3q26 Amplifications in Cervical Squamous Carcinomas
Abstract
:1. Introduction
2. Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key Molecular Events in Cervical Cancer Development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, R.; Nene, B.M.; Shastri, S.S.; Jayant, K.; Muwonge, R.; Budukh, A.M.; Hingmire, S.; Malvi, S.G.; Thorat, R.; Kothari, A.; et al. HPV screening for cervical cancer in rural India. N. Engl. J. Med. 2009, 360, 1385–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, K.S.; Sill, M.W.; Long, H.J., 3rd; Penson, R.T.; Huang, H.; Ramondetta, L.M.; Landrum, L.M.; Oaknin, A.; Reid, T.J.; Leitao, M.M.; et al. Improved survival with bevacizumab in advanced cervical cancer. N. Engl. J. Med. 2014, 370, 734–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. PI3KCA Mutations in Uterine Cervix Carcinoma. J. Clin. Med. 2021, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Pathogenesis of colorectal carcinoma and therapeutic implications: The roles of the ubiquitin-proteasome system and Cox-2. J. Cell Mol. Med. 2007, 11, 252–285. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. SOLAR-1 Study Group. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Narayan, P.; Prowell, T.M.; Gao, J.J.; Fernandes, L.L.; Li, E.; Jiang, X.; Qiu, J.; Fan, J.; Song, P.; Yu, J.; et al. FDA Approval Summary: Alpelisib Plus Fulvestrant for Patients with HR-positive, HER2-negative, PIK3CA-mutated, Advanced or Metastatic Breast Cancer. Clin. Cancer Res. 2021, 27, 1842–1849. