Siglec-15 as a New Perspective Therapy Target in Human Giant Cell Tumor of Bone
Abstract
1. Introduction
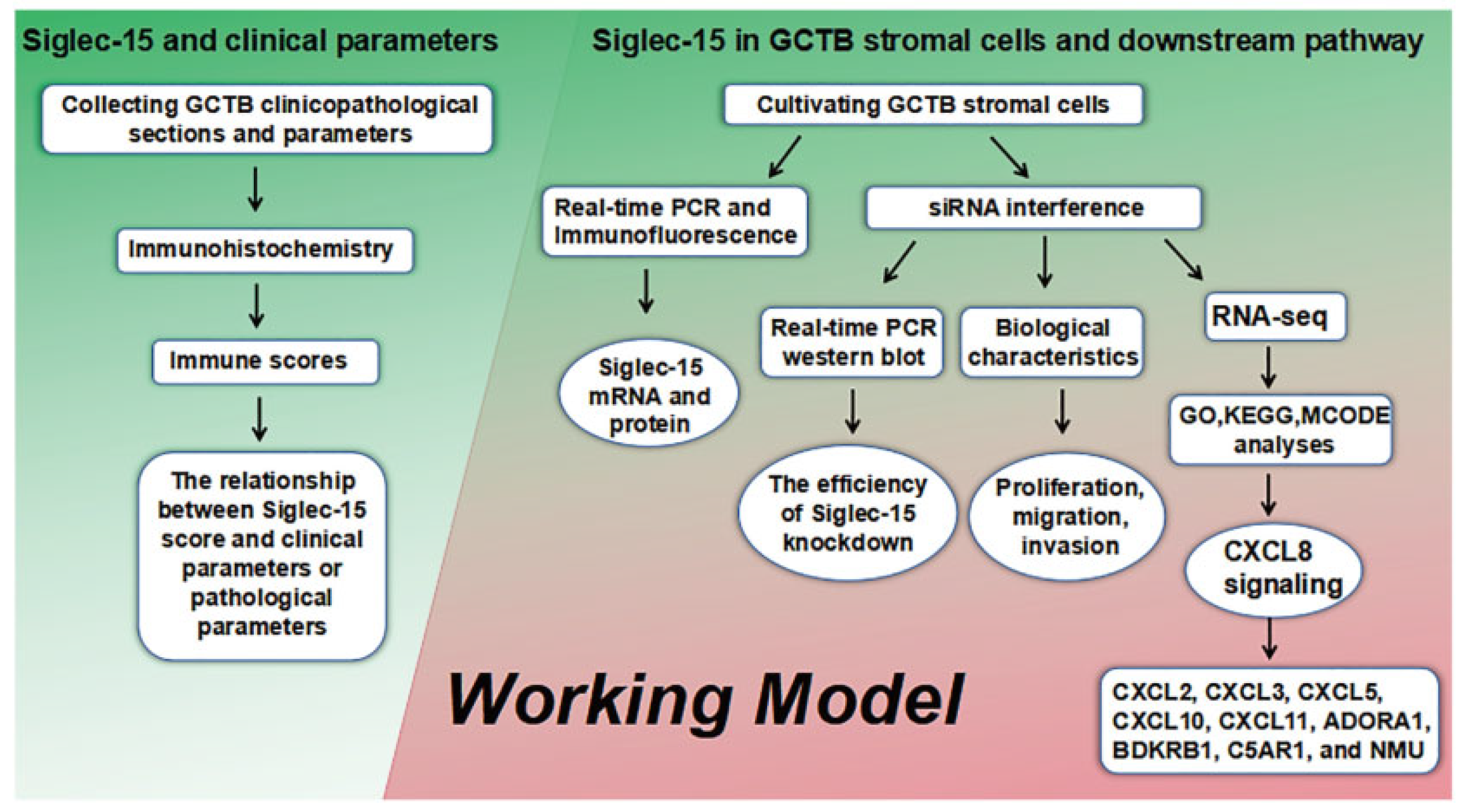
2. Materials and Methods
2.1. Patient and Samples
2.2. Cell Culture
2.3. Antibodies and Major Reagents
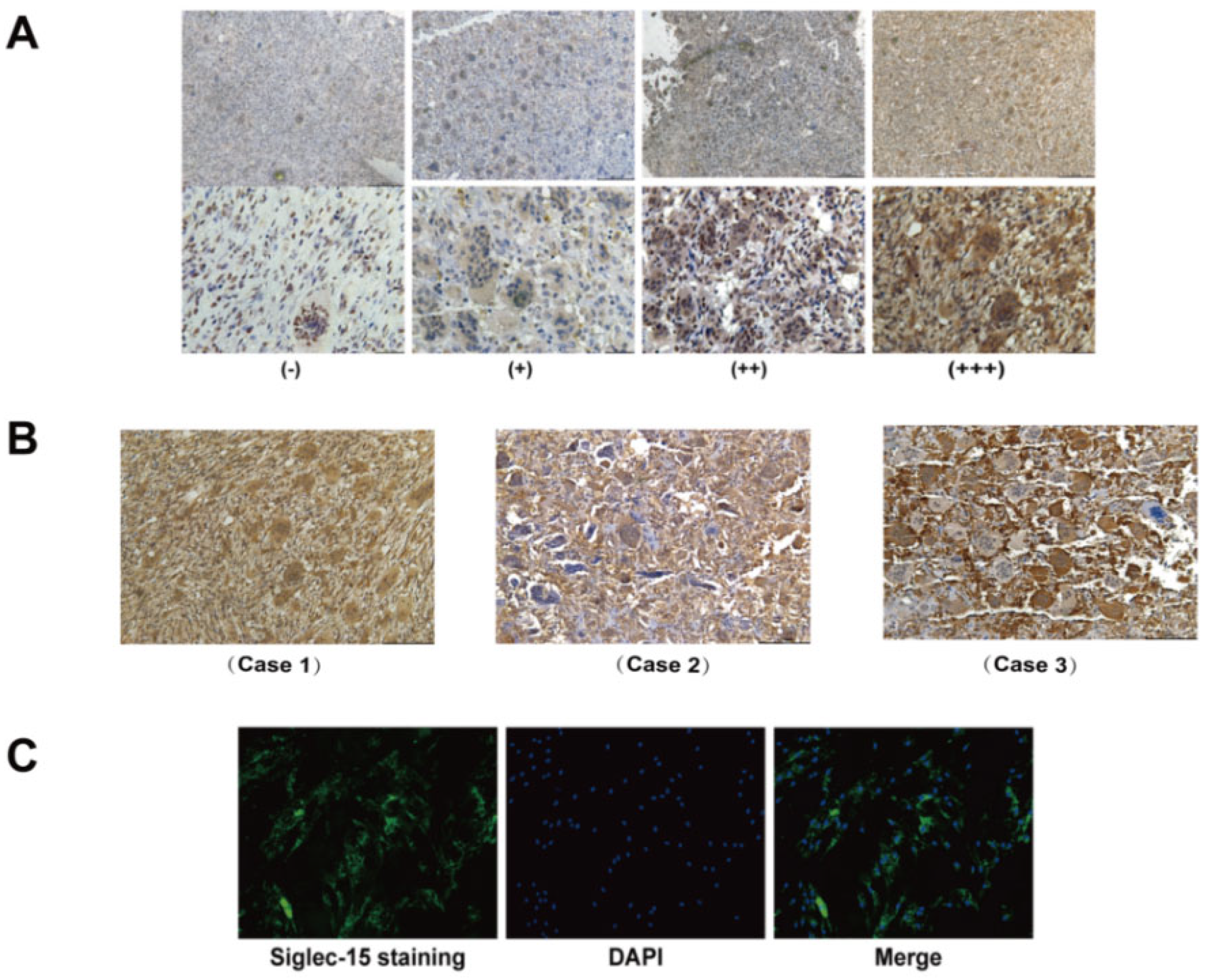
2.4. Immunohistochemistry (IHC)
2.5. Immunofluorescence Assay (IF)
2.6. RNA Interference (RNAi) and Cell Transfection
2.7. RT-PCR
2.8. Western Blotting
2.9. Cell Proliferation and Colony Formation Assays
2.10. Wound Healing Assay
2.11. Transwell Assays
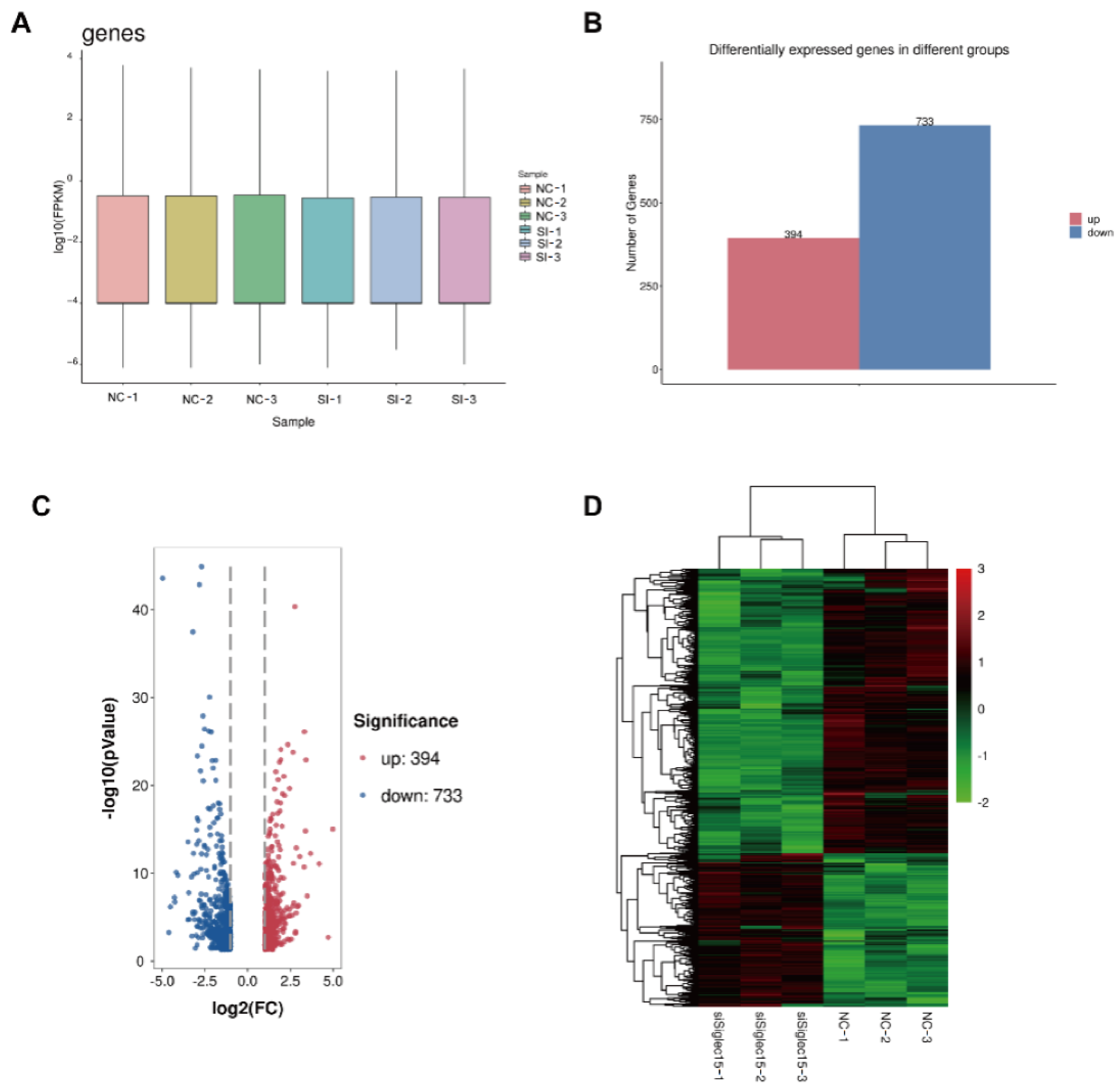
2.12. RNA Sequencing Assay
2.13. STRING Analysis and MCODE Analysis
2.14. Statistical Analysis
2.15. Ethical Approval
3. Results
3.1. Expression of Siglec-15 in Human GCTB Tissues and Stromal Cells
3.2. Knockdown of Siglec-15 Expression in GCTB Stromal Cells
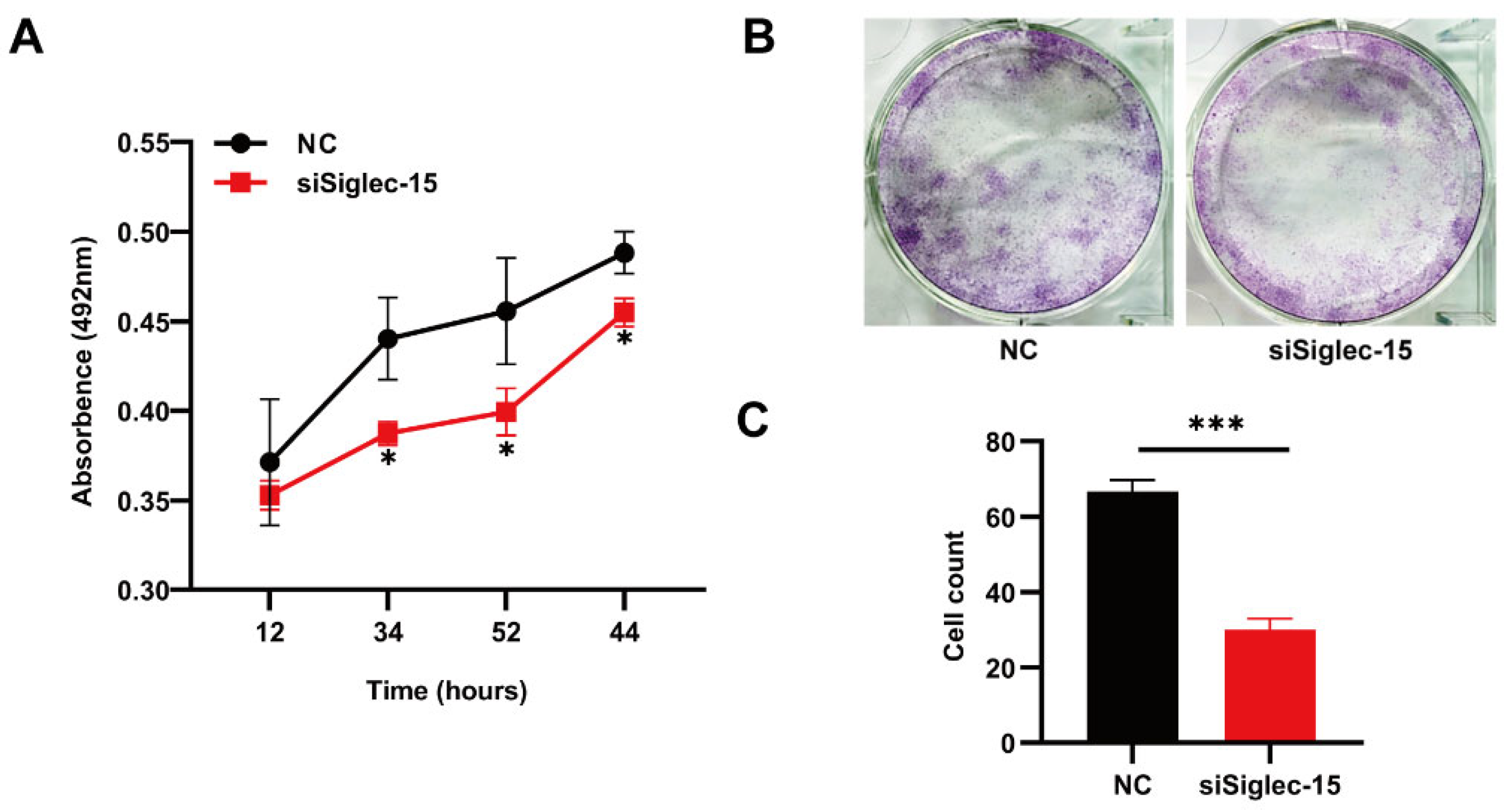
3.3. Knockdown of Siglec-15 Expression Decreases the Proliferation and Clonal Formation of Hs737. T Cells
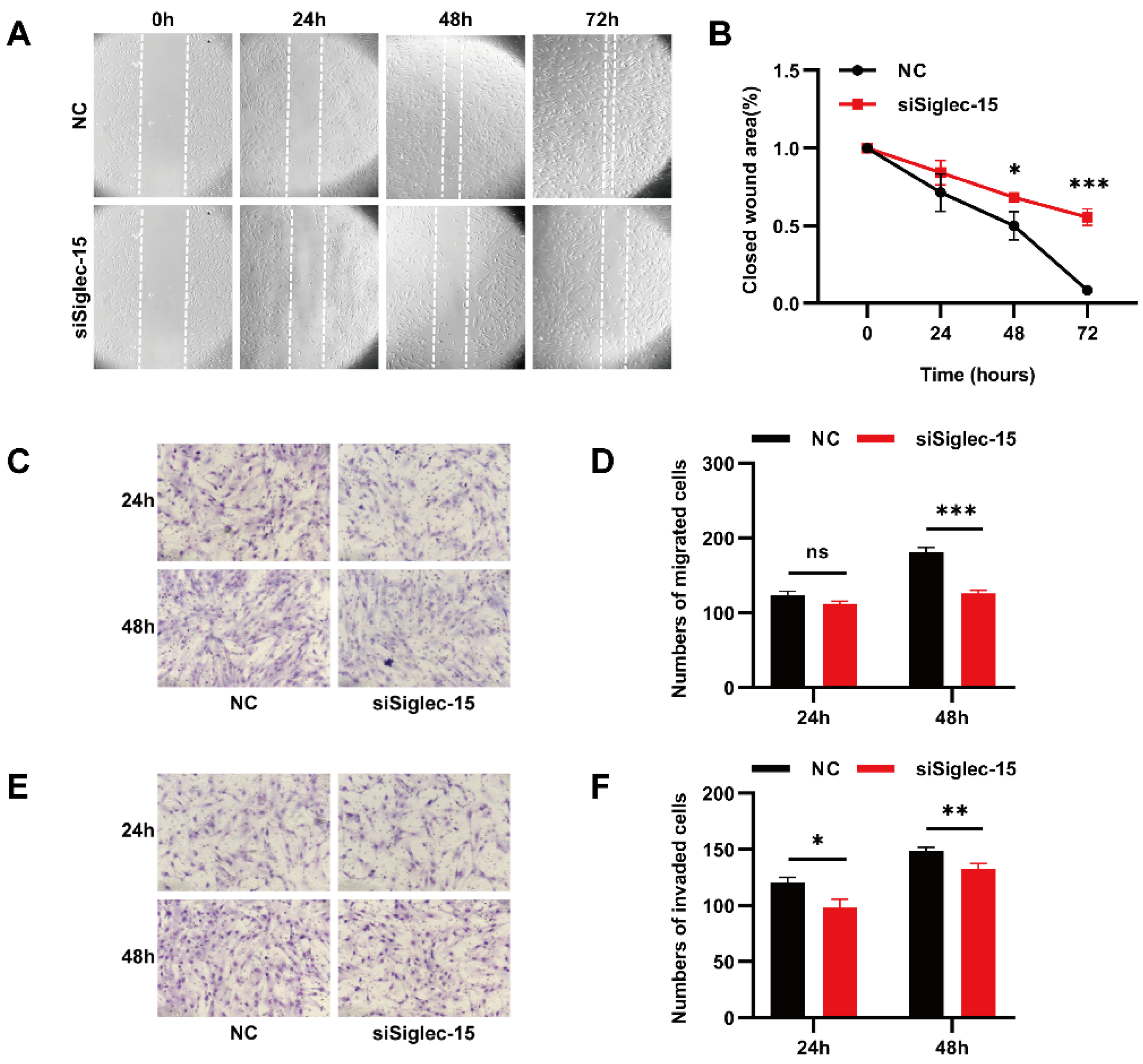
3.4. Knockdown of Siglec-15 Expression Decreases the Migration and Invasion of Hs737. T Cells
3.5. RNA Sequencing Analysis
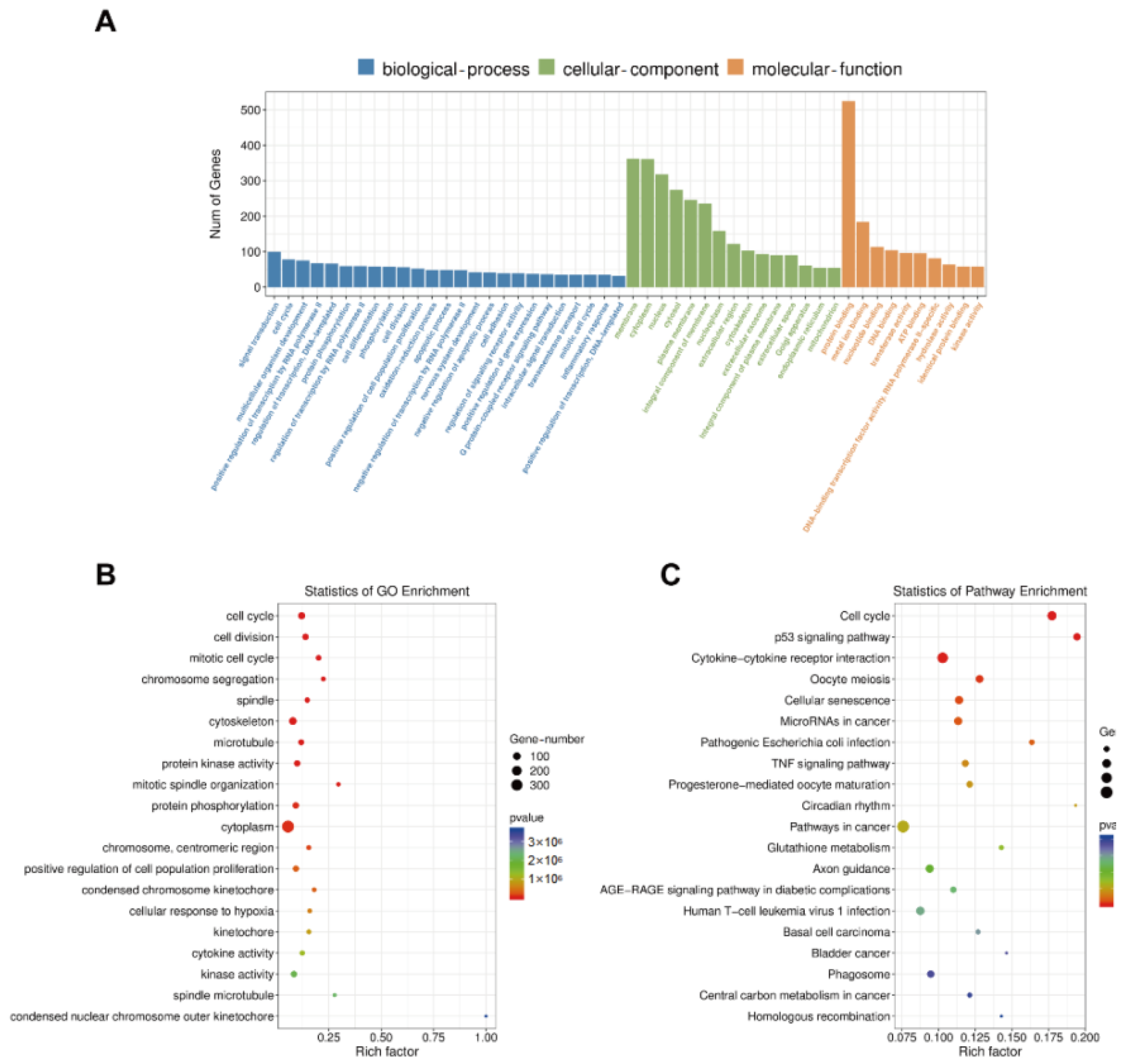
3.6. GO Enrichment, KEGG Pathway, STRING, and MCODE Analyses
3.7. CXCL8 as a Potential Target Regulated by Siglec-15 in Hs737. T Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, P.; Lin, N.; Teng, W.; Wang, S.-D.; Pan, W.-B.; Huang, X.; Yan, X.-B.; Liu, M.; Li, H.-Y.; Li, B.-H.; et al. Recurrence of Giant Cell Tumor of the Spine after Resection: A Report of 10 Cases. Orthop. Surg. 2018, 10, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Herr, I.; Sähr, H.; Zhao, Z.; Yin, L.; Omlor, G.; Lehner, B.; Fellenberg, J. MiR-127 and miR-376a act as tumor suppressors by in vivo targeting of COA1 and PDIA6 in giant cell tumor of bone. Cancer Lett. 2017, 409, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Muheremu, A.; Niu, X. Pulmonary metastasis of giant cell tumor of bones. World J. Surg. Oncol. 2014, 12, 261. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.; Couch, C.; Emory, C.L.; Nicholas, R. Giant Cell Tumor of Bone: Review of Current Literature, Evaluation, and Treatment Options. J. Knee Surg. 2019, 32, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-F.; Tang, J.-Y.; Li, K.-H. RANK pathway in giant cell tumor of bone: Pathogenesis and therapeutic aspects. Tumor Biol. 2015, 36, 495–501. [Google Scholar] [CrossRef]
- Quattrini, I.; Pollino, S.; Pazzaglia, L.; Conti, A.; Novello, C.; Ferrari, C.; Pignotti, E.; Picci, P.; Benassi, M.S. Prognostic role of nuclear factor/IB and bone remodeling proteins in metastatic giant cell tumor of bone: A retrospective study. J. Orthop. Res. 2015, 33, 1205–1211. [Google Scholar] [CrossRef]
