Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease—Type 2 Diabetes Mellitus Comorbidity Models in Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Animals
2.3. Neuronal Cultures
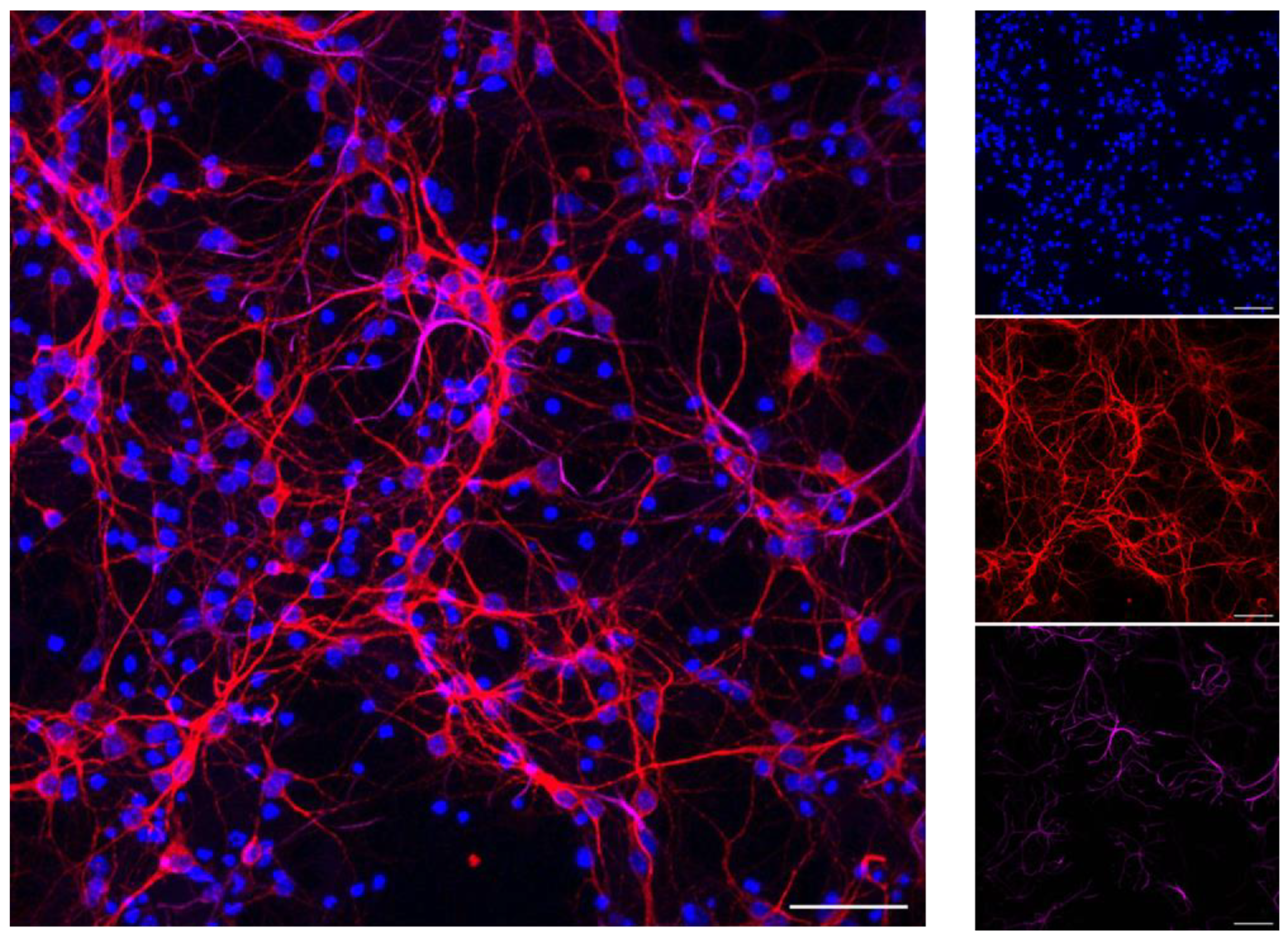
2.4. Primary Cortical Neurons
2.5. Insulin Resistance
2.6. Quantitative Real-Time PCR
2.7. Western Blot
2.8. Immunofluorescence
2.9. Statistical Analysis
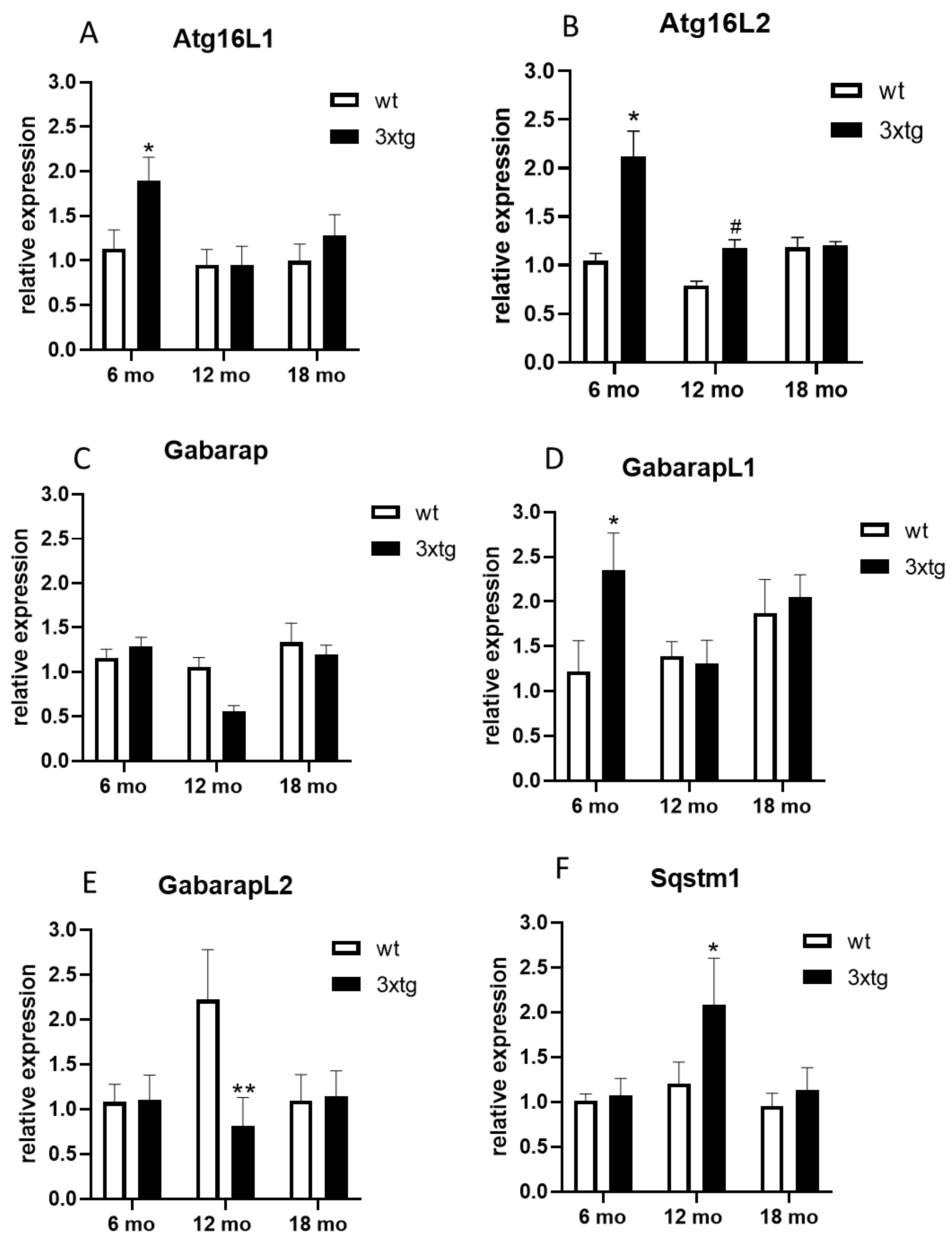
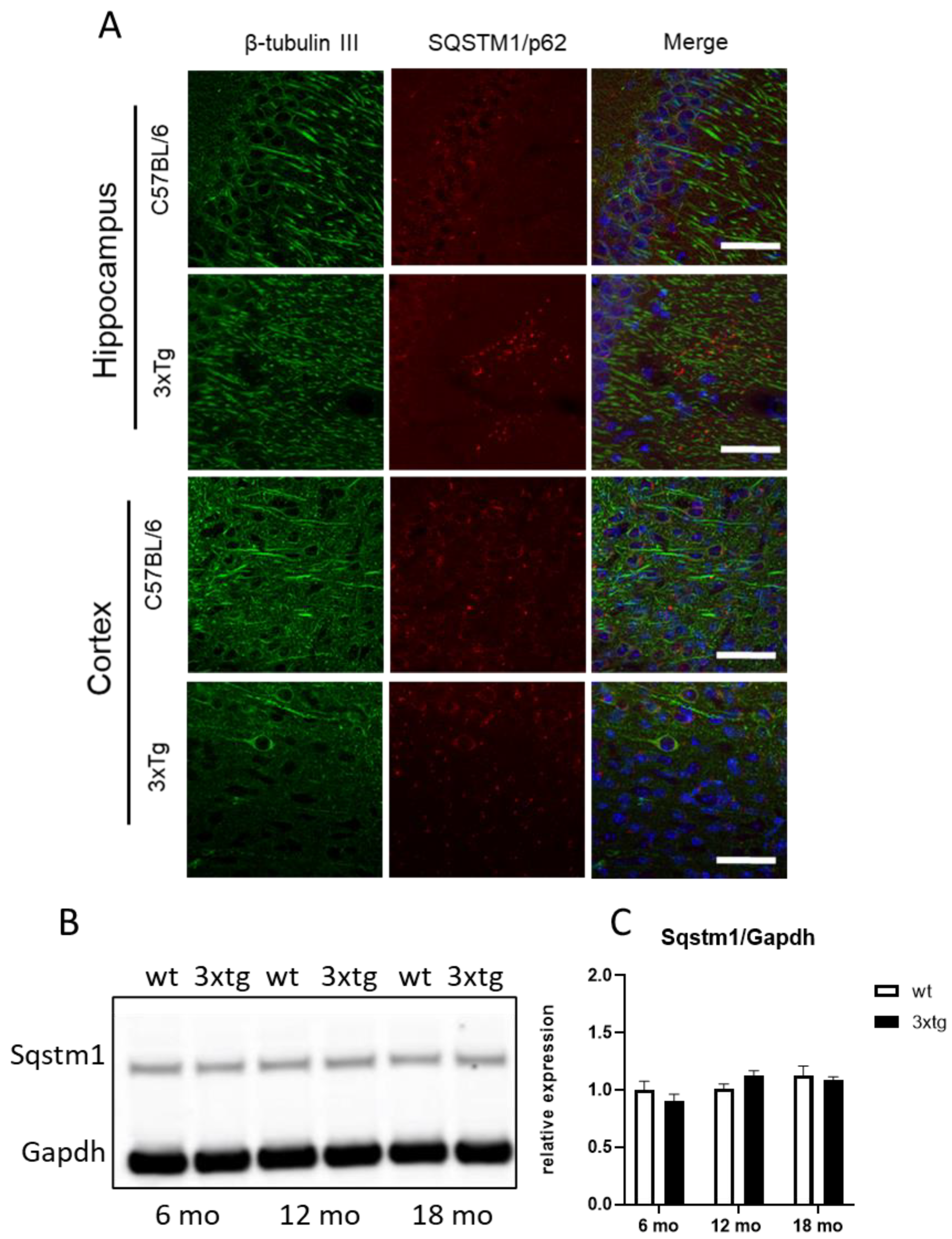
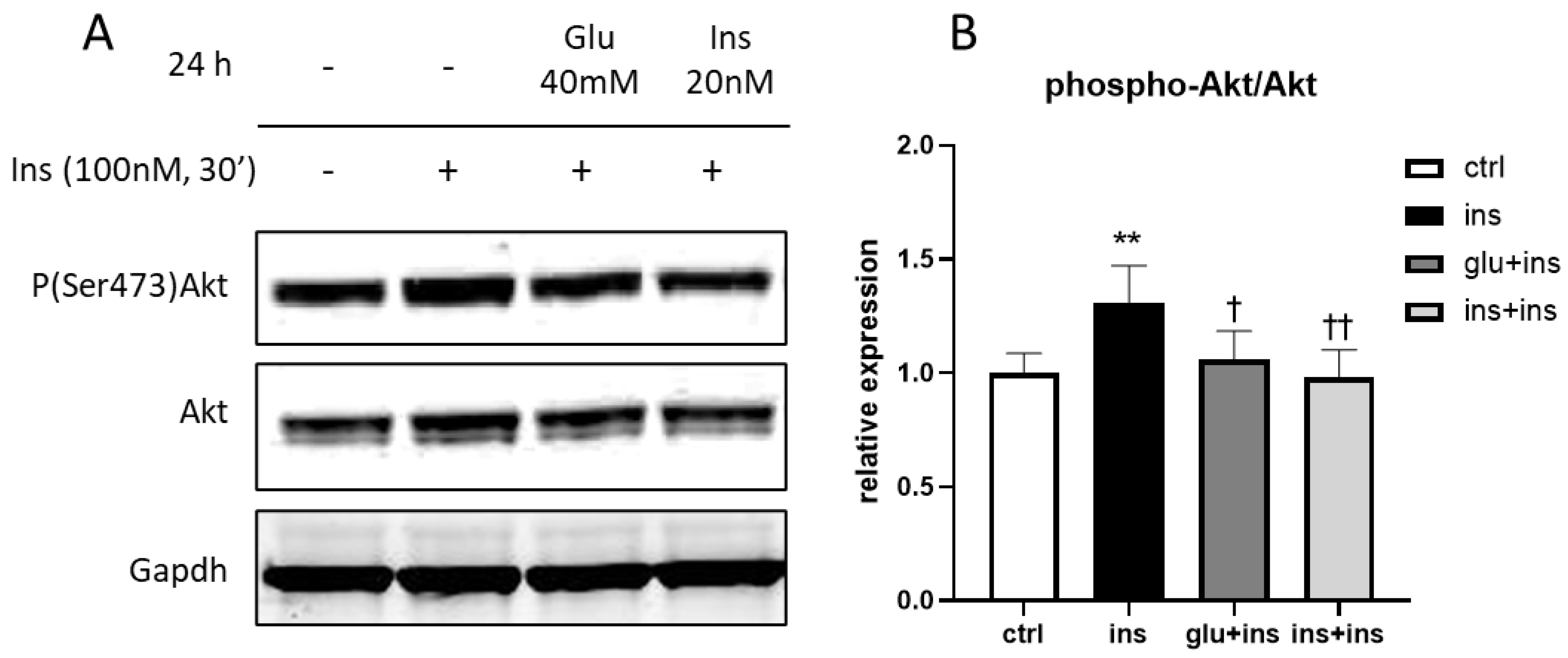
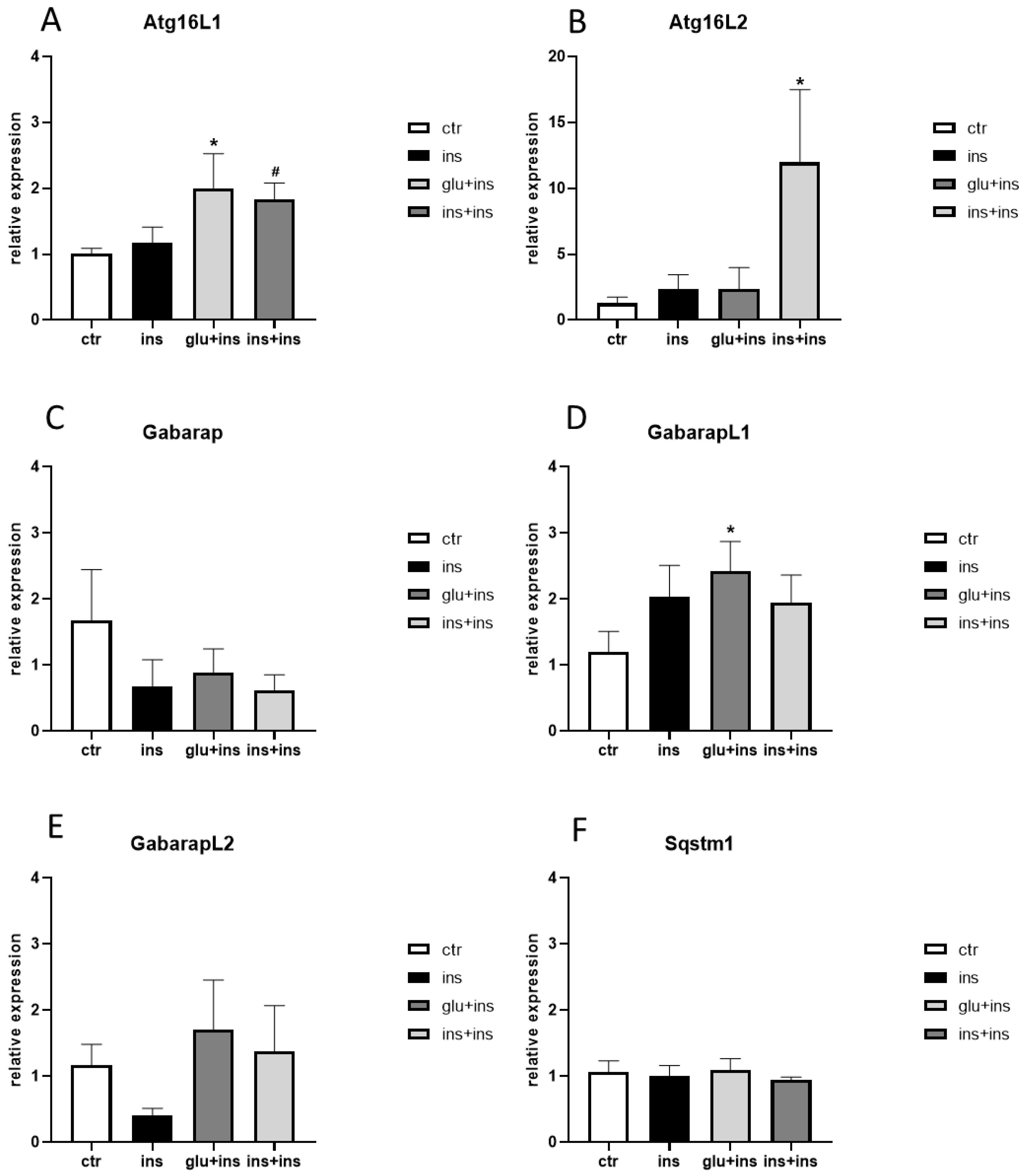
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Livingston, G.; Huntley, J.; Sommerlad, A.; Ames, D.; Ballard, C.; Banerjee, S.; Brayne, C.; Burns, A.; Cohen-Mansfield, J.; Cooper, C.; et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet 2020, 396, 413–446. [Google Scholar] [CrossRef] [PubMed]
- Dementia: Assessment, Management and Support for People Living with Dementia and Their Carers; National Institute for Health and Care Excellence (NICE): London, UK, 2018.
- van Dyck, C.H.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; Kanekiyo, M.; Li, D.; Reyderman, L.; Cohen, S.; et al. Lecanemab in Early Alzheimer’s Disease. N. Engl. J. Med. 2022, 388, 9–21. [Google Scholar] [CrossRef]
- Dunn, B.; Stein, P.; Cavazzoni, P. Approval of Aducanumab for Alzheimer Disease-The FDA’s Perspective. JAMA Intern. Med. 2021, 181, 1276–1278. [Google Scholar] [CrossRef]
- Alexander, G.C.; Knopman, D.S.; Emerson, S.S.; Ovbiagele, B.; Kryscio, R.J.; Perlmutter, J.S.; Kesselheim, A.S. Revisiting FDA Approval of Aducanumab. N. Engl. J. Med. 2021, 385, 769–771. [Google Scholar] [CrossRef]
- The Lancet Lecanemab for Alzheimer’s disease: Tempering hype and hope. Lancet 2022, 400, 1899. [CrossRef]
- Ross, E.L.; Weinberg, M.S.; Arnold, S.E. Cost-effectiveness of Aducanumab and Donanemab for Early Alzheimer Disease in the US. JAMA Neurol. 2022, 79, 478–487. [Google Scholar] [CrossRef]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Prim. 2021, 7, 33. [Google Scholar] [CrossRef]
- Chatterjee, S.; Peters, S.A.E.; Woodward, M.; Arango, S.M.; Batty, G.D.; Beckett, N.; Beiser, A.; Borenstein, A.R.; Crane, P.K.; Haan, M.; et al. Type 2diabetes as a risk factor for dementia in women compared with men: A pooled analysis of 2.3 million people comprising more than 100,000 cases of dementia. Diabetes Care 2016, 39, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Gudala, K.; Bansal, D.; Schifano, F.; Bhansali, A. Diabetes mellitus and risk of dementia: A meta-analysis of prospective observational studies. J. Diabetes Investig. 2013, 4, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Savelieff, M.G.; Chen, K.S.; Elzinga, S.E.; Feldman, E.L. Diabetes and dementia: Clinical perspective, innovation, knowledge gaps. J. Diabetes Complicat. 2022, 36, 108333. [Google Scholar] [CrossRef]