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule phosphatidylinositol 3-kinase inhibitors prescribed for the treatment of malignancies. Pharmacol. Res. 2021, 168, 105579. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Gogineni, S.K.; Sacks, P.G.; Shaha, A.R.; Shah, J.P.; Stoffel, A.; Rao, P.H. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001, 61, 4506–4513. [Google Scholar] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Boehm, J.S.; Garnett, M.J.; Adams, D.J.; Francies, H.E.; Golub, T.R.; Hahn, W.C.; Iorio, F.; McFarland, J.M.; Parts, L.; Vazquez, F. Cancer research needs a better map. Nature 2021, 589, 514–516. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Amplification of 8p11.23 in cancers and the role of amplicon genes. Life Sci. 2021, 264, 118729. [Google Scholar] [CrossRef]
- Wang, J.; Qian, J.; Hoeksema, M.D.; Zou, Y.; Espinosa, A.V.; Rahman, S.M.; Zhang, B.; Massion, P.P. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clin. Cancer Res. 2013, 19, 5580–5590. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Singh, B. Coamplification and cooperation: Toward identifying biologically relevant oncogenes. Clin. Cancer Res. 2013, 19, 5549–5551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Towe, C.W.; Choi, L.; Yonekawa, Y.; Bommeljé, C.C.; Bains, S.; Rechler, W.; Hao, B.; Ramanathan, Y.; Singh, B. The