, A.P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019, 125, 4139–4147. [Google Scholar] [CrossRef]
- Shuford, R.A.; Cairns, A.L.; Moaven, O. Precision Approaches in the Management of Colorectal Cancer: Current Evidence and Latest Advancements Towards Individualizing the Treatment. Cancers 2020, 12, 3481. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Hurwitz, H.; Raghav, K.P.S.; McWilliams, R.R.; Fakih, M.; VanderWalde, A.; Swanton, C.; Kurzrock, R.; Burris, H.; Sweeney, C.; et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): An updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019, 20, 518–530. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Lonardi, S.; Martino, C.; Fenocchio, E.; Tosi, F.; Ghezzi, S.; Leone, F.; Bergamo, F.; Zagonel, V.; Ciardiello, F.; et al. Pertuzumab and trastuzumab emtansine in patients with HER2-amplified metastatic colorectal cancer: The phase II HERACLES-B trial. ESMO Open 2020, 5, e000911. [Google Scholar] [CrossRef]
- Siena, S.; Di Bartolomeo, M.; Raghav, K.; Masuishi, T.; Loupakis, F.; Kawakami, H.; Yamaguchi, K.; Nishina, T.; Fakih, M.; Elez, E.; et al. Trastuzumab deruxtecan (DS-8201) in patients with HER2-expressing metastatic colorectal cancer (DESTINY-CRC01): A multicentre, open-label, phase 2 trial. Lancet Oncol. 2021, 22, 779–789. [Google Scholar] [CrossRef]
- Strickler, J.H.; Zemla, T.; Ou, F.S.; Cercek, A.; Wu, C.; Sanchez, F.A.; Hubbard, J.; Jaszewski, B.; Bandel, L.; Schweitzer, B.; et al. Trastuzumab and tucatinib for the treatment of HER2 amplified metastatic colorectal cancer (mCRC): Initial results from the MOUNTAINEER trial. Ann. Oncol. 2019, 30, v200. [Google Scholar] [CrossRef]
- Tosi, F.; Sartore-Bianchi, A.; Lonardi, S.; Amatu, A.; Leone, F.; Ghezzi, S.; Martino, C.; Bencardino, K.; Bonazzina, E.; Bergamo, F.; et al. Long-term Clinical Outcome of Trastuzumab and Lapatinib for HER2-positive Metastatic Colorectal Cancer. Clin. Colorectal. Cancer 2020, 19, 256–262.e252. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B., 3rd; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal-Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020, 17, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Taniguchi, H.; Ikeda, M.; Bando, H.; Kato, K.; Morizane, C.; Esaki, T.; Komatsu, Y.; Kawamoto, Y.; Takahashi, N.; et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 2020, 26, 1859–1864. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, J.I.; Vincent, J.J.; Mortimer, S.; Vowles, J.V.; Ulrich, B.C.; Banks, K.C.; Fairclough, S.R.; Zill, O.A.; Sikora, M.; Mokhtari, R.; et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin. Cancer Res. 2018, 24, 3539–3549. [Google Scholar] [CrossRef] [Green Version]
- Guardant Health Inc. Guardant360 CDx Technical Information, D-001590 R3, July 2021. Available online: https://guardant360cdx.com/technicalinfo/ (accessed on 11 April 2022).