- Palmerini, E.; Chawla, N.; Ferrari, S.; Sudan, M.; Picci, P.; Marchesi, E.; Leopardi, M.P.; Syed, I.; Sankhala, K.; Parthasarathy, P.; et al. Denosumab in advanced/unresectable giant-cell tumour of bone (GCTB): For how long? Eur. J. Cancer 2017, 76, 118–124. [Google Scholar] [CrossRef]
- Mohaidat, Z.M.; Al-Jamal, H.Z.; Bany-Khalaf, A.M.; Radaideh, A.M.; Audat, Z. Giant cell tumor of bone: Unusual features of a rare tumor. Rare Tumors 2019, 11, 2036361319878894. [Google Scholar] [CrossRef]
- Amanatullah, D.F.; Clark, T.R.; Lopez, M.J.; Borys, D.; Tamurian, R.M. Giant Cell Tumor of Bone. Orthopedics 2014, 37, 112–120. [Google Scholar] [CrossRef]
- Skubitz, K.M. Giant Cell Tumor of Bone: Current Treatment Options. Curr. Treat. Options Oncol. 2014, 15, 507–518. [Google Scholar] [CrossRef]
- Teixeira, L.E.M.; Vilela, J.C.S.; Miranda, R.H.; Gomes, A.H.; Costa, F.A.; De Faria, V.C. Giant cell tumors of bone: Nonsurgical factors associated with local recurrence. Acta Orthop. Traumatol. Turc. 2014, 48, 136–140. [Google Scholar] [CrossRef]
- Kuruogl, u.D.; Rizzo, M.; Rose, P.S.; Moran, S.L.; Houdek, M.T. Treatment of giant cell tumors of the distal radius: A long-term patient-reported outcomes study. J. Surg. Oncol. 2022, 126, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Chakarun, C.J.; Forrester, D.M.; Gottsegen, C.; Patel, D.B.; White, E.A.; Matcuk, G.R., Jr. Giant Cell Tumor of Bone: Review, Mimics, and New Developments in Treatment. RadioGraphics 2013, 33, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Uday, S.; Gaston, C.L.; Rogers, L.; Parry, M.; Joffe, J.; Pearson, J.; Sutton, D.; Grimer, R.; Högler, W. Osteonecrosis of the Jaw and Rebound Hypercalcemia in Young People Treated with Denosumab for Giant Cell Tumor of Bone. J. Clin. Endocrinol. Metab. 2018, 103, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, J.; Wood, D.J.; Zheng, M.H. Gene Expression of Osteoprotegerin Ligand, Osteoprotegerin, and Receptor Activator of NF-κB in Giant Cell Tumor of Bone: Possible involvement in tumor cell-induced osteoclast-like cell formation. Am. J. Pathol. 2000, 156, 761–767. [Google Scholar] [CrossRef]
- Wülling, M.; Engels, C.; Jesse, N.; Werner, M.; Delling, G.; Kaiser, E. The nature of giant cell tumor of bone. J. Cancer Res. Clin. Oncol. 2001, 127, 467–474. [Google Scholar] [CrossRef]
- Liu, S.; Ye, F.; Li, D.; He, C.; He, H.; Zhang, J. p62 overexpression promotes neoplastic stromal cell proliferation and is associated with the recurrence of giant cell tumor of bone. Oncol. Lett. 2020, 20, 86. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, Y.; Wang, X.; Hu, J.; Kuang, M.; Wang, Z.; Li, S.; Xu, W.; Xiao, J. ALCAM+ stromal cells: Role in giant cell tumor of bone progression. Cell Death Dis. 2018, 9, 299. [Google Scholar] [CrossRef]
