Comprehensive Genomic Profiling of Circulating Cell-Free DNA Distinguishes Focal MET Amplification from Aneuploidy in Diverse Advanced Cancers
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Next-Generation Sequencing
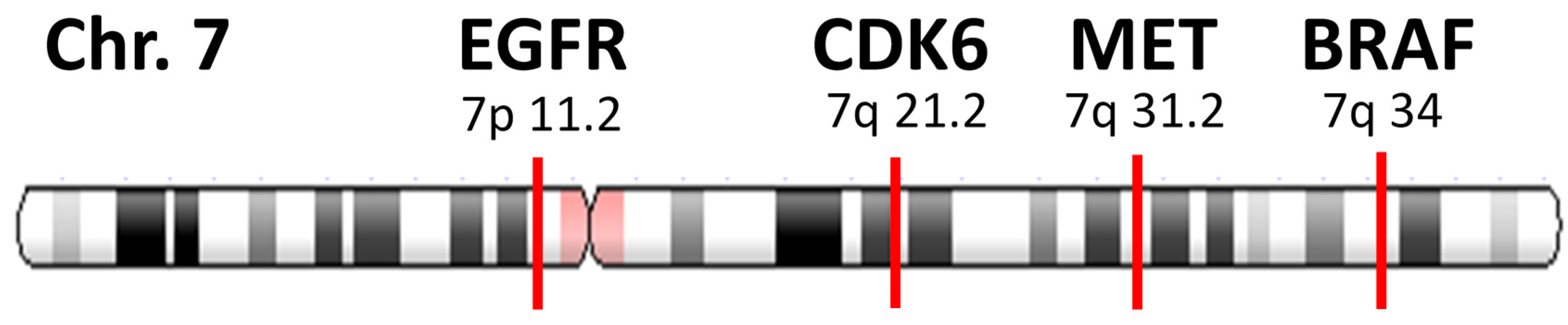
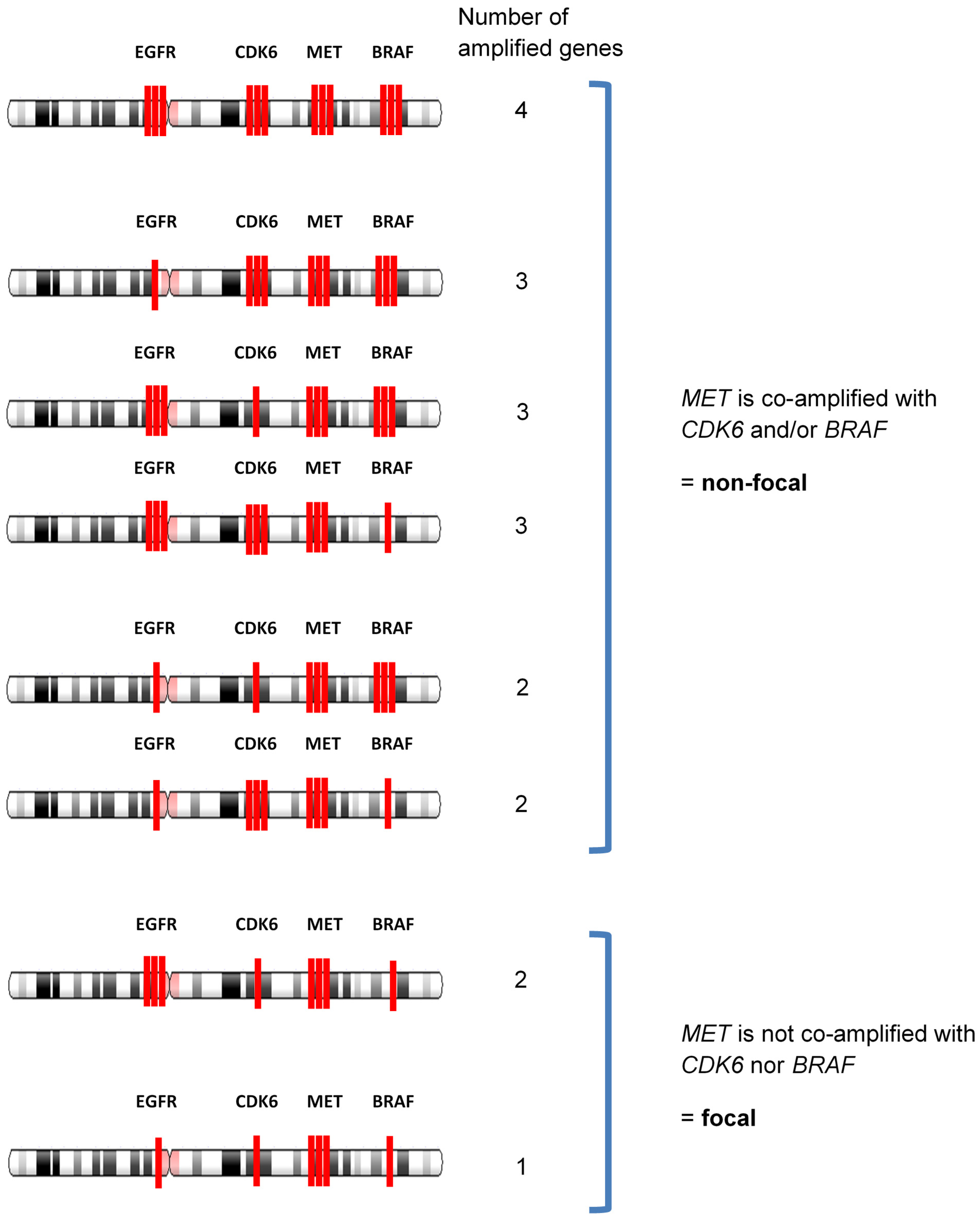
2.3. Determination of Focal Amplification
- (a)
- MET copy number ≥2.2.
- (b)
- MET gene is amplified without co-amplification of CDK6 and BRAF. Co-amplification status was defined as “increased together” when the copy number of other gene (CDK6 or BRAF) ≥2.2, and the difference with MET amplification is within +/−0.5.
- (c)
- MET amplification that satisfies both (a) and (b) is defined as focal.
2.4. Statistical and Outcome Analysis
3. Results
3.1. Patient Demographic Characteristics
3.2. MET Alterations and Associations with Patient Characteristics
3.3. MET Alteration Types and Focal vs. Non-Focal MET Amp
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Organ, S.L.; Tsao, M.-S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011, 3, S7–S19. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Tang, C.; Gagliato, D.D.; Falchook, G.S.; Hess, K.; Janku, F.; Fu, S.; Wheler, J.J.; Zinner, R.G.; Naing, A.; et al. Analysis of 1,115 Patients Tested for MET Amplification and Therapy Response in the MD Anderson Phase I Clinic. Clin. Cancer Res. 2014, 20, 6336–6345. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Frampton, G.M.; Suh, J.; Chalmers, Z.R.; Rosenzweig, M.; Erlich, R.L.; Halmos, B.; Goldman, J.; Forde, P.; Leuenberger, K.; et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 2016, 11, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Okamoto, I.; Okamoto, W.; Tanizaki, J.; Nakagawa, K.; Nishio, K. Targeting MET Amplification as a New Oncogenic Driver. Cancers 2014, 6, 1540–1552. [Google Scholar] [CrossRef] [PubMed]
- Caparica, R.; Yen, C.T.; Coudry, R.; Ou, S.-H.I.; Varella-Garcia, M.; Camidge, D.R.; de Castro, G. Responses to Crizotinib Can Occur in High-Level MET -Amplified Non–Small Cell Lung Cancer Independent of MET Exon 14 Alterations. J. Thorac. Oncol. 2017, 12, 141–144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non–Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef] [PubMed]
