Risk Association of TOX3 and MMP7 Gene Polymorphisms with Sporadic Breast Cancer in Mexican Women
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Primer Design and Restriction Enzymes Selection
2.3. Genomic DNA Isolation and Genotyping
2.4. Statistical Analysis
3. Results
3.1. Clinical and Demographic Characteristics
3.2. PCR-RFLP Assays
3.3. Genotyping and Allelic Distributions in BC Cases and Controls
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iacoviello, L.; Bonaccio, M.; de Gaetano, G.; Donati, M.B. Epidemiology of breast cancer, a paradigm of the “common soil” hypothesis. Semin. Cancer Biol. 2021, 72, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Sharma, R. Global, regional, national burden of breast cancer in 185 countries: Evidence from GLOBOCAN 2018. Breast Cancer Res. Treat. 2021, 187, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, M.; Ciszewski, T.; Lopacka-Szatan, K.; Miotla, P.; Staroslawska, E. Breast cancer risk factors. Prz Menopauzalny 2015, 14, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Eliyatkin, N.; Yalcin, E.; Zengel, B.; Aktas, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; DMRh Aman, H.A. The Associations of Common Genetic Susceptibility Variants with Breast Cancer in Jordanian Arabs: A Case-Control Study. Asian Pac. J. Cancer Prev. 2020, 21, 3045–3054. [Google Scholar] [CrossRef]
- Zavala, V.A.; Serrano-Gomez, S.J.; Dutil, J.; Fejerman, L. Genetic Epidemiology of Breast Cancer in Latin America. Genes 2019, 10, 153. [Google Scholar] [CrossRef]
- Thanh, N.T.N.; Lan, N.T.T.; Phat, P.T.; Giang, N.D.T.; Hue, N.T. Two polymorphisms, rs2046210 and rs3803662, are associated with breast cancer risk in a Vietnamese case-control cohort. Genes Genet. Syst. 2018, 93, 101–109. [Google Scholar] [CrossRef]
- Zhang, L.; Long, X. Association of three SNPs in TOX3 and breast cancer risk: Evidence from 97275 cases and 128,686 controls. Sci. Rep. 2015, 5, 12773. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, Y.; Wang, W.; Liu, G.; Yu, Y.; Liao, M. Association of single nucleotide polymorphism rs3803662 with the risk of breast cancer. Sci. Rep. 2016, 6, 29008. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Yu, S.; Qian, P.; Guo, R.; Zhang, R.; Ao, Z.; Li, Q.; Wu, G.; Chen, Y.; Li, J.; et al. The breast cancer susceptibility-related polymorphisms at the TOX3/LOC643714 locus associated with lung cancer risk in a Han Chinese population. Oncotarget 2016, 7, 59742–59753. [Google Scholar] [CrossRef]
- Tajbakhsh, A.; Javan, F.A.; Rivandi, M.; Moezzi, A.; Abedini, S.; Asghari, M.; Farjami, Z.; Soltanian, H.; Shandiz, F.H.; Kooshyar, M.M.; et al. Significant association of TOX3/LOC643714 locus-rs3803662 and breast cancer risk in a cohort of Iranian population. Mol. Biol. Rep. 2019, 46, 805–811. [Google Scholar] [CrossRef]
- Liao, J.; Chen, Y.; Zhu, J.; Wang, Q.; Mo, Z. Polymorphisms in the TOX3/LOC643714 and risk of breast cancer in south China. Int. J. Biol. Markers 2018, 33, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Guo, G.; Wang, F.; Lv, P.; Zhu, M.; Gu, Y.; Han, M.; Pei, X. TOX high mobility group box family member 3 rs3803662 and breast cancer risk: A meta-analysis. J. Cancer Res. Ther. 2018, 14, S208–S212. [Google Scholar]
- Udler, M.S.; Ahmed, S.; Healey, C.S.; Meyer, K.; Struewing, J.; Maranian, M.; Kwon, E.M.; Zhang, J.; Tyrer, J.; Karlins, E.; et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet. 2010, 19, 2507–2515. [Google Scholar] [CrossRef]
- Seksenyan, A.; Kadavallore, A.; Walts, A.E.; de la Torre, B.; Berel, D.; Strom, S.P.; Aliahmad, P.; Funari, V.; Kaye, J. TOX3 is expressed in mammary ER(+) epithelial cells and regulates ER target genes in luminal breast cancer. BMC Cancer 2015, 15, 22. [Google Scholar] [CrossRef]
- Liang, C.; Huang, S.; Zhao, Y.; Chen, S.; Li, Y. TOX as a potential target for immunotherapy in lymphocytic malignancies. Biomark. Res. 2021, 9, 20. [Google Scholar] [CrossRef]
- NCBI The National Center for Biotechnology Information. MMP7 Matrix Metallopeptidase 7 [Homo Sapiens (Human)] Bethesda MD, USA. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=4316 (accessed on 1 August 2021).