- Lau, C.P.Y.; Fung, C.S.H.; Wong, K.C.; Wang, Y.; Huang, L.; Tsui, S.K.W.; Lee, O.K.; Kumta, S.M. Simvastatin Possesses Antitumor and Differentiation-Promoting Properties That Affect Stromal Cells in Giant Cell Tumor of Bone. J. Orthop. Res. 2020, 38, 297–310. [Google Scholar] [CrossRef]
- Won, K.Y.; Kalil, R.K.; Kim, Y.W.; Park, Y.-K. RANK signalling in bone lesions with osteoclast-like giant cells. Pathology 2011, 43, 318–321. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Kang, F.-B.; Chen, W.; Wang, L.; Zhang, Y.-Z. The diverse functions of Siglec-15 in bone remodeling and antitumor responses. Pharmacol. Res. 2020, 155, 104728. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, M.B.; Nakamura, M.C. A Comprehensive Review of Immunoreceptor Regulation of Osteoclasts. Clin. Rev. Allergy Immunol. 2016, 51, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Ishida-Kitagawa, N.; Tanaka, K.; Bao, X.; Kimura, T.; Miura, T.; Kitaoka, Y.; Hayashi, K.; Sato, M.; Maruoka, M.; Ogawa, T.; et al. Siglec-15 Protein Regulates Formation of Functional Osteoclasts in Concert with DNAX-activating Protein of 12 kDa (DAP12). J. Biol. Chem. 2012, 287, 17493–17502. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, Y.; Hirai, T.; Tsuda, E. Siglec-15, a member of the sialic acid-binding lectin, is a novel regulator for osteoclast differentiation. Biochem. Biophys. Res. Commun. 2011, 409, 424–429. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef]
- Fan, M.-K.; Zhang, G.-C.; Chen, W.; Qi, L.-L.; Xie, M.-F.; Zhang, Y.-Y.; Wang, L.; Zhang, Q. Siglec-15 Promotes Tumor Progression in Osteosarcoma via DUSP1/MAPK Pathway. Front. Oncol. 2021, 11, 710689. [Google Scholar] [CrossRef]
- Xiao, W.-Y.; Zong, Z.; Qiu, M.-L.; Chen, X.-Y.; Shen, H.-X.; Lao, L.-F. Paclitaxel Induce Apoptosis of Giant Cells Tumor of Bone via TP53INP1 Signaling. Orthop. Surg. 2018, 11, 126–134. [Google Scholar] [CrossRef]
- Qi, Y.; Li, Y.; Man, X.; Sui, H.; Zhao, X.; Zhang, P.; Qu, X.; Zhang, H.; Wang, B.; Li, J.; et al. CXCL3 overexpression promotes the tumorigenic potential of uterine cervical cancer cells via the MAPK/ERK pathway. J. Cell. Physiol. 2020, 235, 4756–4765. [Google Scholar] [CrossRef]