ubiquitin-associated (UBA) domain of SCCRO/DCUN1D1 protein serves as a feedback regulator of biochemical and oncogenic activity. J. Biol. Chem. 2015, 290, 296–309. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.M.; Redon, C.E.; Thakur, B.L.; Bahta, M.K.; Aladjem, M.I. Regulation of cell cycle drivers by Cullin-RING ubiquitin ligases. Exp. Mol. Med. 2020, 52, 1637–1651. [Google Scholar] [CrossRef]
- Li, J.Y.; Daniels, G.; Wang, J.; Zhang, X. TBL1XR1 in physiological and pathological states. Am. J. Clin. Exp. Urol. 2015, 3, 13–23. [Google Scholar]
- Choi, H.K.; Choi, K.C.; Yoo, J.Y.; Song, M.; Ko, S.J.; Kim, C.H.; Ahn, J.H.; Chun, K.H.; Yook, J.I.; Yoon, H.G. Reversible SUMOylation of TBL1-TBLR1 regulates β-catenin-mediated Wnt signaling. Mol. Cell 2011, 43, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lin, C.; Liang, W.; Wu, S.; Liu, A.; Wu, J.; Zhang, X.; Ren, P.; Li, M.; Song, L. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut 2015, 64, 26–36. [Google Scholar] [CrossRef]
- Lu, J.; Bang, H.; Kim, S.M.; Cho, S.J.; Ashktorab, H.; Smoot, D.T.; Zheng, C.H.; Ryeom, S.W.; Yoon, S.S.; Yoon, C.; et al. Lymphatic metastasis-related TBL1XR1 enhances stemness and metastasis in gastric cancer stem-like cells by activating ERK1/2-SOX2 signaling. Oncogene 2021, 40, 922–936. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Chopra, S.; Deodhar, K.; Pai, V.; Pant, S.; Rathod, N.; Goda, J.S.; Sudhalkar, N.; Pandey, P.; Waghmare, S.; Engineer, R.; et al. Cancer Stem Cells, CD44, and Outcomes Following Chemoradiation in Locally Advanced Cervical Cancer: Results from a Prospective Study. Int. J. Radiat. Oncol. Biol. Phys. 2019, 103, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Rekhtman, N.; Ladanyi, M.; Paik, P. Squamous-cell carcinomas of the lung: Emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012, 13, e418–e426. [Google Scholar] [CrossRef]
- Kim, B.R.; Van de Laar, E.; Cabanero, M.; Tarumi, S.; Hasenoeder, S.; Wang, D.; Virtanen, C.; Suzuki, T.; Bandarchi, B.; Sakashita, S.; et al. SOX2 and PI3K Cooperate to Induce and Stabilize a Squamous-Committed Stem Cell Injury State during Lung Squamous Cell Carcinoma Pathogenesis. PLoS Biol. 2016, 14, e1002581. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Diaz-Meco, M.T.; Moscat, J. The Dual Roles of the Atypical Protein Kinase Cs in Cancer. Cancer Cell 2019, 36, 218–235. [Google Scholar] [CrossRef]
- Justilien, V.; Walsh, M.P.; Ali, S.A.; Thompson, E.A.; Murray, N.R.; Fields, A.P. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 2014, 25, 139–151. [Google Scholar] [CrossRef] [Green Version]