- Noonan, S.A.; Berry, L.; Lu, X.; Gao, D.; Barón, A.E.; Chesnut, P.; Sheren, J.; Aisner, D.L.; Merrick, D.; Doebele, R.C.; et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number–Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. J. Thorac. Oncol. 2016, 11, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Schwaederle, M.; Mohindra, M.; Jardim, D.L.; Kurzrock, R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J. Hematol. Oncol. 2018, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Cheng, Y.; Zhou, J.; Lu, S.; Zhang, Y.; Zhao, J.; Kim, D.-W.; Soo, R.A.; Kim, S.-W.; Pan, H.; et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): An open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir. Med. 2020, 8, 1132–1143. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.; Tan, D.S.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Sequist, L.V.; Han, J.-Y.; Ahn, M.-J.; Cho, B.C.; Yu, H.; Kim, S.-W.; Yang, J.C.-H.; Lee, J.S.; Su, W.-C.; Kowalski, D.; et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: Interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020, 21, 373–386. [Google Scholar] [CrossRef]
- Yun, J.; Lee, S.-H.; Kim, S.-Y.; Jeong, S.-Y.; Kim, J.-H.; Pyo, K.-H.; Park, C.-W.; Heo, S.G.; Yun, M.R.; Lim, S.; et al. Antitumor Activity of Amivantamab (JNJ-61186372), an EGFR-cMet Bispecific Antibody, in Diverse Models of EGFR Exon 20 Insertion-Driven NSCLC. Cancer Discov. 2020, 10, 1194–1209. [Google Scholar] [CrossRef] [PubMed]
- Lai, G.G.Y.; Lim, T.H.; Lim, J.; Liew, P.J.R.; Kwang, X.L.; Nahar, R.; Aung, Z.W.; Takano, A.; Lee, Y.Y.; Lau, D.P.X.; et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor–Mutant Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2019, 37, 876–884. [Google Scholar] [CrossRef]
- Roper, N.; Brown, A.-L.; Wei, J.S.; Pack, S.; Trindade, C.; Kim, C.; Restifo, O.; Gao, S.; Sindiri, S.; Mehrabadi, F.; et al. Clonal Evolution and Heterogeneity of Osimertinib Acquired Resistance Mechanisms in EGFR Mutant Lung Cancer. Cell Rep. Med. 2020, 1, 100007. [Google Scholar] [CrossRef]