- Mandelli, G.E.; Missale, F.; Bresciani, D.; Gatta, L.B.; Scapini, P.; Caveggion, E.; Roca, E.; Bugatti, M.; Monti, M.; Cristinelli, L.; et al. Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer. Cells 2020, 9, 291. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Guan, Z.; Chen, J.; Xie, H.; Yang, Z.; Fan, J.; Wang, X.; Li, L. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int. J. Oncol. 2015, 47, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Zandueta, C.; Corrales, L.; Moreno, H.; Pajares, M.J.; Ortiz-Espinosa, S.; Martínez-Terroba, E.; Perurena, N.; de Miguel, F.J.; Jantus-Lewintre, E.; et al. Blockade of the Complement C5a/C5aR1 Axis Impairs Lung Cancer Bone Metastasis by CXCL16-mediated Effects. Am. J. Respir. Crit. Care Med. 2018, 197, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Brondani, L.A.; Crispim, D.; Pisco, J.; Guimarães, J.A.; Berger, M. The G Allele of the rs12050217 Polymorphism in the BDKRB1 Gene Is Associated with Protection for Diabetic Retinopathy. Curr. Eye Res. 2019, 44, 994–999. [Google Scholar] [CrossRef]
- Huang, J.; Chen, M.-N.; Du, J.; Liu, H.; He, Y.-J.; Li, G.-L.; Li, S.-Y.; Liu, W.-P.; Long, X.-Y. Differential Expression of Adenosine P1 Receptor ADORA1 and ADORA2A Associated with Glioma Development and Tumor-Associated Epilepsy. Neurochem. Res. 2016, 41, 1774–1783. [Google Scholar] [CrossRef]
- Gao, Y.J.; Liu, D.L.; Li, S.; Yuan, G.F.; Li, L.; Zhu, H.Y.; Cao, G.Y. Down-regulation of CXCL 11 inhibits colorectal cancer cell growth and epithelial-mesenchymal transition. OncoTargets Ther. 2018, 11, 7333–7343. [Google Scholar] [CrossRef]
- Doron, H.; Amer, M.; Ershaid, N.; Blazquez, R.; Shani, O.; Lahav, T.G.; Cohen, N.; Adler, O.; Hakim, Z.; Pozzi, S.; et al. Inflammatory Activation of Astrocytes Facilitates Melanoma Brain Tropism via the CXCL10-CXCR3 Signaling Axis. Cell Rep. 2019, 28, 1785–1798. [Google Scholar] [CrossRef]
- Natsume, M.; Shimura, T.; Iwasaki, H.; Okuda, Y.; Hayashi, K.; Takahashi, S.; Kataoka, H. Omental adipocytes promote peritoneal metastasis of gastric cancer through the CXCL2–VEGFA axis. Br. J. Cancer 2020, 123, 459–470. [Google Scholar] [CrossRef]
- Frankel, T.; Lanfranca, M.P.; Zou, W. The Role of Tumor Microenvironment in Cancer Immunotherapy. Adv. Exp. Med. Biol. 2017, 1036, 51–64. [Google Scholar] [CrossRef]
- Yang, Y. Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Investig. 2015, 125, 3335–3337. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Nizet, V. Siglecs at the Host-Pathogen Interface. Adv. Exp. Med. Biol. 2020, 1204, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Griciuc, A.; Patel, S.; Federico, A.N.; Choi, S.H.; Innes, B.J.; Oram, M.K.; Cereghetti, G.; McGinty, D.; Anselmo, A.; Sadreyev, R.I.; et al. TREM2 Acts Downstream of CD33 in Modulating Microglial Pathology in Alzheimer’s Disease. Neuron 2019, 103, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Läubli, H.; Varki, A. Sialic acid–binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell. Mol. Life Sci. 2020, 77, 593–605. [Google Scholar] [CrossRef]