- Tokinaga-Uchiyama, A.; Mizushima, T.; Akimoto, K.; Nagashima, Y.; Sasaki, K.; Nakaya, M.A.; Ohashi, K.; Kubota, K.; Maruyama, Y.; Kato, H.; et al. Aberrant Nuclear Localization of aPKCλ/ι is Associated With Poorer Prognosis in Uterine Cervical Cancer. Int. J. Gynecol. Pathol. 2019, 38, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, T.; Asai-Sato, M.; Akimoto, K.; Nagashima, Y.; Taguri, M.; Sasaki, K.; Nakaya, M.A.; Asano, R.; Tokinaga, A.; Kiyono, T.; et al. Aberrant Expression of the Cell Polarity Regulator aPKCλ/ι is Associated with Disease Progression in Cervical Intraepithelial Neoplasia (CIN): A Possible Marker for Predicting CIN Prognosis. Int. J. Gynecol. Pathol. 2016, 35, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Hopman, A.H.; Theelen, W.; Hommelberg, P.P.; Kamps, M.A.; Herrington, C.S.; Morrison, L.E.; Speel, E.J.; Smedts, F.; Ramaekers, F.C. Genomic integration of oncogenic HPV and gain of the human telomerase gene TERC at 3q26 are strongly associated events in the progression of uterine cervical dysplasia to invasive cancer. J. Pathol. 2006, 210, 412–419. [Google Scholar] [CrossRef] [PubMed]
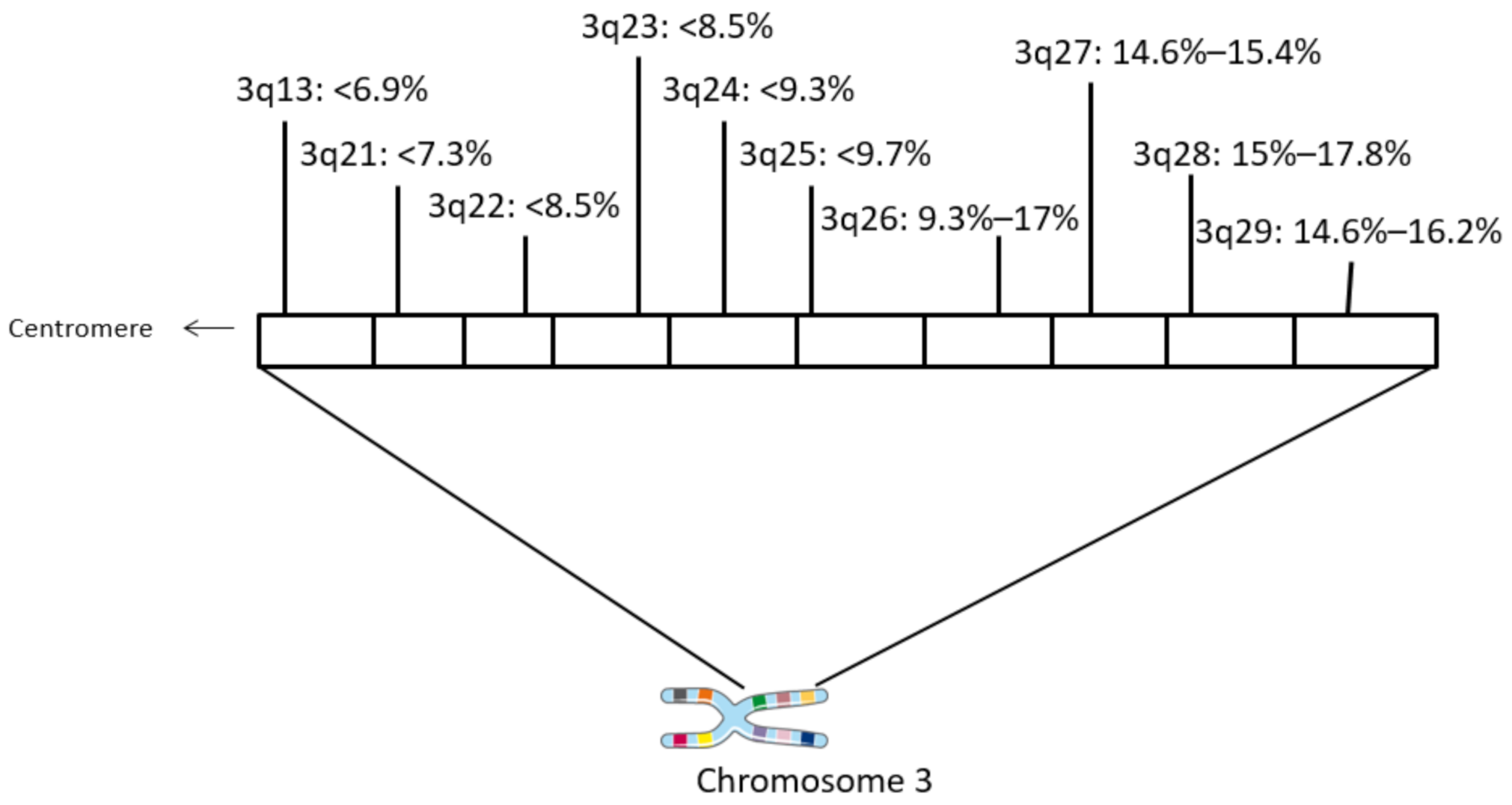
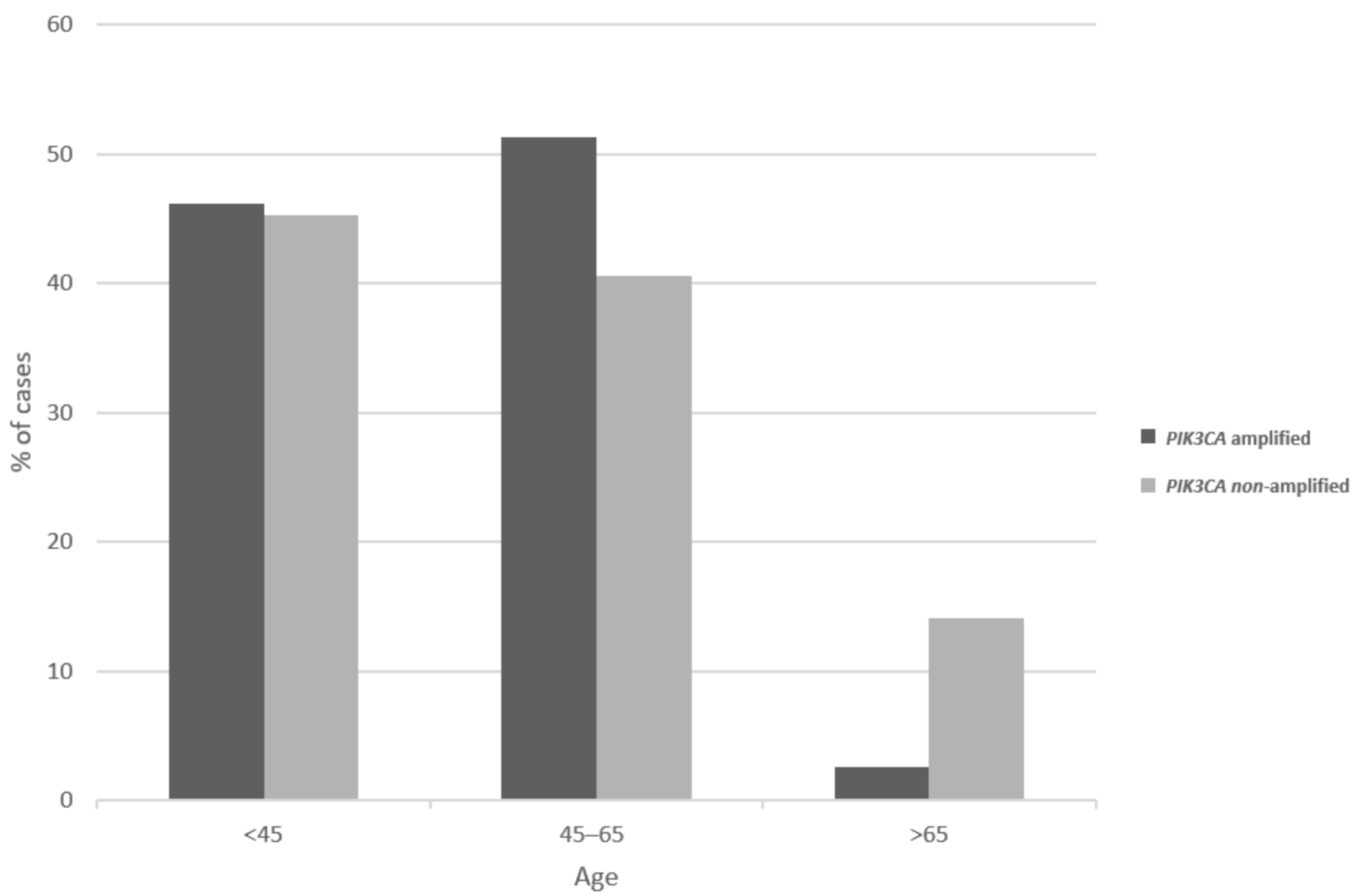
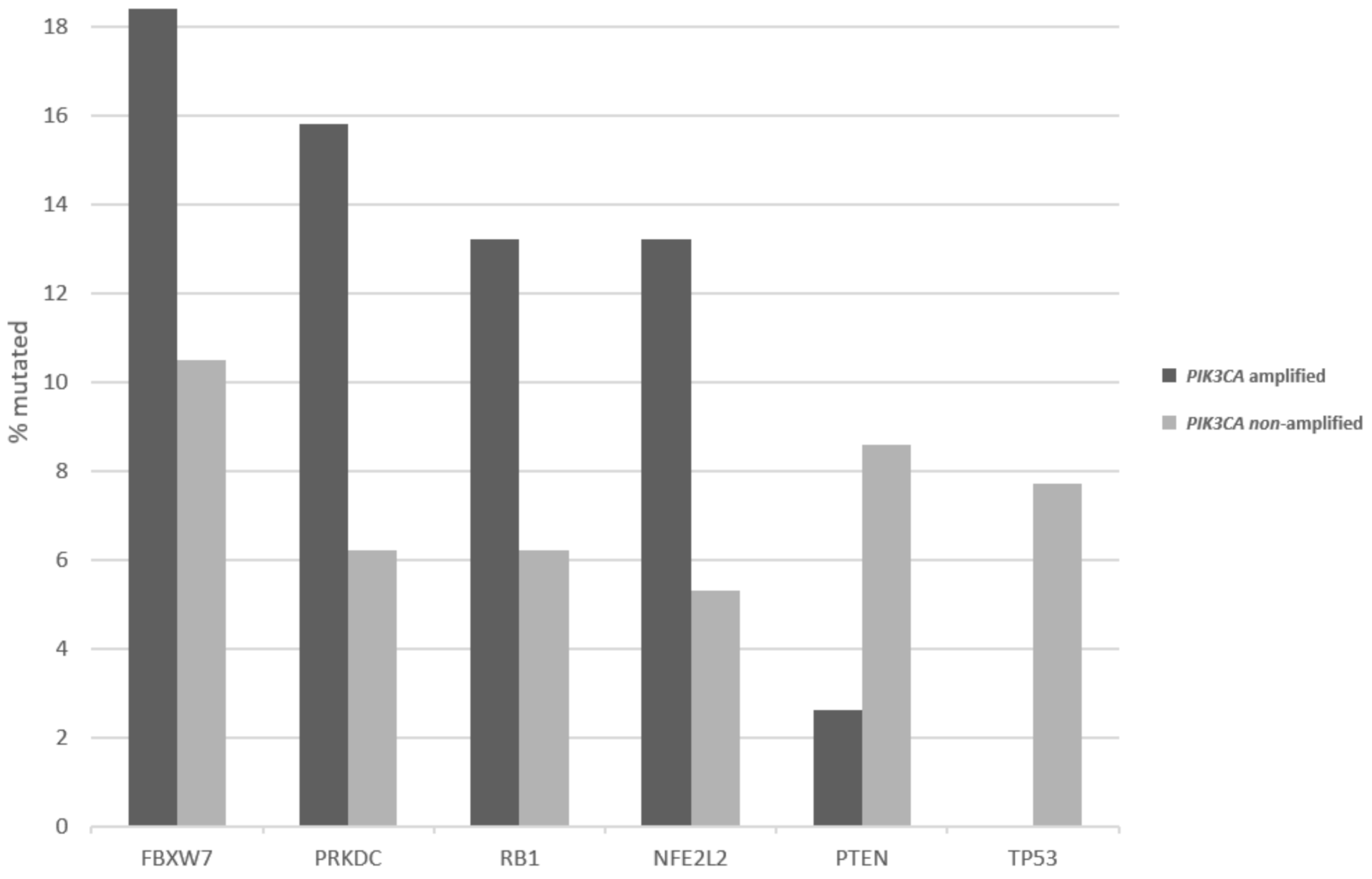
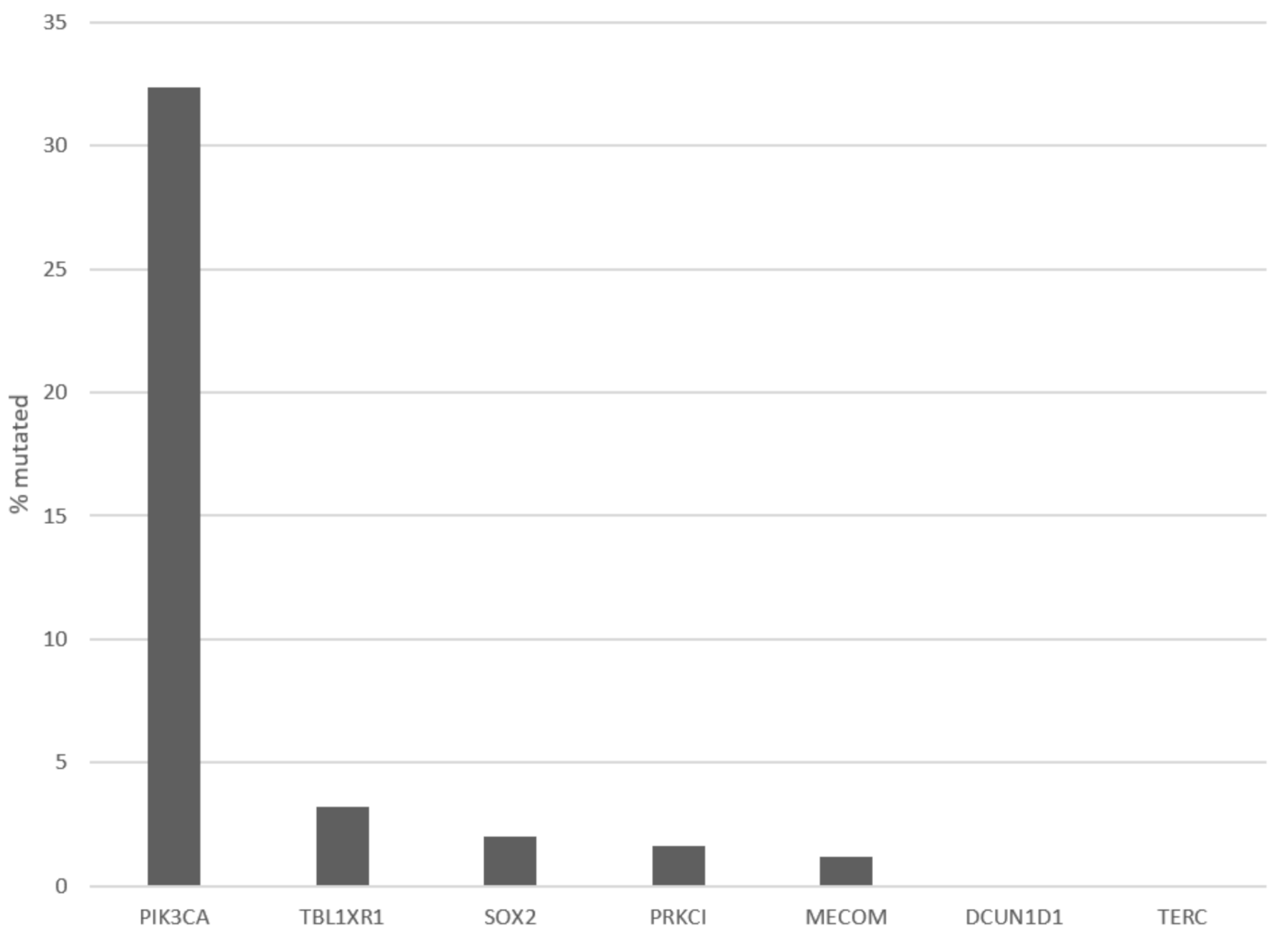
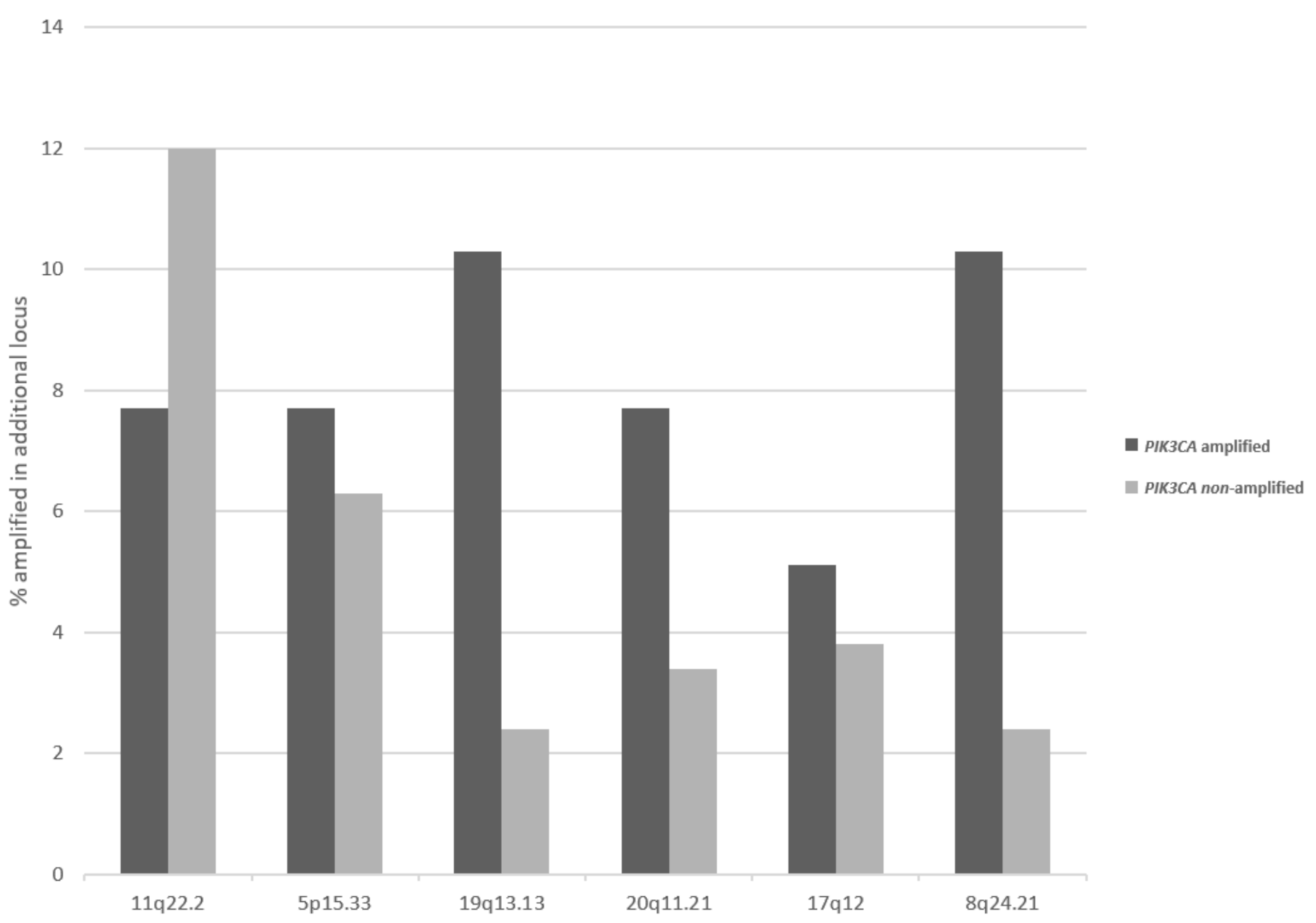
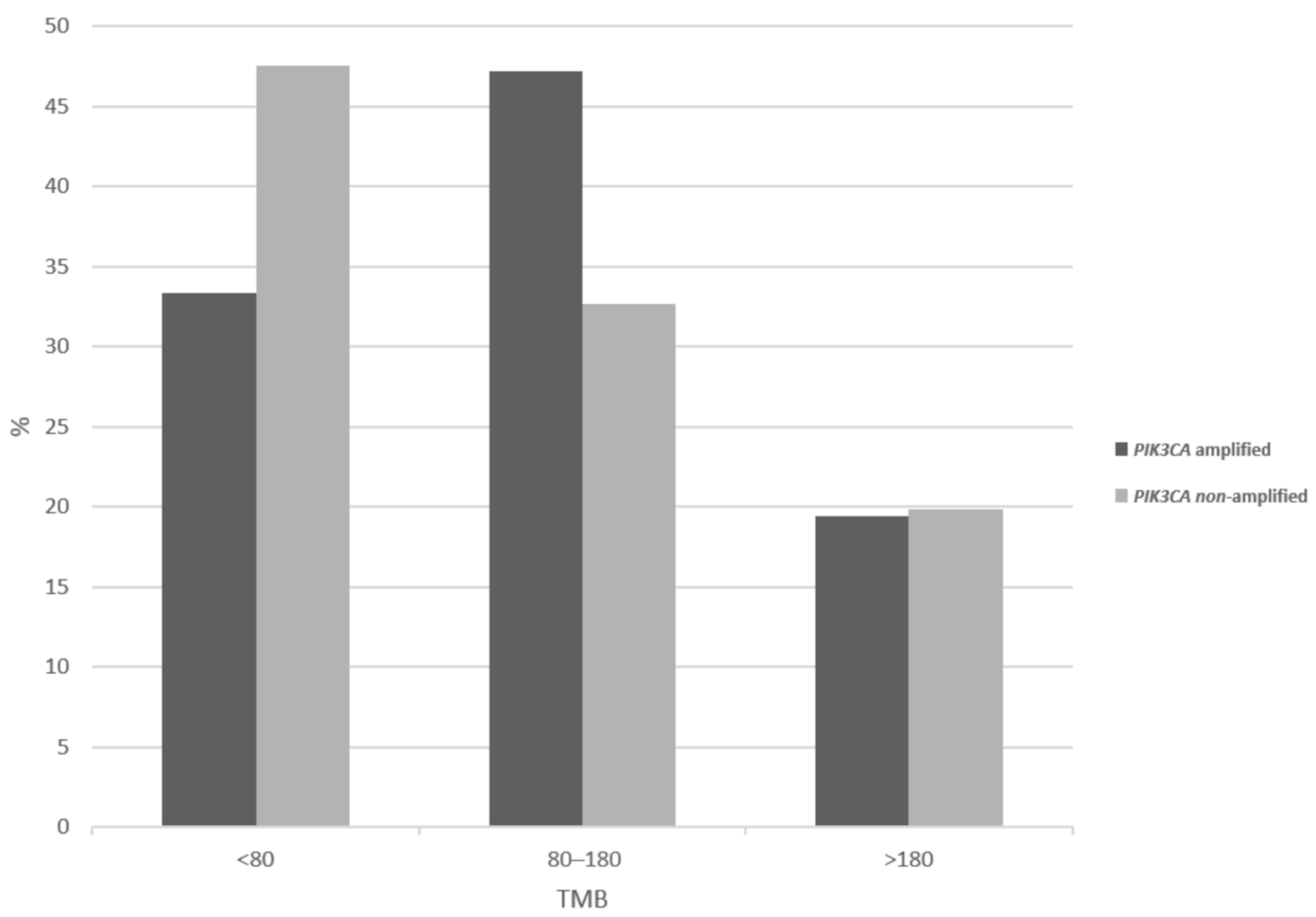
| Locus | Gene | Overall Cases (%) | In PIK3CA Amplified (%) | In MECOM Amplified (%) | In TP63 Amplified (%) |
|---|---|---|---|---|---|
