Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
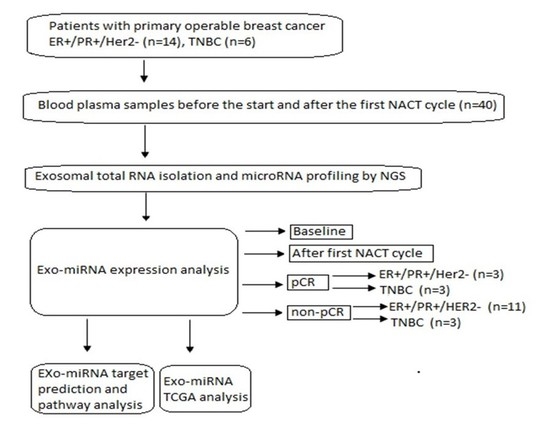
2.1. Study Population
2.2. Plasma Exosomal miRNA Isolation and Next-Generation Sequencing (NGS)
2.3. Data Analysis
2.4. Target Predictions
2.5. cBioportal and UALCAN Databases Analysis
3. Results
3.1. Demographic Characteristics of the Study Participants
3.2. Differentially Expressed Exosomal miRNAs
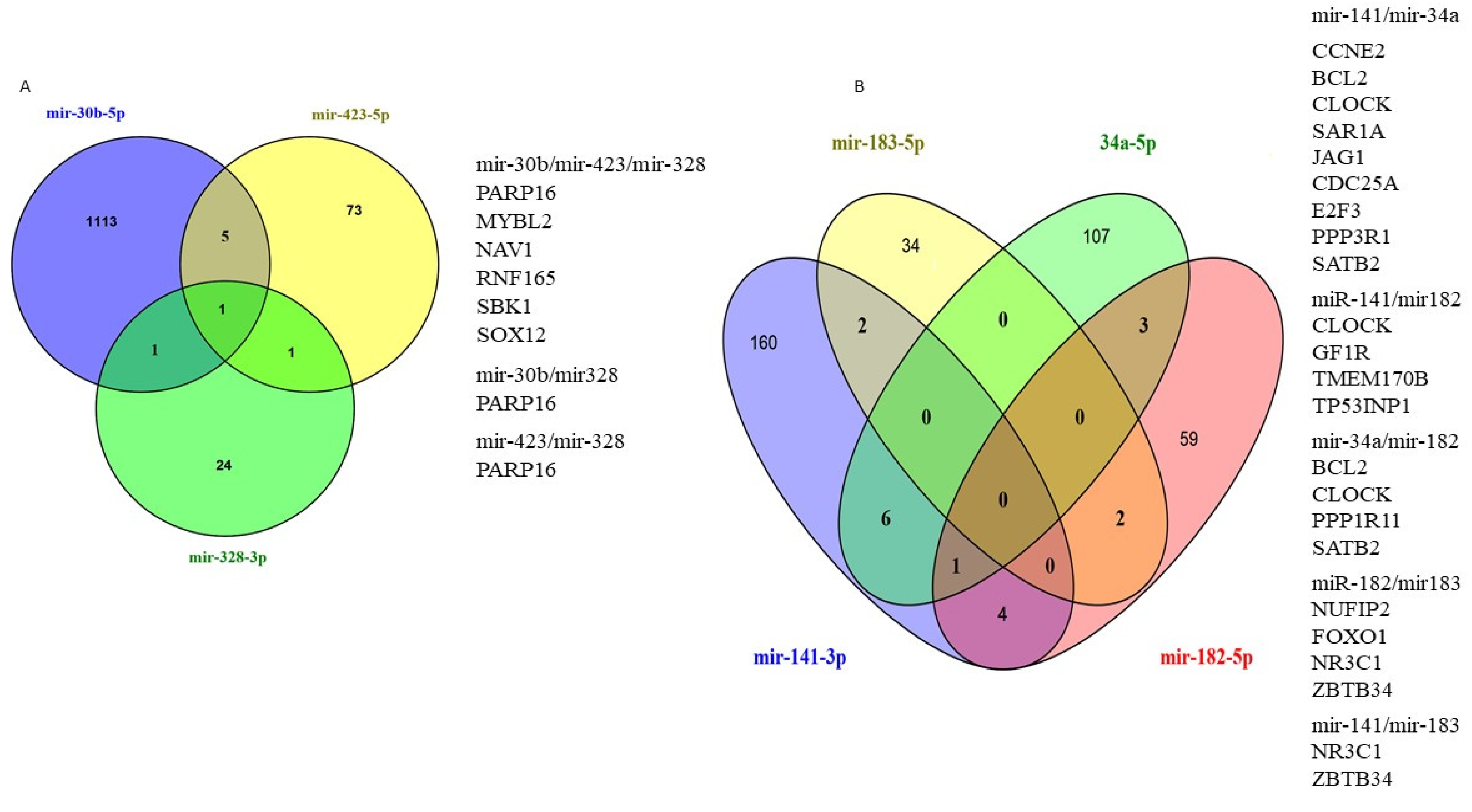
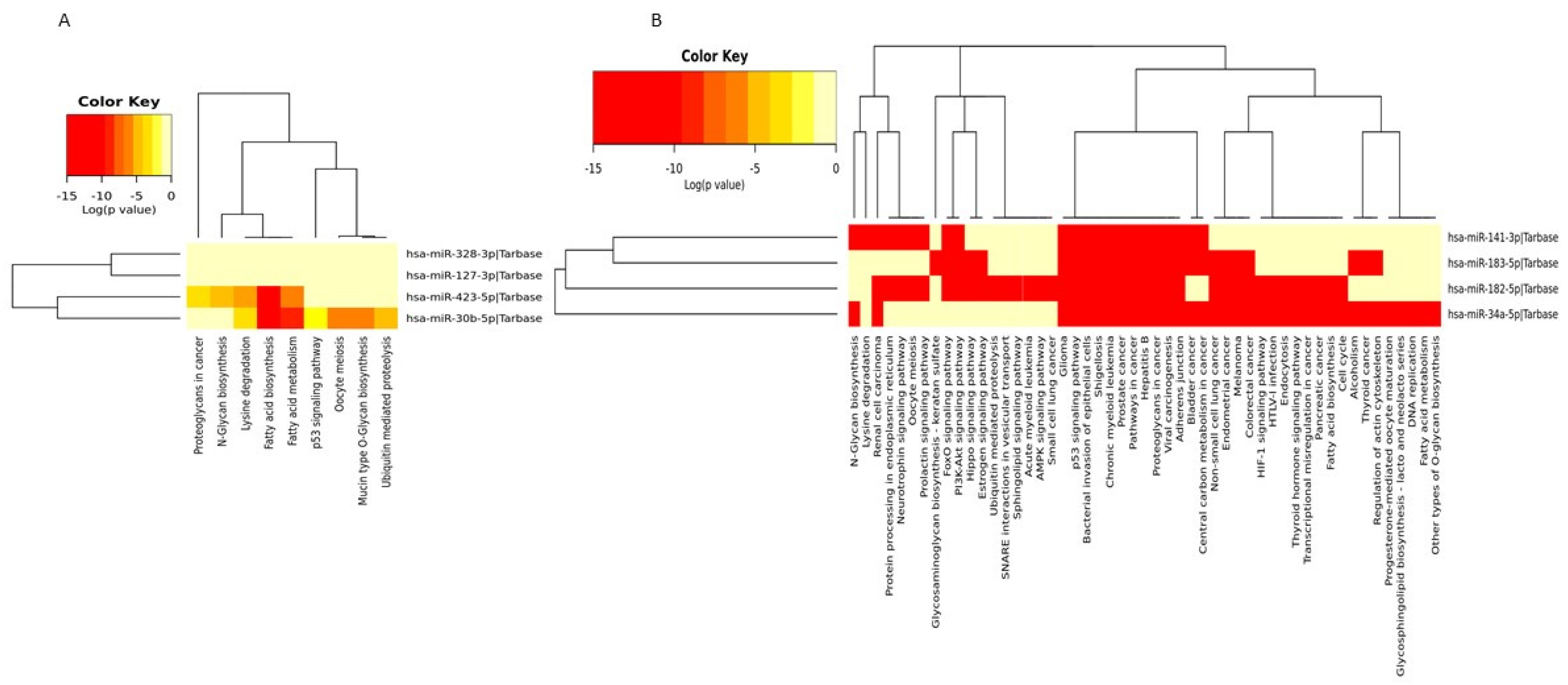
3.3. In Silico Target Prediction and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forouzanfar, M.H.; Foreman, K.J.; Delossantos, A.M.; Lozano, R.; Lopez, A.D.; Murray, C.J.L.; Naghavi, M. Breast and cervical cancer in 187 countries between 1980 and 2010: A systematic analysis. Lancet 2011, 9801, 1461–1484. [Google Scholar] [CrossRef]
- Rastogi, P.; Anderson, S.J.; Bear, H.D.; Geyer, C.E.; Kahlenberg, M.S.; Robidoux, A.; Margolese, R.G.; Hoehn, J.L.; Vogel, V.G.; Dakhil, S.R.; et al. Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J. Clin. Oncol. 2008, 26, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Tryfonidis, K.; Senkus, E.; Cardoso, M.J.; Cardoso, F. Management of locally advanced breast cancer—perspectives and future directions. Nat. Rev. Clin. Oncol. 2015, 12, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, H.; Li, J.; Zhang, J.; Wu, J.; Li, Y.; Piao, Y.; Pan, L.; Xiang, R.; Yue, S. FZR1 as a novel biomarker for breast cancer neoadjuvant chemotherapy prediction. Cell Death Dis. 2020, 11, 804. [Google Scholar] [CrossRef] [PubMed]
- Pathological Complete Response in Neoadjuvant Treatment of High-Risk Early-Stage Breast Cancer: Use as an Endpoint to Support Accelerated Approval. Available online: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm305501.pdf (accessed on 10 January 2022).