- Roy, R.; Morad, G.; Jedinak, A.; Moses, M.A. Metalloproteinases and their roles in human cancer. Anat. Rec. (Hoboken) 2020, 303, 1557–1572. [Google Scholar] [CrossRef]
- Zhang, M.; Teng, X.D.; Guo, X.X.; Li, Z.G.; Han, J.G.; Yao, L. Expression of tissue levels of matrix metalloproteinases and their inhibitors in breast cancer. Breast 2013, 22, 330–334. [Google Scholar] [CrossRef] [PubMed]
- NCBI. National Center for Biotechnology Information dbSNP. Bethesda, MD, USA. Available online: https://www.ncbi.nlm.nih.gov/snp/ (accessed on 1 August 2021).
- Ensembl. EMBL’s European Bioinformatics Institute. Ensembl. BLAST/BLAT. Cambridgeshire, UK. Available online: https://www.ensembl.org/Multi/Tools/Blast (accessed on 1 August 2021).
- NEB. New England Biolabs Inc.- NEBcutter V2.0, Ipswich, Massachusetts, USA. Available online: http://nc2.neb.com/NEBcutter2/ (accessed on 1 August 2021).
- Nature Education. Scitable by Nature Education. Available online: https://www.nature.com/scitable/definition/hardy-weinberg-equilibrium-122/ (accessed on 1 January 2021).
- Elematore, I.; Gonzalez-Hormazabal, P.; Reyes, J.M.; Blanco, R.; Bravo, T.; Peralta, O.; Gomez, F.; Waugh, E.; Margarit, S.; Ibañez, G.; et al. Association of genetic variants at TOX3, 2q35 and 8q24 with the risk of familial and early-onset breast cancer in a South-American population. Mol. Biol. Rep. 2014, 41, 3715–3722. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, N.Y.; Cao, X.M.; Sun, X.; Shen, D.; Yuan, M.; Chen, J. Increased risk of breast cancer in individuals carrying the TNRC9 rs3803662 C > T polymorphism: A meta-analysis of case-control studies. Genet. Mol. Res. 2016, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Baumgartner, K.B.; Giuliano, A.R.; Byers, T.; Herrick, J.S.; Wolff, R.K. Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res. Treat. 2011, 129, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.C.; Tu, S.H.; Su, C.T.; Cho, E.C.; Wu, C.H.; Hsieh, M.C.; Lin, S.-Y.; Liu, Y.-R.; Hung, C.-S.; Chiou, H.-Y. A polygenic risk score for breast cancer risk in a Taiwanese population. Breast Cancer Res. Treat. 2017, 163, 131–138. [Google Scholar] [CrossRef]
