Genomic Landscape of Non-Small Cell Lung Cancer (NSCLC) in East Asia Using Circulating Tumor DNA (ctDNA) in Clinical Practice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Cell-Free Next Generation Sequencing
2.3. Statistical Analyses
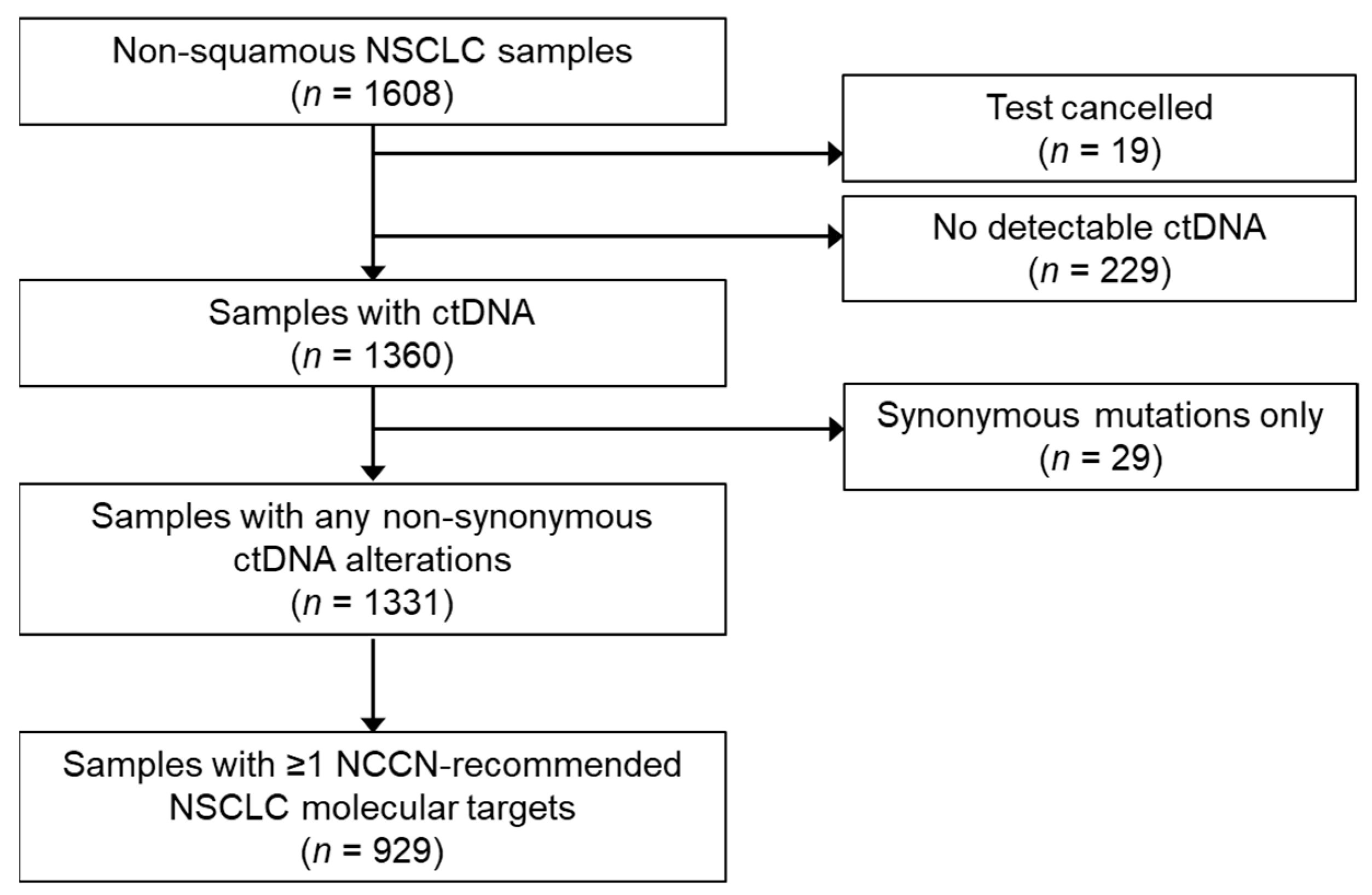
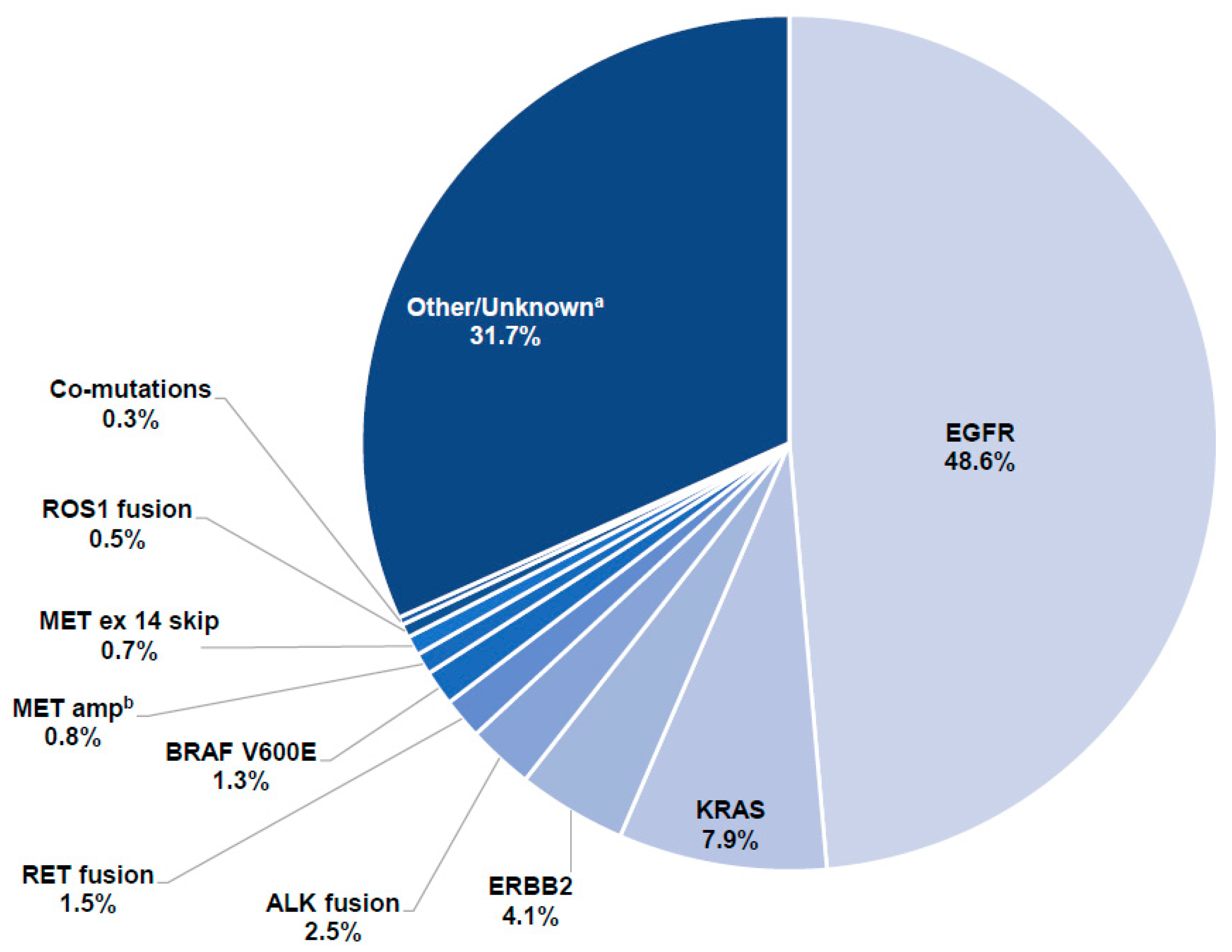
3. Results
3.1. Sample Characteristics