- Albanese, E.; Launer, L.J.; Egger, M.; Prince, M.J.; Giannakopoulos, P.; Wolters, F.J.; Egan, K. Body mass index in midlife and dementia: Systematic review and meta-regression analysis of 589,649 men and women followed in longitudinal studies. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2017, 8, 165–178. [Google Scholar] [CrossRef]
- Campos-Peña, V.; Toral-Rios, D.; Becerril-Pérez, F.; Sánchez-Torres, C.; Delgado-Namorado, Y.; Torres-Ossorio, E.; Franco-Bocanegra, D.; Carvajal, K. Metabolic Syndrome as a Risk Factor for Alzheimer’s Disease: Is Aβ a Crucial Factor in Both Pathologies? Antioxid. Redox Signal. 2017, 26, 542–560. [Google Scholar] [CrossRef] [PubMed]
- Nowell, J.; Blunt, E.; Edison, P. Incretin and insulin signaling as novel therapeutic targets for Alzheimer’s and Parkinson’s disease. Mol. Psychiatry 2022, 28, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Areosa Sastre, A.; Vernooij, R.W.; González-Colaço Harmand, M.; Martínez, G. Effect of the treatment of Type 2 diabetes mellitus on the development of cognitive impairment and dementia. Cochrane Database Syst. Rev. 2017, 6, CD003804. [Google Scholar] [CrossRef]
- Patel, V.N.; Chorawala, M.R.; Shah, M.B.; Shah, K.C.; Dave, B.P.; Shah, M.P.; Patel, T.M. Emerging Pathophysiological Mechanisms Linking Diabetes Mellitus and Alzheimer’s Disease: An Old Wine in a New Bottle. J. Alzheimer’s Dis. Rep. 2022, 6, 349–357. [Google Scholar] [CrossRef]
- Hamzé, R.; Delangre, E.; Tolu, S.; Moreau, M.; Janel, N.; Bailbé, D.; Movassat, J. Type 2 Diabetes Mellitus and Alzheimer’s Disease: Shared Molecular Mechanisms and Potential Common Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 15287. [Google Scholar] [CrossRef] [PubMed]
- Pugazhenthi, S.; Qin, L.; Reddy, P.H. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer’s disease. Biochim. Biophys. Acta—Mol. Basis Dis. 2017, 1863, 1037–1045. [Google Scholar] [CrossRef]
- González, A.; Calfío, C.; Churruca, M.; Maccioni, R.B. Glucose metabolism and AD: Evidence for a potential diabetes type 3. Alzheimer’s Res. Ther. 2022, 14, 56. [Google Scholar] [CrossRef]
- Frisardi, V.; Solfrizzi, V.; Seripa, D.; Capurso, C.; Santamato, A.; Sancarlo, D.; Vendemiale, G.; Pilotto, A.; Panza, F. Metabolic-cognitive syndrome: A cross-talk between metabolic syndrome and Alzheimer’s disease. Ageing Res. Rev. 2010, 9, 399–417. [Google Scholar] [CrossRef]