- Ozgoz, A.; Icduygu, F.M.; Yukselturk, A.; Saml, I.H.; Ozturk, K.H.; Baskan, Z. Low-penetrance susceptibility variants and postmenopausal oestrogen receptor positive breast cancer. J. Genet. 2020, 99, 15. [Google Scholar] [CrossRef]
- Chen, M.B.; Wu, X.Y.; Shen, W.; Wei, M.X.; Li, C.; Cai, B.; Tao, G.-Q.; Lu, P. Association between polymorphisms of trinucleotide repeat containing 9 gene and breast cancer risk: Evidence from 62,005 subjects. Breast Cancer Res. Treat. 2011, 126, 177–183. [Google Scholar] [CrossRef]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef]
- Ruiz-Narvaez, E.A.; Rosenberg, L.; Cozier, Y.C.; Cupples, L.A.; Adams-Campbell, L.L.; Palmer, J.R. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1320–1327. [Google Scholar] [CrossRef]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Rafnar, T.; Gudmundsson, J.; Gudjonsson, S.A.; Masson, G.; Jakobsdottir, M.; Thorlacius, S.; Helgason, A.; et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007, 39, 865–869. [Google Scholar] [CrossRef]
- He, X.; Yao, G.; Li, F.; Li, M.; Yang, X. Risk-association of five SNPs in TOX3/LOC643714 with breast cancer in southern China. Int. J. Mol. Sci. 2014, 15, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, X.; Huang, Z.; Liu, Z.; Song, M.; Guo, Z. TNRC9/LOC643714 polymorphisms are not associated with breast cancer risk in Chinese women. Eur. J. Cancer Prev. 2009, 18, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Chen, P.; Hu, Z.; Shen, H.; Wang, F.; Chen, L.; Li, M.; Tang, J.; Wang, H.; Shen, H. Genetic variants in trinucleotide repeat-containing 9 (TNRC9) are associated with risk of estrogen receptor positive breast cancer in a Chinese population. Breast Cancer Res. Treat. 2010, 124, 237–241. [Google Scholar] [CrossRef]
- Figueroa-Gonzalez, G.; Arellano-Gutierrez, C.V.; Cortes, H.; Leyva-Gomez, G.; Carmen, M.G.; Bustamante-Montes, L.P.; Rodríguez-Morales, M.; López-Reyes, I.; Alcaraz-Estrada, S.L. Breast cancer-related single-nucleotide polymorphism and their risk contribution in Mexican women. J. Cancer Res. Ther. 2020, 16, 1279–1286. [Google Scholar] [PubMed]