| 3q29 | TFRC | 38 (15.4) | 34 (87.2) | 34 (81) | 36 (81.8) |
| 3q28–29 | FGF12 | 37 (15) | 34 (87.2) | 34 (81) | 36 (81.8) |
| 3q28 | TP63 | 44 (17.8) | 35 (89.7) | 34 (81) | 44 (100) |
| 3q27–28 | LPP | 38 (15.4) | 36 (92.3) | 35 (83.3) | 37 (84.1) |
| 3q27.3 | EIF4A2 | 37 (15) | 36 (92.3) | 35 (83.3) | 36 (81.8) |
| BCL6 | 36 (14.6) | 35 (89.7) | 34 (81) | 36 (81.8) | |
| 3q27.2 | MAP3K13 | 36 (14.6) | 36 (92.3) | 35 (83.3) | 35 (79.5) |
| ETV5 | 37 (15) | 37 (94.9) | 36 (85.7) | 35 (79.5) | |
| 3q27.1 | KLHL6 | 37 (15) | 37 (94.9) | 35 (83.3) | 35 (79.5) |
| 3q26.33 | SOX2 | 38 (15.4) | 38 (97.4) | 36 (85.7) | 35 (79.5) |
| DCUN1D1 | 38 (15.4) | 38 (97.4) | 36 (85.7) | 35 (79.5) | |
| 3q26.32 | PIK3CA | 39 (15.8) | 39 (100) | 37 (88.1) | 35 (79.5) |
| TBL1XR1 | 40 (16.2) | 39 (100) | 38 (90.5) | 35 (79.5) | |
| 3q26.2 | TERC | 40 (16.2) | 37 (94.9) | 40 (95.2) | 34 (77.3) |
| PRKCI | 41 (16.6) | 37 (94.9) | 40 (95.2) | 34 (77.3) | |
| MECOM | 42 (17) | 37 (94.9) | 42 (100) | 34 (77.3) |
| Locus | Gene | Cervical Squamous (n = 247) | Cervical Adenocarcinomas (n = 46) | Head and Neck Cancer (n = 517) | Lung Squamous Carcinomas (n = 487) | Lung Adenocarcinomas (n = 511) | Esophageal Squamous Carcinomas (n = 95) | GE Junction Adenocarcinomas (n = 87) |
|---|---|---|---|---|---|---|---|---|
| 3q29 | TFRC | 15.4% | 8.7% | 13% | 30% | 2% | 28.4% | 8% |
| 3q28–29 | FGF12 | 15% | 8.7% | 13.2% | 30% | 2% | 27.4% | 5.7% |