- Steen, E.; Terry, B.M.; Rivera, E.J.; Cannon, J.L.; Neely, T.R.; Tavares, R.; Xu, X.J.; Wands, J.R.; de la Monte, S.M. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease--is this type 3 diabetes? J. Alzheimers. Dis. 2005, 7, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Craft, S. Insulin resistance and Alzheimer’s disease pathogenesis: Potential mechanisms and implications for treatment. Curr. Alzheimer Res. 2007, 4, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Moreira, P.I. Sweet Mitochondria: A Shortcut to Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 62, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.C.; Santos, R.X.; Carvalho, C.; Cardoso, S.; Candeias, E.; Santos, M.S.; Oliveira, C.R.; Moreira, P.I. Insulin signaling, glucose metabolism and mitochondria: Major players in Alzheimer’s disease and diabetes interrelation. Brain Res. 2012, 1441, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Caberlotto, L.; Nguyen, T.-P.; Lauria, M.; Priami, C.; Rimondini, R.; Maioli, S.; Cedazo-Minguez, A.; Sita, G.; Morroni, F.; Corsi, M.; et al. Cross-disease analysis of Alzheimer’s disease and type-2 Diabetes highlights the role of autophagy in the pathophysiology of two highly comorbid diseases. Sci. Rep. 2019, 9, 3965. [Google Scholar] [CrossRef]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef]
- Boland, B.; Yu, W.H.; Corti, O.; Mollereau, B.; Henriques, A.; Bezard, E.; Pastores, G.M.; Rubinsztein, D.C.; Nixon, R.A.; Duchen, M.R.; et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018, 17, 660–688. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Caricasole, A.; Bento, C.F.; Andrews, S.P.; Ashkenazi, A.; Füllgrabe, J.; Jackson, A.; Jimenez Sanchez, M.; Karabiyik, C.; et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017, 93, 1015–1034. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Kitada, M.; Koya, D. Autophagy in metabolic disease and ageing. Nat. Rev. Endocrinol. 2021, 17, 647–661. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; Emerson, M.; et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020, 18, e3000411. [Google Scholar] [CrossRef] [PubMed]
- Capaldi, S.; Suku, E.; Antolini, M.; Di Giacobbe, M.; Giorgetti, A.; Buffelli, M. Allosteric sodium binding cavity in GPR3: A novel player in modulation of Aβ production. Sci. Rep. 2018, 8, 11102. [Google Scholar] [CrossRef]
- Beaudoin, G.M.J.; Lee, S.-H.; Singh, D.; Yuan, Y.; Ng, Y.-G.; Reichardt, L.F.; Arikkath, J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 2012, 7, 1741–1754. [Google Scholar] [CrossRef]