- Shan, J.; Dsouza, S.P.; Bakhru, S.; Al-Azwani, E.K.; Ascierto, M.L.; Sastry, K.S.; Bedri, S.; Kizhakayil, D.; Aigha, I.I.; Malek, J.; et al. TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness. Cancer Res. 2013, 73, 2840–2849. [Google Scholar] [CrossRef]
- Tapper, W.; Hammond, V.; Gerty, S.; Ennis, S.; Simmonds, P.; Collins, A.; Eccles, D. The influence of genetic variation in 30 selected genes on the clinical characteristics of early onset breast cancer. Breast Cancer Res. 2008, 10, R108. [Google Scholar] [CrossRef] [PubMed]
- Piskor, B.M.; Przylipiak, A.; Dabrowska, E.; Sidorkiewicz, I.; Niczyporuk, M.; Szmitkowski, M.; Ławicki, S. Plasma Concentrations of Matrilysins MMP-7 and MMP-26 as Diagnostic Biomarkers in Breast Cancer. J. Clin. Med. 2021, 10, 1436. [Google Scholar] [CrossRef]
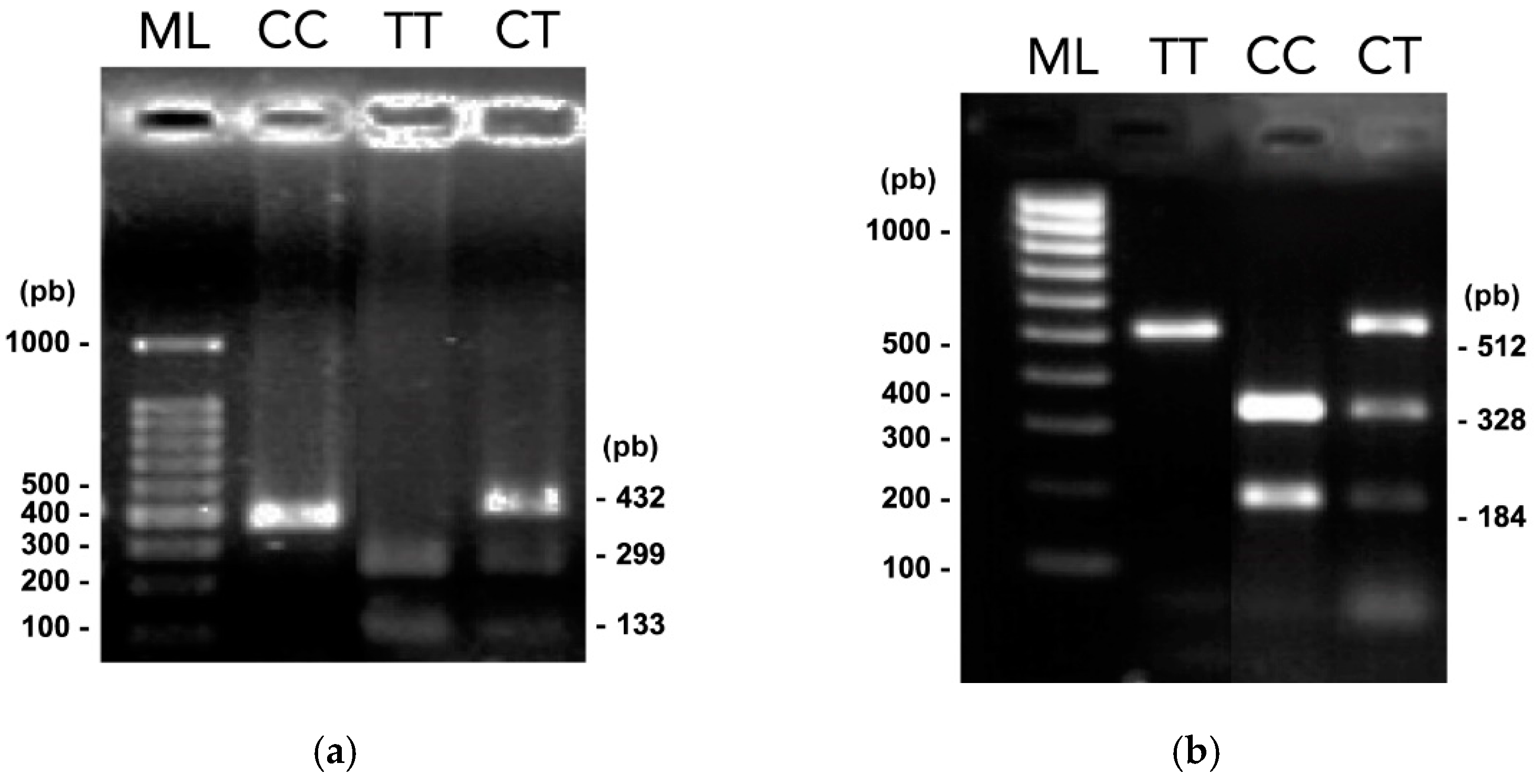
| Gene/SNP | Localization (NT Position) * | Alleles (WT/Var) | Primer Design | RE | Recognition site (5′- 3′) ** | WT Amplicon | |
|---|---|---|---|---|---|---|---|
| NT Sequence | Amplicon (bp) | ||||||
| TOX3/ rs3803662 | Chr 16 (52552429) | C/T | F -5′ AGTCCTTGGCTGTTCTGTG 3′ | 465 | Bpu10I | CCTNAGC | 298,167 |
| R -5′ GTCCAGACAGTCTTCAGCAG 3′ | |||||||
| MMP7/ rs1943779 | Chr 11 (102536460) | C/T | F -5′ CTGTGCTTCAAAAACACTGC 3′ | 514 | HpyCH4IV | ACGT | 328,184 |
| R -5′ TTTCTGTGGGTTGTCTTTCAC 3′ | |||||||
| Characteristics | Cases n (%) n = 212 | Controls n (%) n = 212 | p-Value |
|---|---|---|---|
| Age (media ± SD) | 54.22 ± 12.06 | 52.10 ± 28.95 | 0.327 * |
| Body Mass Index <30 >30 | |||
| 115 (54.2%) | 155 (73.1%) | >0.001 | |
| 97 (45.8%) | 57 (26.9%) | ||
| Menarche (media ± SD) | 12.72 ± 1.54 * | 12.78 ± 1.45 | 0.708 * |
| Oral contraceptives Consumers Non-consumers | |||
| 54 (25.5%) | 52 (24.5%) | 0.911 | |
| 158 (74.5%) | 160 (75.5%) | ||
| Hormonal Replace Therapy Consumers Non-consumers | |||
| 18 (8.5%) | 17 (8.0%) | 0.862 | |
| 193 (91.5%) | 195 (92.0%) | ||
| Age at the first child (media ± SD) | 22.13 ± 5.04 | 22.26 ± 5.03 | 0.796 |
| Menopause | |||
| Pre-menopause | 74 (34.9%) | 133 (62.7%) | >0.001 |