3.2. Detection of Driver Mutations
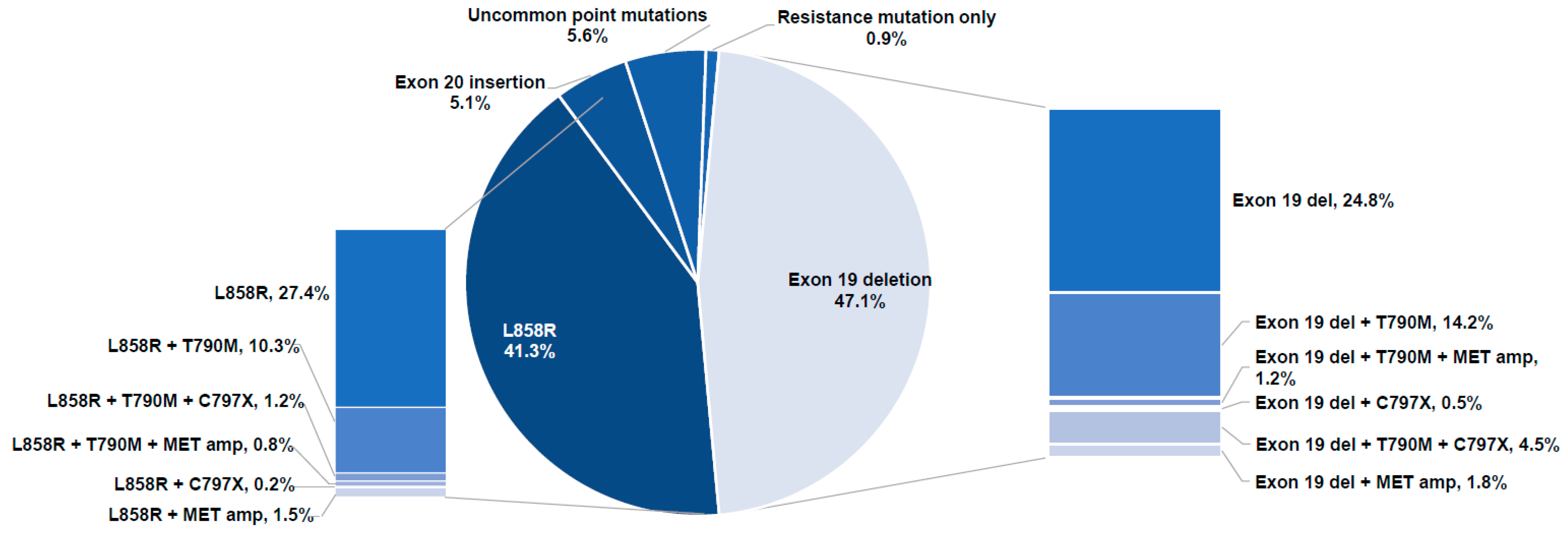
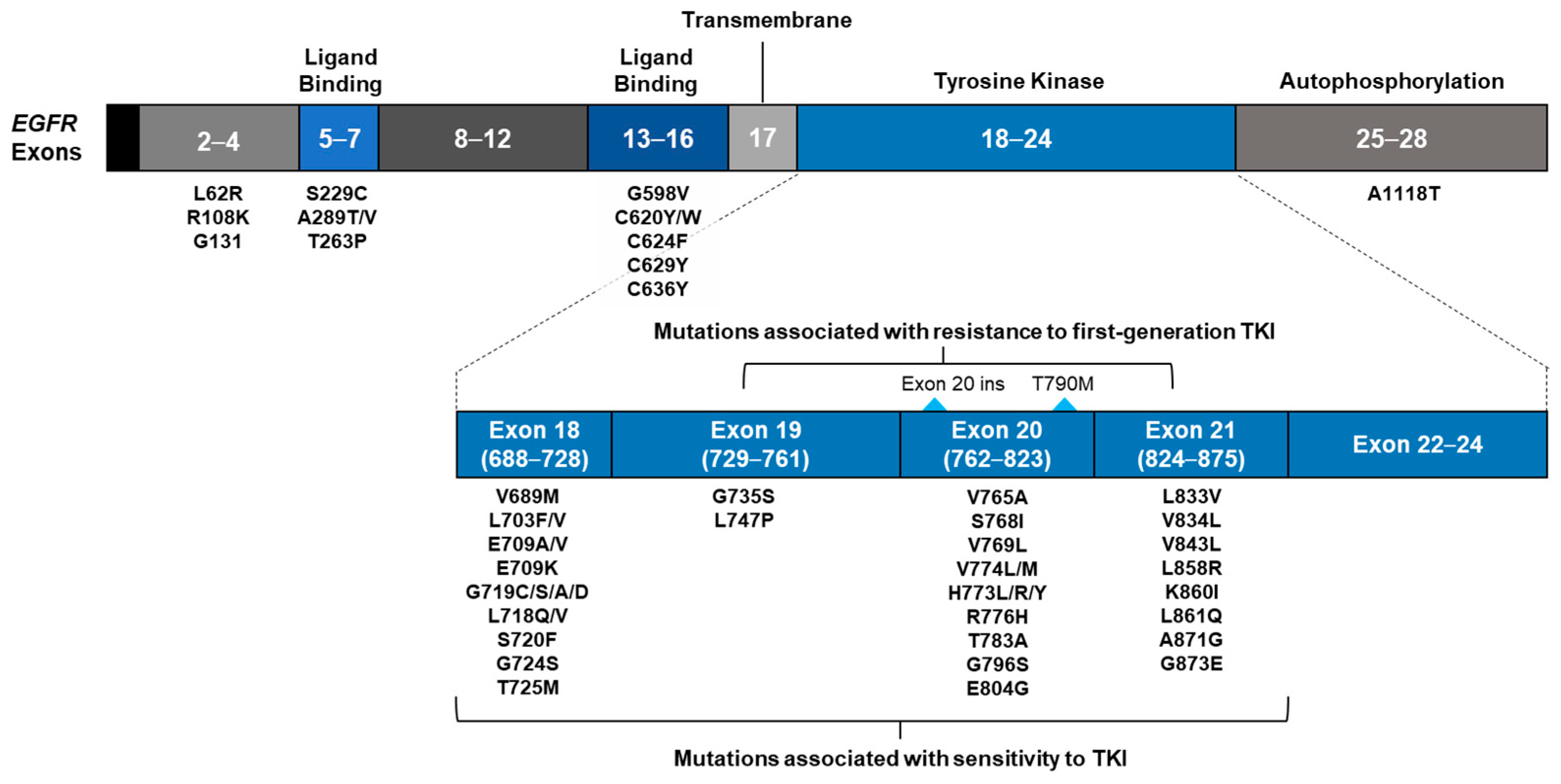
3.3. EGFR Mutations
3.3.1. Classification of Samples with EGFR Alterations by Driver Mutation
3.3.2. Classification of Samples with EGFR Mutations by Presence of EGFR TKI Resistance Alterations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. International Agency for Research on Cancer 2021: Estimated Number of New Cases in 2020, Worldwide, Both Sexes, All Ages; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Esposito Abate, R.; Frezzetti, D.; Maiello, M.R.; Gallo, M.; Camerlingo, R.; De Luca, A.; De Cecio, R.; Morabito, A.; Normanno, N. Next Generation Sequencing-Based Profiling of Cell Free DNA in Patients with Advanced Non-Small Cell Lung Cancer: Advantages and Pitfalls. Cancers 2020, 12, 3084. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network. In Proceedings of the NCCN Clinical Practice Guidelines in Oncology—Non-Small Cell Lung Cancer, Plymouth, PA, USA, 15 June 2021. Version 5.