- Yin, S.-S.; Gao, F.-H. Molecular Mechanism of Tumor Cell Immune Escape Mediated by CD24/Siglec-10. Front. Immunol. 2020, 11, 1324. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; Varki, N.M.; Varki, A. A Second Uniquely Human Mutation Affecting Sialic Acid Biology. J. Biol. Chem. 2001, 276, 40282–40287. [Google Scholar] [CrossRef]
- He, Y.; Zhang, J.; Ding, X. Prognosis of local recurrence in giant cell tumour of bone: What can we do? Radiol. Med. 2017, 122, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chan, C.M.; Gong, L.; Bui, M.M.; Han, G.; Letson, G.D.; Yang, Y.; Niu, X. Malignancy in giant cell tumor of bone in the extremities. J. Bone Oncol. 2020, 26, 100334. [Google Scholar] [CrossRef]
- van der Heijden, L.; Dijkstra, P.S.; Blay, J.-Y.; Gelderblom, H. Giant cell tumour of bone in the denosumab era. Eur. J. Cancer 2017, 77, 75–83. [Google Scholar] [CrossRef]
- López-Pousa, A.; Broto, J.M.; Garrido, T.; Vázquez, J. Giant cell tumour of bone: New treatments in development. Clin. Transl. Oncol. 2015, 17, 419–430. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A New Classification System and Their Role in Immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Edderkaoui, B. Potential Role of Chemokines in Fracture Repair. Front. Endocrinol. 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Sokol, C.L.; Luster, A.D. The Chemokine System in Innate Immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016303. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, A.; Tian, Y.; Wu, J.D.; Liu, Y.; Li, T.; Chen, Y.; Han, X.; Wu, K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016, 31, 61–71. [Google Scholar] [CrossRef]
- Ruffini, P.A. The CXCL8-CXCR1/2 Axis as a Therapeutic Target in Breast Cancer Stem-Like Cells. Front. Oncol. 2019, 9, 40. [Google Scholar] [CrossRef]
- Baci, D.; Bruno, A.; Cascini, C.; Gallazzi, M.; Mortara, L.; Sessa, F.; Pelosi, G.; Albini, A.; Noonan, D.M. Acetyl-L-Carnitine downregulates invasion (CXCR4/CXCL12, MMP-9) and angiogenesis (VEGF, CXCL8) pathways in prostate cancer cells: Rationale for prevention and interception strategies. J. Exp. Clin. Cancer Res. 2019, 38, 464. [Google Scholar] [CrossRef]
- Chao, C.-C.; Lee, C.-W.; Chang, T.-M.; Chen, P.-C.; Liu, J.-F. CXCL1/CXCR2 Paracrine Axis Contributes to Lung Metastasis in Osteosarcoma. Cancers 2020, 12, 459. [Google Scholar] [CrossRef]
- Ogawa, R.; Yamamoto, T.; Hirai, H.; Hanada, K.; Kiyasu, Y.; Nishikawa, G.; Mizuno, R.; Inamoto, S.; Itatani, Y.; Sakai, Y.; et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8–CXCR2 Axis. Clin. Cancer Res. 2019, 25, 2887–2899. [Google Scholar] [CrossRef]