| Post-menopause | 138 (65.1%) | 79 (37.3%) | |
| Smoking | |||
| Smokers | 34 (16.1%) | 44 (21.3%) | 0.209 |
| Non-smokers | 177 (83.9%) | 163 (78.7%) | |
| Alcohol | |||
| Yes | 15 (7.1%) | 29 (14.0%) | 0.025 |
| No | 196 (92.9%) | 178 (86.0%) |
| IHC Status | ER (%) | PR (%) | HER2 (%) |
|---|---|---|---|
| Positive | 147 (69.3%) | 145 (68.4%) | 53 (25.0%) |
| Negative | 65 (30.7%) | 67 (31.6%) | 159 (75.0%) |
| SNP | Genotype | Cases n (%) n = 212 | Controls (%) n = 212 | p-Value | OR | 95% CI |
|---|---|---|---|---|---|---|
| TOX3 rs3803662 (C > T) | CC | 46 (21.7) | 69 (32.5) | 0.034 | 1 | Reference |
| CT | 122 (57.5) | 110 (51.9) | 1.164 | 1.057–2.618 | ||
| TT | 44 (20.8) | 33 (15.6) | 2.000 | 1.114–3.592 | ||
| X2 | 4.84 | 0.9961 | ||||
| p-value HWE | 0.0278 | 0.3118 | ||||
| Allele | ||||||
| C | 214 (50.5) | 248 (58.5) | 0.011 | 1 | Reference | |
| T | 210 (49.5) | 176 (41.5) | 1.38 | 1.054–1.813 | ||
| MMP7 rs1943779 (C > T) | CC | 20 (9.4) | 40 (18.8) | 0.006 | 1 | Reference |
| CT | 70 (33.0) | 71 (33.5) | 2.165 | 1.119–4.190 | ||
| TT | 122 (57.5) | 101 (47.6) | 2.703 | 1.442–5.067 | ||
| X2 | 3.779 | 14.922 | ||||
| p-value HWE | 0.0956 | 0.0003 | ||||
| Allele | ||||||
| C | 110 (26) | 151 (35.6) | 0.002 | 1 | Reference | |
| T | 314 (74) | 273 (64.3) | 1.527 | 1.138–2.824 |
| IHC Status | Allele | TOX3 rs3803662 | MMP7 rs1943779 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CT/TT vs. CC | CT/TT vs. CC | ||||||||
| Positive n (%) | Negative n (%) | OR | 95% CI | Positive n (%) | Negative n (%) | OR | 95% CI | ||
| ER | C | 35 (79.5) | 9 (20.5) | 1 | 10 (58.8) | 7 (41.2) | 1 | ||
| T | 112 (66.7) | 56 (33.3) | 1.944 | 0.874–4.326 | 137 (70.3) | 58 (29.7) | 0.605 | 0.220–1.666 | |
| PR | C | 36 (81.8) | 8 (18.2) | 1 | 9 (52.9) | 8 (47.1) | 1 | ||
| T | 109 (64.9) | 59 (35.1) | 2.436 | 1.063–5.580 | 136 (69.7) | 59 (30.3) | 0.488 | 0.180–1.327 | |
| HER2 | C | 8 (18.2) | 36 (81.8) | 1 | 4 (23.5) | 13 (76.5) | 1 | ||
| T | 45 (26.8) | 123 (73.2) | 0.607 | 0.263–1.405 | 49 (25.1) | 146 (74.9) | 0.917 | 0.286–2.943 | |