- Bolognin, S.; Zatta, P.; Lorenzetto, E.; Valenti, M.T.; Buffelli, M. β-Amyloid-aluminum complex alters cytoskeletal stability and increases ROS production in cortical neurons. Neurochem. Int. 2013, 62, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, J.J.; Shi, L.; Tang, Y.; Gao, D.; Long, J.; Liu, J.J. Mitochondrial dysfunction precedes depression of AMPK/AKT signaling in insulin resistance induced by high glucose in primary cortical neurons. J. Neurochem. 2016, 137, 701–713. [Google Scholar] [CrossRef]
- Carboni, L.; Pischedda, F.; Piccoli, G.; Lauria, M.; Musazzi, L.; Popoli, M.; Mathé, A.A.; Domenici, E. Depression-Associated Gene Negr1-Fgfr2 Pathway Is Altered by Antidepressant Treatment. Cells 2020, 9, 1818. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Carboni, L.; Ponzoni, L.; Braida, D.; Sala, M.; Gotti, C.; Zoli, M. Altered mrna levels of stress-related peptides in mouse hippocampus and caudate-putamen in withdrawal after long-term intermittent exposure to tobacco smoke or electronic cigarette vapour. Int. J. Mol. Sci. 2021, 22, 599. [Google Scholar] [CrossRef]
- Borin, M.; Saraceno, C.; Catania, M.; Lorenzetto, E.; Pontelli, V.; Paterlini, A.; Fostinelli, S.; Avesani, A.; Di Fede, G.; Zanusso, G.; et al. Rac1 activation links tau hyperphosphorylation and Aβ dysmetabolism in Alzheimer’s disease. Acta Neuropathol. Commun. 2018, 6, 61. [Google Scholar] [CrossRef]
- Laperchia, C.; Allegra Mascaro, A.L.; Sacconi, L.; Andrioli, A.; Mattè, A.; De Franceschi, L.; Grassi-Zucconi, G.; Bentivoglio, M.; Buffelli, M.; Pavone, F.S. Two-photon microscopy imaging of thy1GFP-M transgenic mice: A novel animal model to investigate brain dendritic cell subsets in vivo. PLoS ONE 2013, 8, e56144. [Google Scholar] [CrossRef]
- Bate, S.T.; Clark, R.A. The Design and Statistical Analysis of Animal Experiments; Cambridge University Press: Cambridge, UK, 2014; ISBN 978-1-107-69094-3. [Google Scholar]
- Clark, R.A.; Shoaib, M.; Hewitt, K.N.; Stanford, S.C.; Bate, S.T. A comparison of InVivoStat with other statistical software packages for analysis of data generated from animal experiments. J. Psychopharmacol. 2012, 26, 1136–1142. [Google Scholar] [CrossRef]
- Karki, R.; Kodamullil, A.T.; Hofmann-Apitius, M. Comorbidity Analysis between Alzheimer’s Disease and Type 2 Diabetes Mellitus (T2DM) Based on Shared Pathways and the Role of T2DM Drugs. J. Alzheimers. Dis. 2017, 60, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-transgenic model of Alzheimer’s Disease with plaques and tangles: Intracellular Aβ and synaptic dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Kitazawa, M.; Tseng, B.P.; LaFerla, F.M. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiol. Aging 2003, 24, 1063–1070. [Google Scholar] [CrossRef]