| 3q28 | TP63 | 17.8% | 8.7% | 16.1% | 31.6% | 2% | 33.7% | 5.7% |
| 3q27–28 | LPP | 15.4% | 8.7% | 14.5% | 31.8% | 1.8% | 28.4% | 9.2% |
| 3q27.3 | EIF4A2 | 15% | 8.7% | 13.9% | 31.8% | 1.8% | 25.3% | 4.6% |
| BCL6 | 14.6% | 8.7% | 13.9% | 31% | 1.6% | 25.3% | 5.7% | |
| 3q27.2 | MAP3K13 | 14.6% | 8.7% | 14.3% | 35.5% | 2% | 24.2% | 4.6% |
| ETV5 | 15% | 8.7% | 14.1% | 33.5% | 1.8% | 25.3% | 4.6% | |
| 3q27.1 | KLHL6 | 15% | 8.7% | 15.1% | 39% | 2% | 25.3% | 4.6% |
| 3q26.33 | SOX2 | 15.4% | 8.7% | 15.7% | 39.8% | 2% | 28.4% | 3.4% |
| DCUN1D1 | 15.4% | 8.7% | 15.5% | 40% | 2% | 26.3% | 3.4% | |
| 3q26.32 | PIK3CA | 15.8% | 10.9% | 15.7% | 37.8% | 1.8% | 29.5% | 4.6% |
| TBL1XR1 | 16.2% | 8.7% | 15.1% | 36.8% | 2% | 30.5% | 6.9% | |
| 3q26.2 | TERC | 16.2% | 10.9% | 13% | 35.5% | 2.7% | 29.5% | 10.3% |
| PRKCI | 16.6% | 10.9% | 13.2% | 35.9% | 2.7% | 28.4% | 10.3% | |
| MECOM | 17% | 10.9% | 13.2% | 35.7% | 2.7% | 28.4% | 10.3% |
| Gene | Samples with z > 2 Whole Series (%) | Samples with z > 2 in PIK3CA Amplified (%) |
|---|---|---|
| SOX2 | 41 (16.5%) | 16 (41%) |
| DCUN1D1 | 106 (42.7%) | 25 (64.1%) |
| PIK3CA | 98 (39.5%) | 25 (64.1%) |
| TBL1XR1 | 104 (41.9%) | 31 (79.5%) |
| TERC | 7 (2.8%) | 0 |
| PRKCI | 76 (30.6%) | 21 (53.8%) |
| MECOM | 4 (1.6%) | 0 |
| Gene | Alternative Name-Function | Dataset | T Statistic | p-Value |
|---|---|---|---|---|