- Ogston, K.N.; Miller, I.D.; Payne, S.; Hutcheon, A.W.; Sarkar, T.K.; Smith, I.; Schofield, A.; Heys, S.D. A new histological grading system to assess response of breast cancers to primary chemotherapy: Prognostic significance and survival. Breast 2003, 12, 320–327. [Google Scholar] [CrossRef]
- Pennisi, A.; Kieber-Emmons, T.; Makhoul, I.; Hutchins, L. Relevance of Pathological Complete Response after Neoadjuvant Therapy for Breast Cancer. Breast Cancer 2016, 10, 103–106. [Google Scholar] [CrossRef]
- Diagnosis and Treatment of Patients with Primary and Metastatic Breast Cancer. Available online: www.ago-online.de (accessed on 2 April 2020).
- Schott, A.F.; Roubidoux, M.A.; Helvie, M.A.; Hayes, D.F.; Kleer, C.G.; Newman, L.A.; Pierce, L.J.; Griffith, K.A.; Murray, S.; Hunt, K.A.; et al. Clinical and radiologic assessments to predict breast cancer pathologic complete response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2005, 92, 231–238. [Google Scholar] [CrossRef]
- Schaefgen, B.; Mati, M.; Sinn, H.P.; Golatta, M.; Stieber, A.; Rauch, G.; Hennigs, A.; Richter, H.; Domschke, C.; Schuetz, F.; et al. Can Routine Imaging After Neoadjuvant Chemotherapy in Breast Cancer Predict Pathologic Complete Response? Ann. Surg. Oncol. 2016, 23, 789–795. [Google Scholar] [CrossRef]
- Chagpar, A.B.; Middleton, L.P.; Sahin, A.A.; Dempsey, P.; Buzdar, A.U.; Mirza, A.N.; Ames, F.C.; Babiera, G.V.; Feig, B.W.; Hunt, K.K.; et al. Accuracy of physical examination, ultrasonography, and mammography in predicting residual pathologic tumor size in patients treated with neoadjuvant chemotherapy. Ann. Surg. 2006, 243, 257–264. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Ritter, A.; Hirschfeld, M.; Berner, K.; Rücker, G.; Jäger, M.; Weiss, D.; Medl, M.; Nöthling, C.; Gassner, S.; Asberger, J.; et al. Circulating non-coding RNA-biomarker potential in neoadjuvant chemotherapy of triple negative breast cancer? Int. J. Oncol. 2020, 56, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Blohmer, J.U.; Costa, S.D.; Denkert, C.; Eidtmann, H.; Eiermann, W.; Gerber, B.; Hanusch, C.; Hilfrich, J.; Huober, J.; et al. Response-guided neoadjuvant chemotherapy for breast cancer. J. Clin. Oncol. 2013, 31, 3623–3630. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Kim, H.H.; Ahn, J.H.; Kim, S.B.; Jung, K.H.; Gong, G.; Son, B.H.; Ahn, S.H. Comparison of mammography, sonography, MRI and clinical examination in patients with locally advanced or inflammatory breast cancer who underwent neoadjuvant chemotherapy. Br. J. Radiol. 2011, 84, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, M.; Ryan, D.; Carpenter, R.; Vinnicombe, S.; Gallagher, C.J. Using MRI to plan breast-conserving surgery following neoadjuvant chemotherapy for early breast cancer. Br. J. Cancer 2008, 98, 289–293. [Google Scholar] [CrossRef][Green Version]