- Billings, L.M.; Oddo, S.; Green, K.N.; McGaugh, J.L.; LaFerla, F.M. Intraneuronal Aβ causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron 2005, 45, 675–688. [Google Scholar] [CrossRef]
- Belfiore, R.; Rodin, A.; Ferreira, E.; Velazquez, R.; Branca, C.; Caccamo, A.; Oddo, S. Temporal and regional progression of Alzheimer’s disease-like pathology in 3xTg-AD mice. Aging Cell 2019, 18, e12873. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Yu, S.-B.; Yu, U.Y.; Jo, S.A.; Ahn, J.-H. Swedish mutation within amyloid precursor protein modulates global gene expression towards the pathogenesis of Alzheimer’s disease. BMB Rep. 2010, 43, 704–709. [Google Scholar] [CrossRef]
- Branca, C.; Sarnico, I.; Ruotolo, R.; Lanzillotta, A.; Viscomi, A.R.; Benarese, M.; Porrini, V.; Lorenzini, L.; Calzà, L.; Imbimbo, B.P.; et al. Pharmacological targeting of the β-amyloid precursor protein intracellular domain. Sci. Rep. 2014, 4, 4618. [Google Scholar] [CrossRef]
- Haugabook, S.J.; Yager, D.M.; Eckman, E.A.; Golde, T.E.; Younkin, S.G.; Eckman, C.B. High throughput screens for the identification of compounds that alter the accumulation of the Alzheimer’s amyloid beta peptide (Abeta). J. Neurosci. Methods 2001, 108, 171–179. [Google Scholar] [CrossRef]
- Kim, B.; McLean, L.L.; Philip, S.S.; Feldman, E.L. Hyperinsulinemia induces insulin resistance in dorsal root ganglion neurons. Endocrinology 2011, 152, 3638–3647. [Google Scholar] [CrossRef] [PubMed]
- Son, S.M.; Song, H.; Byun, J.; Park, K.S.; Jang, H.C.; Park, Y.J.; Mook-Jung, I. Altered APP processing in insulin-resistant conditions is mediated by autophagosome accumulation via the inhibition of mammalian target of rapamycin pathway. Diabetes 2012, 61, 3126–3138. [Google Scholar] [CrossRef] [PubMed]
- Magri, C.; Vitali, E.; Cocco, S.; Giacopuzzi, E.; Rinaudo, M.; Martini, P.; Barbon, A.; Grassi, C.; Gennarelli, M. Whole Blood Transcriptome Characterization of 3xTg-AD Mouse and Its Modulation by Transcranial Direct Current Stimulation (tDCS). Int. J. Mol. Sci. 2021, 22, 7629. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Zhou, F.; Cao, X.; Yang, J.; Bai, Y.; Yan, X.; Cao, J.; Qi, J. Abnormal circadian locomotor rhythms and Per gene expression in six-month-old triple transgenic mice model of Alzheimer’s disease. Neurosci. Lett. 2018, 676, 13–18. [Google Scholar] [CrossRef]
- Piedrahita, D.; Hernández, I.; López-Tobón, A.; Fedorov, D.; Obara, B.; Manjunath, B.S.; Boudreau, R.L.; Davidson, B.; Laferla, F.; Gallego-Gómez, J.C.; et al. Silencing of CDK5 reduces neurofibrillary tangles in transgenic alzheimer’s mice. J. Neurosci. 2010, 30, 13966–13976. [Google Scholar] [CrossRef] [PubMed]