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 2. J. Natl. Compr. Canc. Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Hanna, N.; Johnson, D.; Temin, S.; Baker, S., Jr.; Brahmer, J.; Ellis, P.M.; Giaccone, G.; Hesketh, P.J.; Jaiyesimi, I.; Leighl, N.B.; et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J. Clin. Oncol. 2017, 35, 3484–3515. [Google Scholar] [CrossRef] [PubMed]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [PubMed] [Green Version]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer: A Consensus Statement from The International Association for the Study of Lung Cancer (IASLC). J. Thorac. Oncol. 2021, 10, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, V.M.L.; Carvalho, L. Heterogeneity in Lung Cancer. Pathobiology 2018, 85, 96–107. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Vincent, J.J.; Mortimer, S.; Vowles, J.V.; Ulrich, B.C.; Banks, K.C.; Fairclough, S.R.; Zill, O.A.; Sikora, M.; Mokhtari, R.; et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin. Cancer Res. 2018, 24, 3539–3549. [Google Scholar] [CrossRef] [Green Version]
- Rijavec, E.; Coco, S.; Genova, C.; Rossi, G.; Longo, L.; Grossi, F. Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges. Cancers 2019, 12, 17. [Google Scholar] [CrossRef] [Green Version]
- Bracht, J.W.P.; Mayo-De-Las-Casas, C.; Berenguer, J.; Karachaliou, N.; Rosell, R. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr. Oncol. Rep. 2018, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Burstein, H.J.; Krilov, L.; Aragon-Ching, J.; Baxter, N.N.; Chiorean, E.G.; Chow, W.A.; De Groot, J.F.; Devine, S.M.; Dubois, S.G.; El-Deiry, W.; et al. Clinical Cancer Advances 2017: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 2017, 35, 1341–1367. [Google Scholar] [CrossRef] [PubMed]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.B.; Blau, S.; Collisson, E.A.; Drivers, S.G.; Hoon, D.S.B.; Kopetz, E.S.; et al. Data from: Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhu, M.; Wang, C. Precision oncology of lung cancer: Genetic and genomic differences in Chinese population. npj Precis. Oncol. 2019, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Choi, Y.-L.; Gong, Z.; Liu, X.; Lira, M.; Kan, Z.; Oh, E.; Wang, J.; Ting, J.C.; Ye, X.; et al. Comprehensive Characterization of Oncogenic Drivers in Asian Lung Adenocarcinoma. J. Thorac. Oncol. 2016, 11, 2129–2140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellison, G.; Zhu, G.; Moulis, A.; Dearden, S.; Speake, G.; McCormack, R. EGFR mutation testing in lung cancer: A review of available methods and their use for analysis of tumour tissue and cytology samples. J. Clin. Pathol. 2012, 66, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Keedy, V.L.; Temin, S.; Somerfield, M.R.; Beasley, M.B.; Johnson, D.H.; McShane, L.M.; Milton, D.T.; Strawn, J.R.; Wakelee, H.A.; Giaccone, G. American Society of Clinical Oncology Provisional Clinical Opinion: Epidermal Growth Factor Receptor (EGFR) Mutation Testing for Patients with Advanced Non–Small-Cell Lung Cancer Considering First-Line EGFR Tyrosine Kinase Inhibitor Therapy. J. Clin. Oncol. 2011, 29, 2121–2127. [Google Scholar] [CrossRef] [PubMed]
- Szumera-Ciećkiewicz, A.; Olszewski, W.T.; Tysarowski, A.; Kowalski, D.M.; Głogowski, M.; Krzakowski, M.; A Siedlecki, J.; Wągrodzki, M.; Prochorec-Sobieszek, M. EGFR mutation testing on cytological and histological samples in non-small cell lung cancer: A Polish, single institution study and systematic review of European incidence. Int. J. Clin. Exp. Pathol. 2013, 6, 2800. [Google Scholar]