| ZER1 | Ubquitin ligase involved in meiosis | CRISPR (combined) * | −11.8 | 4 × 10−30 |
| UBE3A | E6-AP | CRISPR (combined) * | −11.1 | 3.36 × 10−27 |
| FBXL5 | F-box component of SCF ligases | CRISPR (combined) * | −5.06 | 4.97 × 10−7 |
| GEN1 | Holliday junction 5′ endonuclease | CRISPR (combined) * | −4.5 | 7.54 × 10−6 |
| RNF145 | Ring finger ligase | CRISPR (combined) * | 4.27 | 2.1 × 10−5 |
| RAB39B | Rab family GTPase vesicle trafficking | CRISPR (combined) | −4.09 | 4.75 × 10−5 |
| SKP2 | FBXL1, F-box component of SCF ligases | CRISPR (combined) * | −3.94 | 8.76 × 10−5 |
| CUL2 | Cullin component of SCF ligases, E7 interacting | CRISPR (combined) | −3.86 | 0.000122 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voutsadakis, I.A. 3q26 Amplifications in Cervical Squamous Carcinomas. Curr. Oncol. 2021, 28, 2868-2880. https://doi.org/10.3390/curroncol28040251
Voutsadakis IA. 3q26 Amplifications in Cervical Squamous Carcinomas. Current Oncology. 2021; 28(4):2868-2880. https://doi.org/10.3390/curroncol28040251
Chicago/Turabian StyleVoutsadakis, Ioannis A. 2021. "3q26 Amplifications in Cervical Squamous Carcinomas" Current Oncology 28, no. 4: 2868-2880. https://doi.org/10.3390/curroncol28040251
APA StyleVoutsadakis, I. A. (2021). 3q26 Amplifications in Cervical Squamous Carcinomas. Current Oncology, 28(4), 2868-2880. https://doi.org/10.3390/curroncol28040251