- Melkonyan, M.M.; Hunanyan, L.; Lourhmati, A.; Layer, N.; Beer-Hammer, S.; Yenkoyan, K.; Schwab, M.; Danielyan, L. Neuroprotective, Neurogenic, and Amyloid Beta Reducing Effect of a Novel Alpha 2-Adrenoblocker, Mesedin, on Astroglia and Neuronal Progenitors upon Hypoxia and Glutamate Exposure. Int. J. Mol. Sci. 2017, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Macauley, S.L.; Stanley, M.; Caesar, E.E.; Yamada, S.A.; Raichle, M.E.; Perez, R.; Mahan, T.E.; Sutphen, C.L.; Holtzman, D.M. Hyperglycemia modulates extracellular amyloid-β concentrations and neuronal activity in vivo. J. Clin. Investig. 2015, 125, 2463–2467. [Google Scholar] [CrossRef]
- Mizushima, N.; Kuma, A.; Kobayashi, Y.; Yamamoto, A.; Matsubae, M.; Takao, T.; Natsume, T.; Ohsumi, Y.; Yoshimori, T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003, 116, 1679–1688. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Mercer, T.J.; Gubas, A.; Tooze, S.A. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 2018, 293, 5386–5395. [Google Scholar] [CrossRef]
- Kaufmann, A.; Beier, V.; Franquelim, H.G.; Wollert, T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell 2014, 156, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Mizushima, N.; Ishihara, N.; Ohsumi, Y. Formation of the ∼350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002, 277, 18619–18625. [Google Scholar] [CrossRef]
- Gammoh, N. The multifaceted functions of ATG16L1 in autophagy and related processes. J. Cell Sci. 2020, 133, jcs249227. [Google Scholar] [CrossRef]
- Hamaoui, D.; Subtil, A. ATG16L1 functions in cell homeostasis beyond autophagy. FEBS J. 2022, 289, 1779–1800. [Google Scholar] [CrossRef]
- Heckmann, B.L.; Teubner, B.J.W.; Boada-Romero, E.; Tummers, B.; Guy, C.; Fitzgerald, P.; Mayer, U.; Carding, S.; Zakharenko, S.S.; Wileman, T.; et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Sci. Adv. 2020, 6, eabb9036. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Fujita, N.; Kanno, E.; Omori, H.; Yoshimori, T.; Itoh, T.; Fukuda, M. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy 2011, 7, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wu, X.; Holzer, R.G.; Lee, J.H.; Todoric, J.; Park, E.J.; Ogata, H.; Gukovskaya, A.S.; Gukovsky, I.; Pizzo, D.P.; et al. Loss of acinar cell IKKα triggers spontaneous pancreatitis in mice. J. Clin. Investig. 2013, 123, 2231–2243. [Google Scholar] [CrossRef]
- Khor, B.; Conway, K.L.; Omar, A.S.; Biton, M.; Haber, A.L.; Rogel, N.; Baxt, L.A.; Begun, J.; Kuballa, P.; Gagnon, J.D.; et al. Distinct Tissue-Specific Roles for the Disease-Associated Autophagy Genes ATG16L2 and ATG16L1. J. Immunol. 2019, 203, 1820–1829. [Google Scholar] [CrossRef]