- Sims, A.H. Bioinformatics and breast cancer: What can high-throughput genomic approaches actually tell us? J. Clin. Pathol. 2009, 62, 879–885. [Google Scholar] [CrossRef]
- Weigelt, B.; Baehner, F.L.; Reis-Filho, J.S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: A retrospective of the last decade. J. Pathol. 2010, 220, 263–280. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Robertson, K.E.; Ravel, P.; Bray, S.E.; Bajard, A.; Purdie, C.A.; Nguyen, C.; Hadad, S.M.; Bieche, I.; Chabaud, S.; et al. A candidate molecular signature associated with tamoxifen failure in primary breast cancer. Breast Cancer Res. 2008, 10, R88–R95. [Google Scholar] [CrossRef]
- Turnbull, A.K.; Arthur, L.M.; Renshaw, L.; Larionov, A.A.; Kay, C.; Dunbier, A.K.; Thomas, J.S.; Dowsett, M.; Sims, A.H.; Dixon, J.M. Accurate prediction and validation of response to endocrine therapy in breast cancer. J. Clin. Oncol. 2015, 33, 2270–2278. [Google Scholar] [CrossRef]
- Bownes, R.J.; Turnbull, A.K.; Martinez-Perez, C.; Cameron, D.A.; Sims, A.H.; Oikonomidou, O. On-treatment biomarkers can improve prediction of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. 2019, 21, 73–81. [Google Scholar] [CrossRef]
- Bartel, P.D. MiRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C. The role of MicroRNAs in human cancer. Sig. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef]
- Schwarzenbach, H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev. Mol. Diagn. 2015, 15, 1159–1169. [Google Scholar] [CrossRef]
- Nagamitsu, Y.; Nishi, H.; Sasaki, T.; Takaesu, Y.; Terauchi, F.; Isaka, K. Profiling analysis of circulating microRNA expression in cervical cancer. Mol. Clin. Oncol. 2016, 5, 189–194. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Qi, P.; Zhou, X.Y.; Du, X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef]
- Zelli, V.; Compagnoni, C.; Capelli, R.; Cannita, K.; Sidoni, T.; Ficorella, C.; Capalbo, C.; Zazzeroni, F.; Tessitore, A.; Alesse, E. Circulating MicroRNAs as Prognostic and Therapeutic Biomarkers in Breast Cancer Molecular Subtypes. J. Pers. Med. 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Davidovic, R.; Bajic, V.; Obradovic, M.; Isenovic, R.E. MicroRNA in breast cancer: The association with BRCA1/2. Cancer Biomark. 