- Zhang, H.; Ye, Y.-L.; Li, M.-X.; Ye, S.-B.; Huang, W.-R.; Cai, T.-T.; He, J.; Peng, J.-Y.; Duan, T.-H.; Cui, J.; et al. CXCL2/MIF-CXCR2 signaling promotes the recruitment of myeloid-derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene 2017, 36, 2095–2104. [Google Scholar] [CrossRef]
- Taki, M.; Abiko, K.; Baba, T.; Hamanishi, J.; Yamaguchi, K.; Murakami, R.; Yamanoi, K.; Horikawa, N.; Hosoe, Y.; Nakamura, E.; et al. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat. Commun. 2018, 9, 1685. [Google Scholar] [CrossRef]
- Chen, M.; Baskaran, R.; Lee, N.; Hsu, H.; Ho, T.; Tu, C.; Lin, Y.; Viswanadha, V.P.; Kuo, W.; Huang, C. CXCL2/CXCR2 axis induces cancer stem cell characteristics in CPT-11-resistant LoVo colon cancer cells via Gαi-2 and Gαq/11. J. Cell. Physiol. 2019, 234, 11822–11834. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.-L.; Teng, L.-C.; Wang, S.-Q.; Liu, S.; Lin, Y.-L.; Zhao, X.-L.; Liu, L.; Sui, H.-Y.; Yang, Y.; Liang, L.-C.; et al. Overexpression of CXCL3 can enhance the oncogenic potential of prostate cancer. Int. Urol. Nephrol. 2016, 48, 701–709. [Google Scholar] [CrossRef] [PubMed]
- See, A.L.P.; Chong, P.K.; Lu, S.-Y.; Lim, Y.P. CXCL3 is a potential target for breast cancer metastasis. Curr. Cancer Drug Targets 2014, 14, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Overman, M.J.; Boutin, A.T.; Shang, X.; Zhao, D.; Dey, P.; Li, J.; Wang, G.; Lan, Z.; Li, J.; et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019, 35, 559–572.e7. [Google Scholar] [CrossRef]
- Mao, Z.; Zhang, J.; Shi, Y.; Li, W.; Shi, H.; Ji, R.; Mao, F.; Qian, H.; Xu, W.; Zhang, X. CXCL5 promotes gastric cancer metastasis by inducing epithelial-mesenchymal transition and activating neutrophils. Oncogenesis 2020, 9, 63. [Google Scholar] [CrossRef]
- Li, L.; Zhu, Y.H.; Li, Y.; Guan, X.Y. Identification of chemokine CXCL10 in tumor microenvironment by antibody array as a prognostic marker in hepatocellular carcinoma. Neoplasma 2017, 64, 778–786. [Google Scholar] [CrossRef]
- Li, C.X.; Ling, C.C.; Shao, Y.; Xu, A.; Li, X.C.; Ng, K.T.-P.; Liu, X.B.; Ma, Y.Y.; Qi, X.; Liu, H.; et al. CXCL10/CXCR3 signaling mobilized-regulatory T cells promote liver tumor recurrence after transplantation. J. Hepatol. 2016, 65, 944–952. [Google Scholar] [CrossRef]
- Hwang, H.J.; Lee, Y.-R.; Kang, D.; Lee, H.C.; Seo, H.R.; Ryu, J.-K.; Kim, Y.-N.; Ko, Y.-G.; Park, H.J.; Lee, J.-S. Endothelial cells under therapy-induced senescence secrete CXCL11, which increases aggressiveness of breast cancer cells. Cancer Lett. 2020, 490, 100–110. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, S.; Chen, X.; Cheng, S.; Zhang, Z.; Li, F.; Huang, L.; Yang, Y.; Zhou, B.; Yue, D.; et al. Cancer-cell-secreted CXCL11 promoted CD8+ T cells infiltration through docetaxel-induced-release of HMGB1 in NSCLC. J. Immunother. Cancer 2019, 7, 42. [Google Scholar] [CrossRef]