- Bean, J.; Brennan, C.; Shih, J.-Y.; Riely, G.; Viale, A.; Wang, L.; Chitale, D.; Motoi, N.; Szoke, J.; Broderick, S.; et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. USA 2007, 104, 20932–20937. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.-M.; Zhao, X.; Christensen, J.; et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Total Patients, n = 1025 (100%) | MET Alt Not Detected, n = 915 (89.3%) | MET Alt Detected, n = 110 (10.7%) | p-Values ** |
|---|---|---|---|---|
| Mean age at time of testing, y | 62.4 | 62.4 | 62.4 | 0.966 |
| (CI 95%), n = 880 known | (61.6–63.3) | (61.5–63.3) | (59.4–65.3) | |
| Gender | 0.00234 | |||
| Men | 482 (47.0%) | 415 (86.1%) | 67 (13.9%) | |
| Women | 543 (53.0%) | 500 (92.1%) | 43 (7.9%) | |
| Type of cancer | 0.0863 | |||
| NSCLC | 510 (49.8%) | 448 (87.8%) | 62 (12.2%) | |
| SCLC | 9 (0.9%) | 6 (66.7%) | 3 (33.3%) | |
| Colorectal | 77 (7.5%) | 71 (92.2%) | 6 (7.8%) | |
| Gastroesophageal | 36 (3.5%) | 29 (80.6%) | 7 (19.4%) | |
| Other gastrointestinal | 158 (15.4%) | 147 (94.2%) | 9 (5.8%) | |
| Breast | 92 (9.0%) | 84 (91.3%) | 8 (8.7%) | |
| Unknown primary | 20 (2.0%) | 18 (94.7%) | 1 (5.3%) | |
| Gynecologic | 46 (4.5%) | 43 (93.5%) | 3 (6.5%) | |
| Prostate | 32 (3.1%) | 27 (84.4%) | 5 (15.6%) | |
| Other | 45 (4.4%) | 42 (87.5%) | 6 (12.5%) * |
| Case | EGFR | CDK6 | MET | BRAF | Feature | Focal/Non-Focal |
|---|---|---|---|---|---|---|
| 1 | 2.2 * | 2.2 | 2.1 | 2.3 | MET copy number is lower than 2.2 | non-focal |
| 2 | 2.2 | 2.1 | 2.1 | 2.3 | non-focal | |
| 3 | 2.2 | 2.2 | 2.1 | 2.0 | non-focal | |
| 4 | 3.1 | 2.9 | 2.8 | 3 | MET copy number is increased together with either CDK6 and/or BRAF | non-focal |
| 5 | 2.9 | 2.8 | 2.6 | 2.8 | non-focal | |
| 6 | 2.6 | 2.6 | 2.6 | 2.5 | non-focal | |
| 7 | 2.2 | 2.3 | 2.2 | 2.2 | non-focal | |
| 8 | 2.8 | 3.6 | 3.3 | 3.6 | non-focal | |
| 9 | 2.4 | 2.3 | 2.6 | 2.5 | non-focal | |
| 10 | 2.0 | 2.6 | 2.8 | 2.6 | non-focal | |
| 11 | 3 | 2.3 | 2.2 | 2.2 | non-focal | |
| 12 | 3.3 | 2.3 | 2.3 | 2.3 | non-focal | |
| 13 | 80 | 2.6 | 2.5 | 2.6 | non-focal | |
| 14 | 2.0 | 2.5 | 2.3 | 2.4 | non-focal | |
| 15 | 2.0 | 2.6 | 2.5 | 2.5 | non-focal | |
| 16 | 2.1 | 2.0 | 2.3 | 2.2 | non-focal | |