| TNBC | C | 4 (9.09) | 40 (90.9) | 1 | 7 (41.17) | 10 (58.8) | 1 | ||
| T | 47 (27.97) | 121 (72.02) | 3.884 | 1.317–11.456 | 151 (77.43) | 44 (22.5) | 0.416 | 0.150–1.157 | |
| Metastasis | C | 35 (79.5) | 9 (20.4) | 1 | 4 (23.5) | 13 (76.4) | 1 | ||
| T | 132 (78.5) | 36 (21.4) | 1.061 | 0.467–2.408 | 41 (21.02) | 154 (78.9) | 0.865 | 0.268–2.795 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solis-Coronado, O.; Villarreal-Vela, M.P.; Rodríguez-Gutiérrez, H.F.; González-Guerrero, J.F.; Cerda-Flores, R.M.; Alcorta-Núñez, F.; Camarillo-Cárdenas, K.P.; Pérez-Ibave, D.C.; Vidal-Gutiérrez, O.; Ramírez-Correa, G.A.; et al. Risk Association of TOX3 and MMP7 Gene Polymorphisms with Sporadic Breast Cancer in Mexican Women. Curr. Oncol. 2022, 29, 1008-1017. https://doi.org/10.3390/curroncol29020086
Solis-Coronado O, Villarreal-Vela MP, Rodríguez-Gutiérrez HF, González-Guerrero JF, Cerda-Flores RM, Alcorta-Núñez F, Camarillo-Cárdenas KP, Pérez-Ibave DC, Vidal-Gutiérrez O, Ramírez-Correa GA, et al. Risk Association of TOX3 and MMP7 Gene Polymorphisms with Sporadic Breast Cancer in Mexican Women. Current Oncology. 2022; 29(2):1008-1017. https://doi.org/10.3390/curroncol29020086
Chicago/Turabian StyleSolis-Coronado, Orlando, Mónica Patricia Villarreal-Vela, Hazyadee Frecia Rodríguez-Gutiérrez, Juan Francisco González-Guerrero, Ricardo M. Cerda-Flores, Fernando Alcorta-Núñez, Karen Paola Camarillo-Cárdenas, Diana Cristina Pérez-Ibave, Oscar Vidal-Gutiérrez, Genaro A. Ramírez-Correa, and et al. 2022. "Risk Association of TOX3 and MMP7 Gene Polymorphisms with Sporadic Breast Cancer in Mexican Women" Current Oncology 29, no. 2: 1008-1017. https://doi.org/10.3390/curroncol29020086
APA StyleSolis-Coronado, O., Villarreal-Vela, M. P., Rodríguez-Gutiérrez, H. F., González-Guerrero, J. F., Cerda-Flores, R. M., Alcorta-Núñez, F., Camarillo-Cárdenas, K. P., Pérez-Ibave, D. C., Vidal-Gutiérrez, O., Ramírez-Correa, G. A., & Garza-Rodríguez, M. L. (2022). Risk Association of TOX3 and MMP7 Gene Polymorphisms with Sporadic Breast Cancer in Mexican Women. Current Oncology, 29(2), 1008-1017. https://doi.org/10.3390/curroncol29020086