- Haskó, G.; Linden, J.; Cronstein, B.; Pacher, P. Adenosine receptors: Therapeutic aspects for inflammatory and immune diseases. Nat. Rev. Drug Discov. 2008, 7, 759–770. [Google Scholar] [CrossRef]
- Ni, S.; Wei, Q.; Yang, L. ADORA1 Promotes Hepatocellular Carcinoma Progression via PI3K/AKT Pathway. OncoTargets Ther. 2020, 13, 12409–12419. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kuang, X.; Zhang, Y.; Ye, Y.; Li, J.; Liang, L.; Xie, Z.; Weng, L.; Guo, J.; Li, H.; et al. ADORA1 Inhibition Promotes Tumor Immune Evasion by Regulating the ATF3-PD-L1 Axis. Cancer Cell 2020, 37, 324–339.e8. [Google Scholar] [CrossRef] [PubMed]
- Chai, K.X.; Ni, A.; Wang, D.; Ward, D.C.; Chao, J.; Chao, L. Genomic DNA Sequence, Expression, and Chromosomal Localization of the Human B1 Bradykinin Receptor Gene BDKRB1. Genomics 1996, 31, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.-P.; Lee, Y.-W.; Chen, J.-T.; Lin, Y.-W.; Chen, R.-M. The Bradykinin-BDKRB1 Axis Regulates Aquaporin 4 Gene Expression and Consequential Migration and Invasion of Malignant Glioblastoma Cells via a Ca2+-MEK1-ERK1/2-NF-κB Mechanism. Cancers 2020, 12, 667. [Google Scholar] [CrossRef]
- Guo, R.-F.; Ward, P.A. ROLE OF C5A IN INFLAMMATORY RESPONSES. Annu. Rev. Immunol. 2005, 23, 821–852. [Google Scholar] [CrossRef]
- Markiewski, M.M.; DeAngelis, R.A.; Benencia, F.; Ricklin-Lichtsteiner, S.K.; Koutoulaki, A.; Gerard, C.; Coukos, G.; Lambris, J.D. Modulation of the antitumor immune response by complement. Nat. Immunol. 2008, 9, 1225–1235. [Google Scholar] [CrossRef]
- Nabizadeh, J.A.; Manthey, H.D.; Panagides, N.; Steyn, F.J.; Lee, J.D.; Li, X.X.; Akhir, F.N.M.; Chen, W.; Boyle, G.M.; Taylor, S.M.; et al. C5a receptors C5aR1 and C5aR2 mediate opposing pathologies in a mouse model of melanoma. FASEB J. 2019, 33, 11060–11071. [Google Scholar] [CrossRef]
- Ajona, D.; Ortiz-Espinosa, S.; Moreno, H.; Lozano, T.; Pajares, M.J.; Agorreta, J.; Bértolo, C.; Lasarte, J.J.; Vicent, S.; Hoehlig, K.; et al. A Combined PD-1/C5a Blockade Synergistically Protects against Lung Cancer Growth and Metastasis. Cancer Discov. 2017, 7, 694–703. [Google Scholar] [CrossRef]
- Hanadaa, R.; Nakazato, M.; Murakamic, N.; Sakiharad, S.; Yoshimatsua, H.; Toshinaib, K.; Hanadae, T.; Sudad, T.; Kangawaf, K.; Matsukurab, S.; et al. A Role for Neuromedin U in Stress Response. Biochem. Biophys. Res. Commun. 2001, 289, 225–228. [Google Scholar] [CrossRef]