| 17 | 2.0 | 2.2 | 2.2 | 2.0 | non-focal | |
| 18 | 3.2 | 2.0 | 3.3 | 2.0 | MET gene is amplified without co-amplification of CDK6 and BRAF | focal |
| 19 | 2.5 | 2.0 | 2.5 | 2.0 | focal | |
| 20 | 2.0 | 2.0 | 2.6 | 2.0 | focal | |
| 21 | 2.0 | 2.5 | 3.7 | 2.0 | focal | |
| 22 | 2.0 | 2.0 | 2.2 | 2.0 | focal | |
| 23 | 2.2 | 2.3 | 6.2 | N/A ** | focal | |
| 24 | 2.0 | 2.0 | 2.3 | 2.0 | focal |
| Cancer Type (n Patients, %w/AMP) | Patients w/Focal MET Amp in ≥1 Sample | Patients w/Only Non-Focal MET Amp | Proportion Focal | p-Value |
|---|---|---|---|---|
| NSCLC (144, 6.9%) | 4 (2.8%) | 6 (4.2%) | 4/10 (40.0%) | 0.140 |
| Gastrointestinal (69, 5.8%) | 1 (1.4%) | 3 (4.3%) | 1/4 (25.0%) | |
| Breast (26, 15.4%) | 2 (7.7%) | 2 (7.7%) | 2/4 (50.0%) | |
| Other (52, 11.5%) | 0 (0.0%) | 6 (11.5%) | 0/6 (0%) | |
| Overall (291, 8.2%) | 7 (2.4%) | 17 (5.8%) | 7/24 (29.2%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaki, Y.; Olsen, S.; Suenaga, M.; Nakagawa, T.; Uetake, H.; Ikeda, S. Comprehensive Genomic Profiling of Circulating Cell-Free DNA Distinguishes Focal MET Amplification from Aneuploidy in Diverse Advanced Cancers. Curr. Oncol. 2021, 28, 3717-3728. https://doi.org/10.3390/curroncol28050317
Kumaki Y, Olsen S, Suenaga M, Nakagawa T, Uetake H, Ikeda S. Comprehensive Genomic Profiling of Circulating Cell-Free DNA Distinguishes Focal MET Amplification from Aneuploidy in Diverse Advanced Cancers. Current Oncology. 2021; 28(5):3717-3728. https://doi.org/10.3390/curroncol28050317
Chicago/Turabian StyleKumaki, Yuichi, Steve Olsen, Mitsukuni Suenaga, Tsuyoshi Nakagawa, Hiroyuki Uetake, and Sadakatsu Ikeda. 2021. "Comprehensive Genomic Profiling of Circulating Cell-Free DNA Distinguishes Focal MET Amplification from Aneuploidy in Diverse Advanced Cancers" Current Oncology 28, no. 5: 3717-3728. https://doi.org/10.3390/curroncol28050317
APA StyleKumaki, Y., Olsen, S., Suenaga, M., Nakagawa, T., Uetake, H., & Ikeda, S. (2021). Comprehensive Genomic Profiling of Circulating Cell-Free DNA Distinguishes Focal MET Amplification from Aneuploidy in Diverse Advanced Cancers. Current Oncology, 28(5), 3717-3728. https://doi.org/10.3390/curroncol28050317