- Luangdilok, S.; Wanchaijiraboon, P.; Chantranuwatana, P.; Teerapakpinyo, C.; Shuangshoti, S.; Sriuranpong, V. Cyclin D1 expression as a potential prognostic factor in advanced KRAS-mutant non-small cell lung cancer. Transl. Lung Cancer Res. 2019, 8, 959–966. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping with the Delivery of Personal-ized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.P.; Wynes, M.W.; Lantuejoul, S.; Soo, R.; Ramalingam, S.S.; Varella-Garcia, M.; Taylor, M.M.; Richeimer, K.; Wood, K.; Howell, K.E.; et al. The International Association for the Study of Lung Cancer Global Survey on Molecular Testing in Lung Cancer. J. Thorac. Oncol. 2020, 15, 1434–1448. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.E.; Choi, K.; Lanman, R.B.; Licitra, E.J.; Skrzypczak, S.M.; Benito, R.P.; Wu, T.; Arunajadai, S.; Kaur, S.; Harper, H.; et al. Genomic Profiling of Advanced Non–Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clin. Lung Cancer 2017, 18, 651–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatabe, Y.; Yoshiki, Y.; Matsumura, K.; Togo, K.; Kikkawa, H.; Iadeluca, L.; Li, B.; Nishio, K. Real-World Evidence of Diagnostic Testing for Driver Oncogene Mutations in Lung Cancer in Japan. JTO Clin. Res. Rep. 2021, 2, 10036. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, Y.; Zhao, J.; Liu, Y.; Gao, H.; Ma, K.; Zhang, S.; Xin, H.; Liu, J.; Han, C.; et al. Real-world EGFR testing in patients with stage IIIB/IV non-small-cell lung cancer in North China: A multicenter, non-interventional study. Thorac. Cancer 2018, 9, 1461–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Li, J.; Zhang, S.; Wang, M.; Yang, S.; Li, N.; Wu, G.; Liu, W.; Liao, G.; Cai, K.; et al. Molecular Epidemiology of EGFR Mutations in Asian Patients with Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology—Mainland China Subset Analysis of the PIONEER study. PLoS ONE 2015, 10, e0143515. [Google Scholar] [CrossRef]
- Lu, S.; Shih, J.Y.; Jang, T.W.; Liam, C.K.; Yu, Y. Afatinib as First-Line Treatment in Asian Patients with EGFR Mutation-Positive NSCLC: A Narrative Review of Real-World Evidence. Adv. Ther. 2021, 38, 2038–2053. [Google Scholar] [CrossRef]
- Gonzalvez, F.; Vincent, S.; Baker, T.E.; Gould, A.E.; Li, S.; Wardwell, S.D.; Nadworny, S.; Ning, Y.; Zhang, S.; Huang, W.S.; et al. Mobocertinib (TAK-788): A Targeted Inhibitor of EGFR Exon 20 Insertion Mutants in Non–Small Cell Lung Cancer. Cancer Discov. 2021, 11, 1672–1687. [Google Scholar] [CrossRef]
- Yun, J.; Lee, S.H.; Kim, S.Y.; Jeong, S.Y.; Kim, J.H.; Pyo, K.H.; Park, C.W.; Heo, S.G.; Yun, M.R.; Lim, S.; et al. Antitumor Activity of Amivantamab (JNJ-61186372), an EGFR-cMet Bispecific Antibody, in Diverse Models of EGFR Exon 20 Insertion-Driven NSCLC. Cancer Discov. 2020, 10, 1194–1209. [Google Scholar] [CrossRef]
- Kashima, K.; Kawauchi, H.; Tanimura, H.; Tachibana, Y.; Chiba, T.; Torizawa, T.; Sakamoto, H. CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation. Mol. Cancer Ther. 2020, 19, 2288–2297. [Google Scholar] [CrossRef]
- Bulbul, A.; Leal, A.; Husain, H. Applications of cell-free circulating tumor DNA detection in EGFR mutant lung cancer. J. Thorac. Dis. 2020, 12, 2877–2882. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Dai, L.; Wang, L.; Wang, W.; Wu, D.; Wang, K.; He, Z.; Wang, A.; Chen, H.; Zhang, P.; et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncol. 2019, 24, e1070–e1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Cong, X.; Liu, Z. Mutation Profile Assessed by Next-Generation Sequencing (NGS) of Circulating Tumor DNA (ctDNA) in Chinese Lung Adenocarcinoma Patients: Analysis of Real-World Data. BioMed Res. Int. 2021, 2021, 8817898. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhan, C.; Li, M.; Yang, X.; Yang, X.; Zhang, Y.; Lin, M.; Xia, Y.; Feng, M.; Wang, Q. Aberrant status and clinicopathologic characteristic associations of 11 target genes in 1,321 Chinese patients with lung adenocarcinoma. J. Thorac. Dis. 2018, 10, 398–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, C.R.; Garassino, M.C.; Nadal, E.; Öhrling, K.; Scheffler, M.; Mazières, J. On target: Rational approaches to KRAS inhibition for treatment of non-small cell lung carcinoma. Lung Cancer 2021, 160, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Liao, W.Y.; Su, K.Y.; Yu, S.L.; Huang, Y.L.; Yu, C.J.; Yang, J.C.H.; Shih, J.Y. Prognostic Characteristics and Immunotherapy Response of Patients with Nonsquamous NSCLC With Kras Mutation in East Asian Populations: A Single-Center Cohort Study in Taiwan. JTO Clin. Res. Rep. 2021, 2, 100140. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Abedpour, N.; Ueckeroth, F.; et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019, 14, 606–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Lim, J.S.; Poh, J.; Bivona, T.G. Targeted ctDNA sequencing analysis reveals concurrent genomic alterations and its impact on TKI and immune checkpoint inhibitor therapy in advanced NSCLC from Asian population. J. Clin. Oncol. 2021, 39, 3042. [Google Scholar] [CrossRef]
- Tran, H.T.; Lam, V.K.; Elamin, Y.Y.; Hong, L.; Colen, R.; Elshafeey, N.A.; Hassan, I.S.A.; Altan, M.; Blumenschein, G.R.; Rinsurongkawong, W.; et al. Clinical Outcomes in Non–Small-Cell Lung Cancer Patients Treated With EGFR-Tyrosine Kinase Inhibitors and Other Targeted Therapies Based on Tumor Versus Plasma Genomic Profiling. JCO Precis. Oncol. 2021, 5, 1241–1249. [Google Scholar] [CrossRef]
- Acuna-Hidalgo, R.; Sengul, H.; Steehouwer, M.; van de Vorst, M.; Vermeulen, S.H.; Kiemeney, L.A.; Veltman, J.A.; Gilissen, C.; Hoischen, A. Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life. Am. J. Hum. Genet. 2017, 101, 50–64. [Google Scholar] [CrossRef] [Green Version]
| Category | Total, n (%) (n = 661) |
|---|---|
| Driver mutation only | 411 (62.2) |
| Exon 19 deletion | |
| L858R | |
| Exon 20 insertion | |
| G719X | |
| Exon 19 insertion | |
| 1 uncommon SNV | |
| Driver mutation + T790M | 165 (25.0) |
| Exon 19 deletion + T790M | |
| L858R + T790M | |
| G719X + E709A + T790M | |
| Driver mutation + T790M + additional resistance mutation | 51 (7.7) |
| Exon 19 deletion + T790M + MET amplification | |
| Exon 19 deletion + T790M + C797X | |
| L858R + T790M + C797X | |
| L858R + T790M + MET amplification | |
| Driver mutation + T790M + other alterations | 0 |
| N/A | |
| Driver mutation + resistance mutation other than T790M | 28 (4.2) |
| Exon 19 deletion + C797X | |
| Exon 19 deletion + MET amplification | |
| L858R + C797X | |
| L858R + MET amplification | |
| Exon 20 insertion + MET amplification | |
| 1 uncommon SNV + C797X | |
| Resistance mutation only | 6 (0.9) |
| T790M alone |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, B.C.; Loong, H.H.F.; Tsai, C.-M.; Teo, M.L.P.; Kim, H.R.; Lim, S.M.; Jain, S.; Olsen, S.; Park, K. Genomic Landscape of Non-Small Cell Lung Cancer (NSCLC) in East Asia Using Circulating Tumor DNA (ctDNA) in Clinical Practice. Curr. Oncol. 2022, 29, 2154-2164. https://doi.org/10.3390/curroncol29030174
Cho BC, Loong HHF, Tsai C-M, Teo MLP, Kim HR, Lim SM, Jain S, Olsen S, Park K. Genomic Landscape of Non-Small Cell Lung Cancer (NSCLC) in East Asia Using Circulating Tumor DNA (ctDNA) in Clinical Practice. Current Oncology. 2022; 29(3):2154-2164. https://doi.org/10.3390/curroncol29030174
Chicago/Turabian StyleCho, Byoung Chul, Herbert H. F. Loong, Chun-Ming Tsai, Man Lung P. Teo, Hye Ryun Kim, Sun Min Lim, Suyog Jain, Steve Olsen, and Keunchil Park. 2022. "Genomic Landscape of Non-Small Cell Lung Cancer (NSCLC) in East Asia Using Circulating Tumor DNA (ctDNA) in Clinical Practice" Current Oncology 29, no. 3: 2154-2164. https://doi.org/10.3390/curroncol29030174
APA StyleCho, B. C., Loong, H. H. F., Tsai, C.-M., Teo, M. L. P., Kim, H. R., Lim, S. M., Jain, S., Olsen, S., & Park, K. (2022). Genomic Landscape of Non-Small Cell Lung Cancer (NSCLC) in East Asia Using Circulating Tumor DNA (ctDNA) in Clinical Practice. Current Oncology, 29(3), 2154-2164. https://doi.org/10.3390/curroncol29030174