- Yoo, W.; Lee, J.; Jun, E.; Noh, K.H.; Lee, S.; Jung, D.; Jung, K.H.; Kim, J.-S.; Park, Y.-Y.; Kim, S.C.; et al. The YAP1–NMU Axis Is Associated with Pancreatic Cancer Progression and Poor Outcome: Identification of a Novel Diagnostic Biomarker and Therapeutic Target. Cancers 2019, 11, 1477. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Wu, F.-J.; Chang, C.-L.; Li, Z.; Luo, C.-W. NMU signaling promotes endometrial cancer cell progression by modulating adhesion signaling. Oncotarget 2016, 7, 10228–10242. [Google Scholar] [CrossRef] [PubMed]


| Clinical Parameters | Cases | Siglec-15 Expression | p Value | ||||
|---|---|---|---|---|---|---|---|
| (Neg) | (+) | (++) | (+++) | ||||
| Gender | Male | 20 | 2 | 9 | 8 | 1 | 0.091 |
| Female | 36 | 3 | 11 | 11 | 11 | ||
| Age (years) | ≤20 | 4 | 2 | 1 | 0 | 1 | 0.126 |
| 20–40 | 34 | 3 | 13 | 13 | 5 | ||
| >40 | 18 | 0 | 6 | 6 | 6 | ||
| Site | Femur | 20 | 0 | 8 | 9 | 3 | 0.657 |
| Tibia | 12 | 1 | 4 | 3 | 4 | ||
| Others | 24 | 4 | 8 | 7 | 5 | ||
| Tumor size (cm) | <5 | 30 | 4 | 8 | 9 | 9 | 0.383 |
| ≥5 | 26 | 1 | 12 | 10 | 3 | ||
| Campanacci’s grade | I | 15 | 4 | 5 | 4 | 2 | 0.018 |
| II | 24 | 0 | 12 | 10 | 2 | ||
| III | 17 | 1 | 3 | 5 | 8 | ||
| Recurrence | Yes | 8 | 0 | 0 | 5 | 3 | 0.015 |
| No | 48 | 5 | 20 | 14 | 9 | ||
| Pathological Parameters | Cases | Siglec-15 Expression | r | p Value | |||
|---|---|---|---|---|---|---|---|
| (Neg) | (+) | (++) | (+++) | ||||
| CD163 (+) | 18 | 1 | 10 | 6 | 1 | −0.334 | 0.119 |
| CD163 (-) | 5 | 4 | 0 | 0 | 1 | ||
| CD34 (+) | 12 | 1 | 5 | 4 | 2 | −0.377 | 0.076 |
| CD34 (-) | 15 | 4 | 8 | 2 | 0 | ||
| CD68 (+) | 22 | 5 | 10 | 5 | 2 | 0.173 | 0.429 |
| CD68 (-) | 1 | 0 | 0 | 1 | 0 | ||
| Ki-67 ≤ 5% | 18 | 2 | 8 | 6 | 2 | −0.462 | 0.027 |
| Ki-67 > 5% | 5 | 3 | 2 | 0 | 0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, M.; Zhang, G.; Xie, M.; Liu, X.; Zhang, Q.; Wang, L. Siglec-15 as a New Perspective Therapy Target in Human Giant Cell Tumor of Bone. Curr. Oncol. 2022, 29, 7655-7671. https://doi.org/10.3390/curroncol29100605
Fan M, Zhang G, Xie M, Liu X, Zhang Q, Wang L. Siglec-15 as a New Perspective Therapy Target in Human Giant Cell Tumor of Bone. Current Oncology. 2022; 29(10):7655-7671. https://doi.org/10.3390/curroncol29100605
Chicago/Turabian StyleFan, Mengke, Guochuan Zhang, Mingfang Xie, Xinbo Liu, Qi Zhang, and Ling Wang. 2022. "Siglec-15 as a New Perspective Therapy Target in Human Giant Cell Tumor of Bone" Current Oncology 29, no. 10: 7655-7671. https://doi.org/10.3390/curroncol29100605
APA StyleFan, M., Zhang, G., Xie, M., Liu, X., Zhang, Q., & Wang, L. (2022). Siglec-15 as a New Perspective Therapy Target in Human Giant Cell Tumor of Bone. Current Oncology, 29(10), 7655-7671. https://doi.org/10.3390/curroncol29100605