2017, 19, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Ghaffari, S.H. Why have microRNA biomarkers not been translated from bench to clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Q.; Zhang, R.; Dai, X.; Chen, W.; Xing, D. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta 2021, 516, 46–54. [Google Scholar] [CrossRef]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS ONE 2015, 10, e0136133. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Pirim, H.; Ekşioğlu, B.; Perkins, A.; Yüceer, C. Clustering of High Throughput Gene Expression Data. Comput. Oper. Res. 2012, 39, 3046–3061. [Google Scholar] [CrossRef]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef]
- Musa, J.; Aynaud, M.M.; Mirabeau, O.; Delattre, O.; Grünewald, T.G. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017, 8, e2895. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Pan, Y.; Lu, H.; Zheng, S.; Lin, H.; Fang, H.; Cao, F. B-myb is a gene implicated in cell cycle and proliferation of breast cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 5819–5827. [Google Scholar] [PubMed]
- Iness, A.N.; Felthousen, J.; Ananthapadmanabhan, V.; Sesay, F.; Saini, S.; Guiley, K.Z.; Rubin, S.M.; Dozmorov, M.; Litovchick, L. The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B-Myb. Oncogene 2019, 38, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Fry, E.A. Novel molecular markers for breast cancer. Biomark. Cancer 2016, 8, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; He, Z.; Lin, C.; Shen, Z. MiR-30b-5p inhibits proliferation and promotes apoptosis of medulloblastoma cells via targeting MYB proto-oncogene like 2 (MYBL2). J. Investig. Med. 2020, 68, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lee, K.-M.; Han, W.; Choi, J.-Y.; Lee, J.-Y.; Kang, G.H.; Park, S.K.; Noh, D.Y.; Yoo, K.Y.; Kang, D. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum. Mol. Genet. 2010, 19, 4273–4277. [Google Scholar] [CrossRef]
- Gudas, J.M.; Payton, M.; Thukral, S.; Chen, E.; Bass, M.; Robinson, M.O.; Coats, S. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol. Cell Biol. 1999, 19, 612–622. [Google Scholar] [CrossRef]
- Payton, M.; Scully, S.; Chung, G.; Coats, S. Deregulation of cyclin E2 expression and associated kinase activity in primary breast tumors. Oncogene 2002, 21, 8529–8534. [Google Scholar] [CrossRef]
- Caldon, C.E.; Sergio, C.M.; Kang, J.; Muthukaruppan, A.; Boersma, M.N.; Stone, A.; Barraclough, J.; Lee, C.S.; Black, M.A.; Miller, L.D.; et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol. Cancer Ther. 2012, 11, 1488–1499. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Look, M.P.; Meijer van Gelder, M.E.; Timmermans, M.; Trapman, A.M.; Garcia, R.R.; Arnold, M.; Goedheer, A.J.; de Weerd, V.; Portengen, H.; et al. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node negative breast cancer patients. Clin. Cancer Res. 2006, 12, 3319–3328. [Google Scholar] [CrossRef]