- Don Wai Luu, L.; Kaakoush, N.O.; Castaño-Rodríguez, N. The role of ATG16L2 in autophagy and disease. Autophagy 2022, 18, 2537–2546. [Google Scholar] [CrossRef]
- Jacquet, M.; Guittaut, M.; Fraichard, A.; Despouy, G. The functions of Atg8-family proteins in autophagy and cancer: Linked or unrelated? Autophagy 2021, 17, 599–611. [Google Scholar] [CrossRef]
- Koutsifeli, P.; Varma, U.; Daniels, L.J.; Annandale, M.; Li, X.; Neale, J.P.H.; Hayes, S.; Weeks, K.L.; James, S.; Delbridge, L.M.D.; et al. Glycogen-autophagy: Molecular machinery and cellular mechanisms of glycophagy. J. Biol. Chem. 2022, 298, 102093. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.; Santos, M.S.; Oliveira, C.R.; Moreira, P.I. Alzheimer’s disease and type 2 diabetes-related alterations in brain mitochondria, autophagy and synaptic markers. Biochim. Biophys. Acta 2015, 1852, 1665–1675. [Google Scholar] [CrossRef] [PubMed]
- Vaillant-Beuchot, L.; Mary, A.; Pardossi-Piquard, R.; Bourgeois, A.; Lauritzen, I.; Eysert, F.; Kinoshita, P.F.; Cazareth, J.; Badot, C.; Fragaki, K.; et al. Accumulation of amyloid precursor protein C-terminal fragments triggers mitochondrial structure, function, and mitophagy defects in Alzheimer’s disease models and human brains. Acta Neuropathol. 2021, 141, 39–65. [Google Scholar] [CrossRef]
- Du, Y.; Wooten, M.C.; Gearing, M.; Wooten, M.W. Age-associated oxidative damage to the p62 promoter: Implications for Alzheimer disease. Free Radic. Biol. Med. 2009, 46, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Broderick, T.L.; Rasool, S.; Li, R.; Zhang, Y.; Anderson, M.; Al-Nakkash, L.; Plochocki, J.H.; Geetha, T.; Babu, J.R. Neuroprotective Effects of Chronic Resveratrol Treatment and Exercise Training in the 3xTg-AD Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 7337. [Google Scholar] [CrossRef]

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Mouse Atg16L1 | GGTCTGTCCGAATCTCCCCT | GCGCATCGAAGACATACGAG |
| Mouse Atg16L2 | CTGTGTGGATGTGGTGAAGG | AGCATTGACCTCAGAGAGAT |
| Mouse Gabarap | TTCTCCCCTTGTTTACCCTCCAT | TCCAATGTCAATCCCTTCCAC |
| Mouse GabarapL1 | CATCGTGGAGAAGGCTCCTA | ATACAGCTGGCCCATGGTAG |
| Mouse GabarapL2 | GCTATTTGCTCCAGGGAACCT | AACTATGCATCAGCCCCTCC |
| Mouse Sqstm1 | GCCAGAGGAACAGATGGAGT | TCCGATTCTG GCATCTGTAG |
| Mouse Gapdh | GCCAAGGTCATCCATGACAACT | GAGGGGCCATCCACAGTCT |
| Mouse Ywhaz | TAGGTCATCGTGGAGGGTCG | GAAGCATTGGGGATCAAGAACTT |
| Human ATG16L1 | ACACAAGAAACGTGGGGAGT | CTCCGTCTCCAGGTCAGAGA |
| Human ATG16L2 | TTCGGGACCGTACGCAAAAG | TCAAGCTCTGACTCCTCCCA |