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 2017, 1–16. [Google Scholar] [CrossRef]
- Brulé, S.Y.; Jonker, D.J.; Karapetis, C.S.; O’Callaghan, C.J.; Moore, M.J.; Wong, R.; Tebbutt, N.C.; Underhill, C.; Yip, D.; Zalcberg, J.R.; et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur. J. Cancer 2015, 51, 1405–1414. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.S.; Fang, H.Y.; Hung, Y.C.; Ke, T.W.; Chang, C.M.; Liu, T.Y.; Chen, Y.C.; Chao, D.S.; Huang, H.Y.; Chang, J.G. Correlation of genomic alterations between tumor tissue and circulating tumor DNA by next-generation sequencing. J. Cancer Res. Clin. Oncol. 2018, 144, 2167–2175. [Google Scholar] [CrossRef]
- Choi, I.S.; Kato, S.; Fanta, P.T.; Leichman, L.; Okamura, R.; Raymond, V.M.; Lanman, R.B.; Lippman, S.M.; Kurzrock, R. Genomic Profiling of Blood-Derived Circulating Tumor DNA from Patients with Colorectal Cancer: Implications for Response and Resistance to Targeted Therapeutics. Mol. Cancer 2019, 18, 1852–1862. [Google Scholar] [CrossRef] [Green Version]
- Kato, S.; Schwaederlé, M.C.; Fanta, P.T.; Okamura, R.; Leichman, L.; Lippman, S.M.; Lanman, R.B.; Raymond, V.M.; Talasaz, A.; Kurzrock, R. Genomic Assessment of Blood-Derived Circulating Tumor DNA in Patients With Colorectal Cancers: Correlation With Tissue Sequencing, Therapeutic Response, and Survival. JCO Precis. Oncol. 2019, 3, 1–16. [Google Scholar] [CrossRef]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar] [CrossRef]
- Innocenti, F.; Ou, F.S.; Qu, X.; Zemla, T.J.; Niedzwiecki, D.; Tam, R.; Mahajan, S.; Goldberg, R.M.; Bertagnolli, M.M.; Blanke, C.D.; et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J. Clin. Oncol. 2019, 37, 1217–1227. [Google Scholar] [CrossRef]
- Fabrizio, D.A.; George, T.J., Jr.; Dunne, R.F.; Frampton, G.; Sun, J.; Gowen, K.; Kennedy, M.; Greenbowe, J.; Schrock, A.B.; Hezel, A.F.; et al. Beyond microsatellite testing: Assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J. Gastrointest. Oncol. 2018, 9, 610–617. [Google Scholar] [CrossRef]
- Pereira, A.A.L.; Morelli, M.P.; Overman, M.; Kee, B.; Fogelman, D.; Vilar, E.; Shureiqi, I.; Raghav, K.; Eng, C.; Manuel, S.; et al. Clinical utility of circulating cell-free DNA in advanced colorectal cancer. PLoS ONE 2017, 12, e0183949. [Google Scholar] [CrossRef] [Green Version]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [Green Version]
- Pishvaian, M.J.; Blais, E.M.; Brody, J.R.; Lyons, E.; DeArbeloa, P.; Hendifar, A.; Mikhail, S.; Chung, V.; Sahai, V.; Sohal, D.P.S.; et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: A retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020, 21, 508–518. [Google Scholar] [CrossRef]
- Schwaederle, M.; Parker, B.A.; Schwab, R.B.; Daniels, G.A.; Piccioni, D.E.; Kesari, S.; Helsten, T.L.; Bazhenova, L.A.; Romero, J.; Fanta, P.T.; et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol. Cancer 2016, 15, 743–752. [Google Scholar] [CrossRef] [Green Version]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; Lazar, V.; Kurzrock, R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J. Clin. Oncol. 2015, 33, 3817–3825. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Iskander, N.G.; Hong, D.S.; Wheler, J.J.; Falchook, G.S.; Fu, S.; Piha-Paul, S.; Naing, A.; Janku, F.; Luthra, R.; et al. Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center initiative. Clin. Cancer Res. 2012, 18, 6373–6383. [Google Scholar] [CrossRef] [Green Version]
- Pairawan, S.; Hess, K.R.; Janku, F.; Sanchez, N.S.; Mills Shaw, K.R.; Eng, C.; Damodaran, S.; Javle, M.; Kaseb, A.O.; Hong, D.S.; et al. Cell-free Circulating Tumor DNA Variant Allele Frequency Associates with Survival in Metastatic Cancer. Clin. Cancer Res. 2020, 26, 1924–1931. [Google Scholar] [CrossRef]
- Wang, F.; Zhao, Q.; Wang, Y.N.; Jin, Y.; He, M.M.; Liu, Z.X.; Xu, R.H. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol. 2019, 5, 1504–1506. [Google Scholar] [CrossRef] [Green Version]
- Inc, M.C. KEYTRUDA® (Pembrolizumab) Injection, for Intravenous Use. Highlights of Prescribing Information, Revised February 2022. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/125514s125lbl.pdf (accessed on 17 February 2022).
- Rousseau, B.; Foote, M.B.; Maron, S.B.; Diplas, B.H.; Lu, S.; Argilés, G.; Cercek, A.; Diaz, L.A., Jr. The Spectrum of Benefit from Checkpoint Blockade in Hypermutated Tumors. N. Engl. J. Med. 2021, 384, 1168–1170. [Google Scholar] [CrossRef]
- Strickler, J.H.; Loree, J.M.; Ahronian, L.G.; Parikh, A.R.; Niedzwiecki, D.; Pereira, A.A.L.; McKinney, M.; Korn, W.M.; Atreya, C.E.; Banks, K.C.; et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov. 2018, 8, 164–173. [Google Scholar] [CrossRef] [Green Version]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e123. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).