- Leung, L.K.; Wang, T.T. Paradoxical regulation of Bcl-2 family proteins by 17beta-oestradiol in human breast cancer cells MCF-7. Br. J. Cancer 1999, 81, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Eom, Y.H.; Kim, H.S.; Lee, A.; Song, B.J.; Chae, B.J. BCL2 as a Subtype-Specific Prognostic Marker for Breast Cancer. J. Breast Cancer 2016, 19, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, C.; van de Sandt, L.; Edlund, K.; Lohr, M.; Hellwig, B.; Marchan, R.; Schmidt, M.; Rahnenführer, J.; Oster, H.; Hengstler, J.G. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle 2014, 13, 3282–3291. [Google Scholar] [CrossRef] [PubMed]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.H.; Bachelier, R.; Allioli, N.; Hong, S.S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef]
- Amorim, M.; Lobo, J.; Fontes-Sousa, M.; Estevão-Pereira, H.; Salta, S.; Lopes, P.; Coimbra, N.; Antunes, L.; Palma de Sousa, S.; Henrique, R.; et al. Predictive and Prognostic Value of Selected MicroRNAs in Luminal Breast Cancer. Front. Genet. 2019, 10, 815. [Google Scholar] [CrossRef]
- Sun, X.; Huang, T.; Zhang, C.; Zhang, S.; Wang, Y.; Zhang, Q.; Liu, Z. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle 2019, 18, 1908–1924. [Google Scholar] [CrossRef]
- Dai, T.; Zhao, X.; Li, Y.; Yu, L.; Li, Y.; Zhou, X.; Gong, Q. miR-423 Promotes Breast Cancer Invasion by Activating NF-κB Signaling. Onco Targets Ther. 2020, 13, 5467–5478. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, A.; Zhang, Z.; Tian, R.; Luo, A.; Li, M.; Zhao, D.; Fu, L.; Fu, L.; Dong, J.T.; et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. 2015, 36, 4763–4771. [Google Scholar] [CrossRef]
- Zeng, F.; Yao, M.; Wang, Y.; Zheng, W.; Liu, S.; Hou, Z.; Cheng, X.; Sun, S.; Li, T.; Zhao, H.; et al. Fatty acid β-oxidation promotes breast cancer stemness and metastasis via the miRNA-328-3p-CPT1A pathway. Cancer Gene Ther. 2021, 37, 1–13. [Google Scholar] [CrossRef]
- Xiao, B.; Chen, D.; Zhou, Q.; Hang, J.; Zhang, W.; Kuang, Z.; Sun, Z.; Li, L. Glutamate metabotropic receptor 4 (GRM4) inhibits cell proliferation, migration and invasion in breast cancer and is regulated by miR-328-3p and miR-370-3p. BMC Cancer 2019, 19, 891. [Google Scholar] [CrossRef]