| Human GABARAP | CTCCCTTATTCAGGACCGGC | TGCCAACTCCACCATTACCC |
| Human GABARAPL1 | TACCTAGTGCCCTCTGACCT | TGGACGGCATCCAGTATTGT |
| Human GABARAPL2 | GCGAAATATCCCGACAGGGT | TCCACATGAACTGAGCCACA |
| Human SQSTM1 | GAGATTCGCCGCTTCAGCTT | GGAAAAGGCAACCAAGTCCC |
| Human GAPDH | CCATGGGGAAGGTGAAGGTC | TGGAATTTGCCATGGGTGGA |
| Human YWHAZ | GGACTACGACGTCCCTCAAA | CCAGTTTGGCCTTCTGAACC |
| Wild-Type | 3xTg-AD | |||||||
|---|---|---|---|---|---|---|---|---|
| ctr | ins | glu + ins | ins + ins | ctr | ins | glu + ins | ins + ins | |
| Atg16L1 | 1.51 ± 0.76 | 2.18 ± 1.25 | 0.15 ± 0.04 | 0.16 ± 0.06 | 0.18 ± 0.11 | 0.95 ± 0.92 | 3.03 ± 1.99 * | 0.77 ± 0.47 |
| Atg16L2 | 1.14 ± 0.34 | 0.41 ± 0.07 | 1.14 ± 0.34 | 1.04 ± 0.54 | 0.88 ± 0.69 | 0.92 ± 0.68 | 1.07 ± 0.54 | 0.44 ± 0.27 |
| Gabarap | 1.13 ± 0.27 | 1.30 ± 0.47 | 0.95 ± 0.17 | 0.76 ± 0.13 | 0.64 ± 0.14 | 0.46 ± 0.15 | 0.63 ± 0.23 | 0.46 ± 0.21 |
| GabarapL1 | 1.05 ± 0.23 | 0.91 ± 0.23 | 0.87 ± 0.07 | 0.86 ± 0.30 | 1.31 ± 0.54 | 1.02 ± 0.45 | 0.81 ± 0.32 | 1.30 ± 0.25 |
| GabarapL2 | 1.17 ± 0.31 | 1.56 ± 0.38 | 1.03 ± 0.19 | 1.49 ± 0.39 | 1.32 ± 0.56 | 0.63 ± 0.29 | 1.51 ± 0.69 | 0.88 ± 0.47 |
| Sqstm1 | 1.00 ± 0.05 | 0.74 ± 0.11 | 0.99 ± 0.15 | 0.66 ± 0.19 | 0.74 ± 0.32 | 0.79 ± 0.39 | 0.83 ± 0.35 | 0.60 ± 0.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vianello, C.; Salluzzo, M.; Anni, D.; Boriero, D.; Buffelli, M.; Carboni, L. Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease—Type 2 Diabetes Mellitus Comorbidity Models in Cells. Int. J. Environ. Res. Public Health 2023, 20, 4540. https://doi.org/10.3390/ijerph20054540
Vianello C, Salluzzo M, Anni D, Boriero D, Buffelli M, Carboni L. Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease—Type 2 Diabetes Mellitus Comorbidity Models in Cells. International Journal of Environmental Research and Public Health. 2023; 20(5):4540. https://doi.org/10.3390/ijerph20054540
Chicago/Turabian StyleVianello, Clara, Marco Salluzzo, Daniela Anni, Diana Boriero, Mario Buffelli, and Lucia Carboni. 2023. "Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease—Type 2 Diabetes Mellitus Comorbidity Models in Cells" International Journal of Environmental Research and Public Health 20, no. 5: 4540. https://doi.org/10.3390/ijerph20054540
APA StyleVianello, C., Salluzzo, M., Anni, D., Boriero, D., Buffelli, M., & Carboni, L. (2023). Increased Expression of Autophagy-Related Genes in Alzheimer’s Disease—Type 2 Diabetes Mellitus Comorbidity Models in Cells. International Journal of Environmental Research and Public Health, 20(5), 4540. https://doi.org/10.3390/ijerph20054540