- Chen, J.; Wang, M.; Guo, M.; Xie, Y.; Cong, Y.-S. miR-127 regulates cell proliferation and senescence by targeting BCL6. PLoS ONE. 2013, 8, e80266. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Kim, H.S.; Jin, T.; Hwang, E.H.; Jung, M.; Moon, W.K. Overexpression of the miR-141/200c cluster promotes the migratory and invasive ability of triple-negative breast cancer cells through the activation of the FAK and PI3K/AKT signaling pathways by secreting VEGF-A. BMC Cancer 2016, 16, 570. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Mitwally, N.; Soliman, A.S.; Yousef, E. Potential Diagnostic and Prognostic Utility of miR-141, miR-181b1, and miR-23b in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 8589. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, J.; Segura, M.F.; Shao, C.; Lee, P.; Gong, Y.; Hernando, E.; Wei, J.J. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J. Pathol. 2012, 228, 204–215. [Google Scholar] [CrossRef]
- Perilli, L.; Vicentini, C.; Agostini, M.; Pizzini, S.; Pizzi, M.; D’Angelo, E.; Bortoluzzi, S.; Mandruzzato, S.; Mammano, E.; Rugge, M.; et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget 2014, 5, 6611–6619. [Google Scholar] [CrossRef]
- Wang, D.; Lu, G.; Shao, Y.; Xu, D. MiR-182 promotes prostate cancer progression through activating Wnt/beta-catenin signal pathway. Biomed. Pharmacother. 2018, 99, 334–339. [Google Scholar] [CrossRef]
- Xiong, D.; Lv, J.; Wei, K.; Feng, Z.; Chen, J.; Liu, K.; Chen, G.; Luo, D. A nine-miRNA signature as a potential diagnostic marker for breast carcinoma: An integrated study of 1,110 cases. Oncol. Rep. 2017, 37, 3297–3304. [Google Scholar] [CrossRef]
- Krishnan, K.; Steptoe, A.L.; Martin, H.C.; Wani, S.; Nones, K.; Waddell, N.; Mariasegaram, M.; Simpson, P.T.; Lakhani, S.R.; Gabrielli, B.; et al. MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 2013, 19, 230–242. [Google Scholar] [CrossRef]
- Lowery, A.J.; Miller, N.; Dwyer, R.M.; Kerin, M.J. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 2010, 10, 502–510. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Kuo, W.H.; Hung, J.H.; Lee, C.Y.; Lee, Y.H.; Chang, Y.C.; Lin, W.C.; Shen, C.Y.; Huang, C.S.; Hsieh, F.J.; et al. Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol. Cancer 2015, 14, 36. [Google Scholar] [CrossRef]
- Chekhun, V.F.; Borikun, T.V.; Bazas, V.M.; Andriiv, A.V.; Klyusov, O.M.; Yalovenko, T.M.; Lukianova, N.Y. Association of circulating miR-21, -205, and -182 with response of luminal breast cancers to neoadjuvant FAC and AC treatment. Exp. Oncol. 2020, 42, 162–166. [Google Scholar] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Castelli, G.; Pelosi, E. Breast Cancer. A Molecularly Heterogenous Disease Needing Subtype-Specific Treatments. Med. Sci. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- Chen, C.; Pan, Y.; Bai, L.; Chen, H.; Duan, Z.; Si, Q.; Zhu, R.; Chuang, T.H.; Luo, Y. MicroRNA-3613-3p functions as a tumor suppressor and represents a novel therapeutic target in breast cancer. Breast Cancer Res. 2021, 23, 12. [Google Scholar] [CrossRef]





| pCR | Non-pCR | |
|---|---|---|
| Number of patients | 6 | 14 |
| Age (years), median (range) | 52 (35–69) | 52 (39–65) |
| Histology | IDC | IDC |
| Race | ||
| White | 4 | 12 |
| Black | 2 | 2 |
| Tumor grade | ||
| Grade I | 1 | 1 |
| Grade II | 1 | 6 |
| Grade III | 4 | 7 |
| Hormone status | ||
| ER+/PR+/Her2- | 3 | 11 |
| ER-/PR-/Her2- | 3 | 3 |
| Before NACT | After First Cycle of NACT | |||||
|---|---|---|---|---|---|---|
| LogFC | p-Value | FDR | LogFC | p-Value | FDR | |
| pCR | ||||||
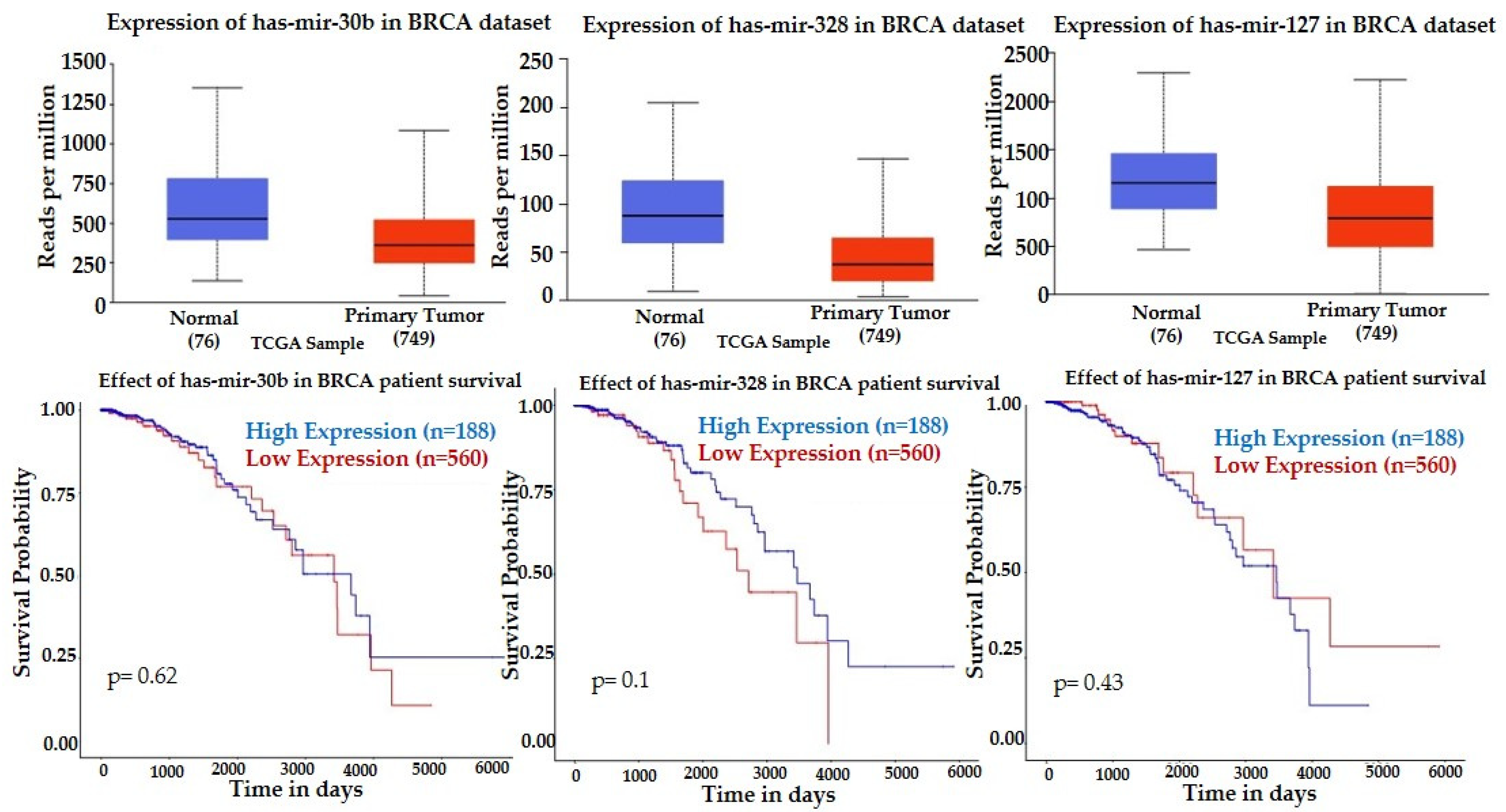
| hsa-miR-30b-5p | 1.2025 | 0.0000 | 0.0039 | −0.0499 | 0.8860 | 0.9996 |
| hsa-miR-328-3p | −1.1040 | 0.0019 | 0.0360 | −0.1081 | 0.7679 | 0.9996 |
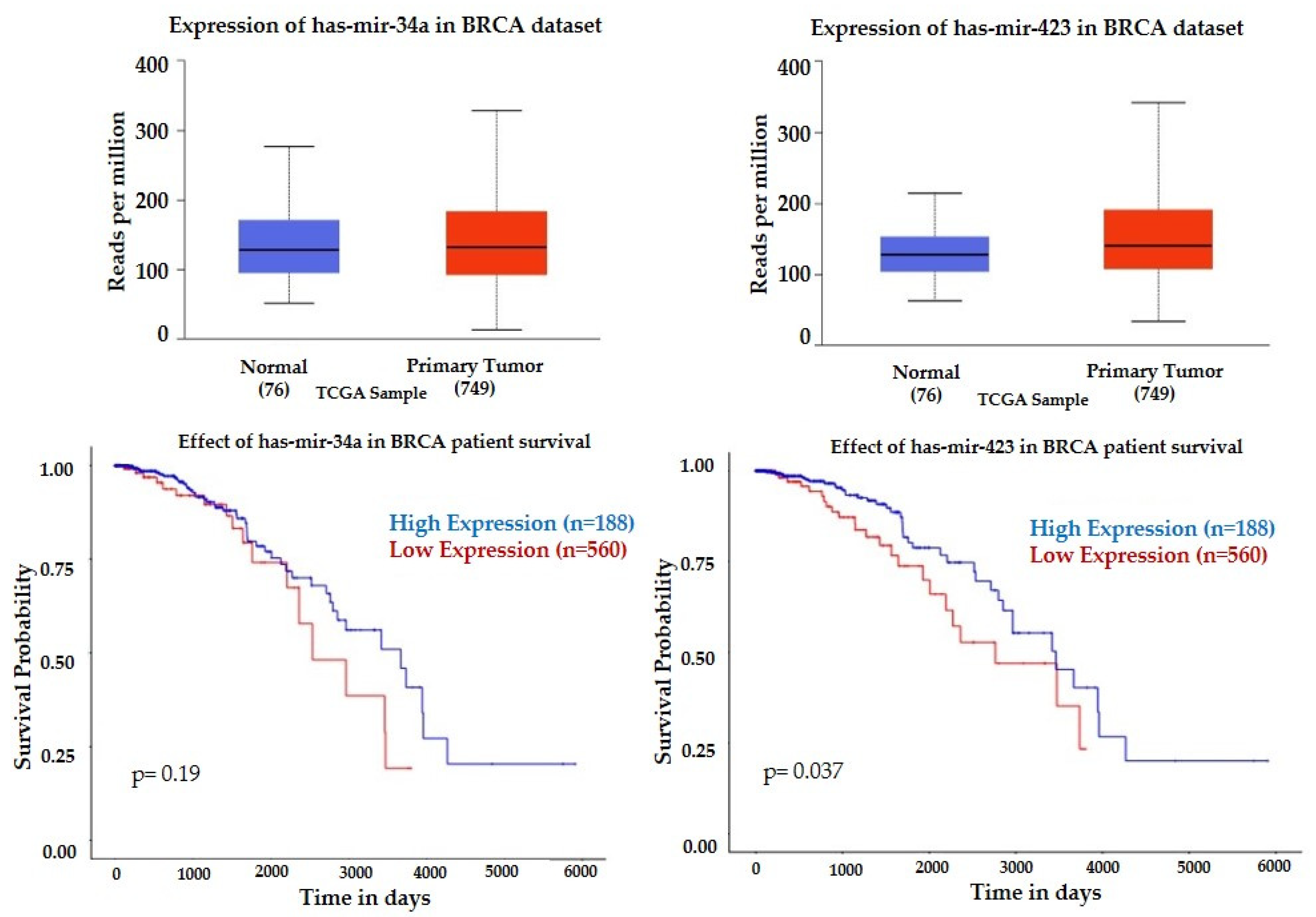
| hsa-miR-423-5p | −1.4271 | 0.0005 | 0.0127 | −0.8390 | 0.0167 | 0.8232 |
| hsa-miR-127-3p | 4.5388 | 0.0000 | 0.0023 | −0.0225 | 0.9514 | 0.9996 |
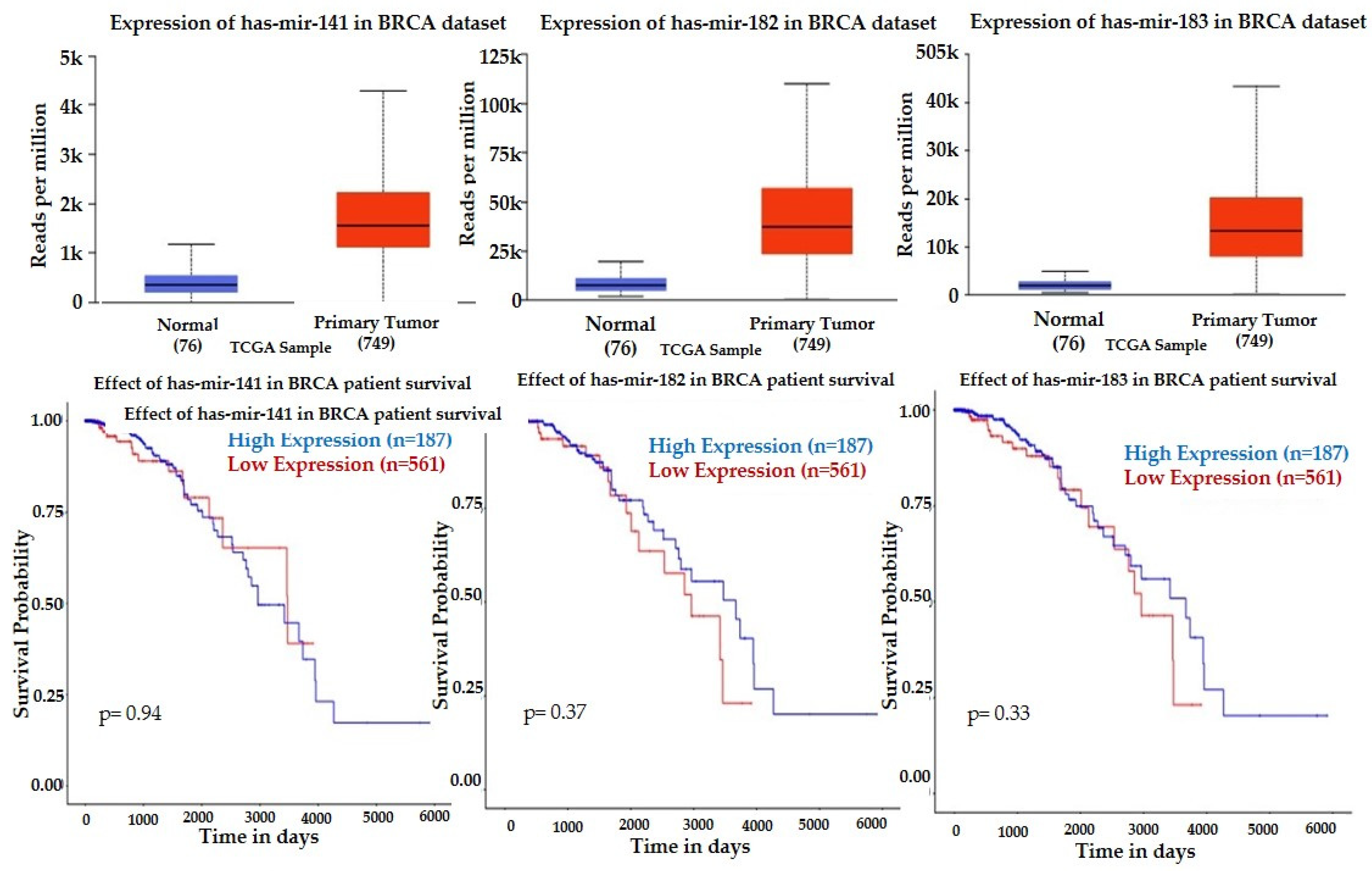
| hsa-mir-141-3p | −0.1823 | 0.8631 | 0.9882 | −2.5652 | 0.0000 | 0.0003 |
| non-pCR | ||||||
| hsa-miR-34a-5p | 2.2219 | 0.0287 | 0.9883 | 3.0152 | 0.0000 | 0.0000 |
| hsa-miR-182-5p | 3.0821 | 0.0022 | 0.4906 | 1.2929 | 0.0000 | 0.0062 |
| hsa-miR-183-5p | 1.6985 | 0.0996 | 0.9883 | 1.7837 | 0.0000 | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorova, V.K.; Byrum, S.D.; Gies, A.J.; Haynie, C.; Smith, H.; Reyna, N.S.; Makhoul, I. Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Curr. Oncol. 2022, 29, 613-630. https://doi.org/10.3390/curroncol29020055
Todorova VK, Byrum SD, Gies AJ, Haynie C, Smith H, Reyna NS, Makhoul I. Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Current Oncology. 2022; 29(2):613-630. https://doi.org/10.3390/curroncol29020055
Chicago/Turabian StyleTodorova, Valentina K., Stephanie D. Byrum, Allen J. Gies, Cade Haynie, Hunter Smith, Nathan S. Reyna, and Issam Makhoul. 2022. "Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer" Current Oncology 29, no. 2: 613-630. https://doi.org/10.3390/curroncol29020055
APA StyleTodorova, V. K., Byrum, S. D., Gies, A. J., Haynie, C., Smith, H., Reyna, N. S., & Makhoul, I. (2022). Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Current Oncology, 29(2), 613-630. https://doi.org/10.3390/curroncol29020055