No Causal Effects Detected in COVID-19 and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Two Sample Mendelian Randomization Study
Abstract
1. Introduction
2. Methods
2.1. Data Sources and Processing
2.2. Selection of the Genetic IVs
2.3. TSMR Analysis
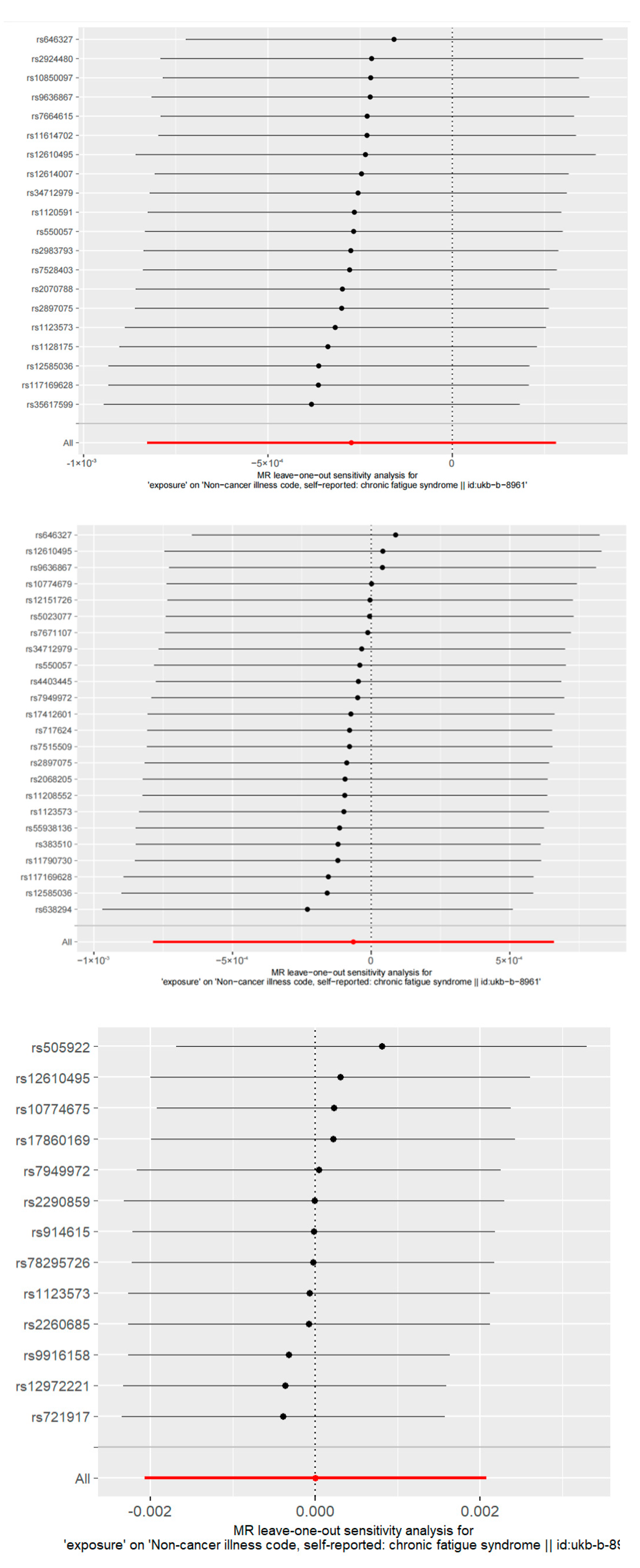
2.4. Sensitivity Analysis
3. Results
3.1. SNPs
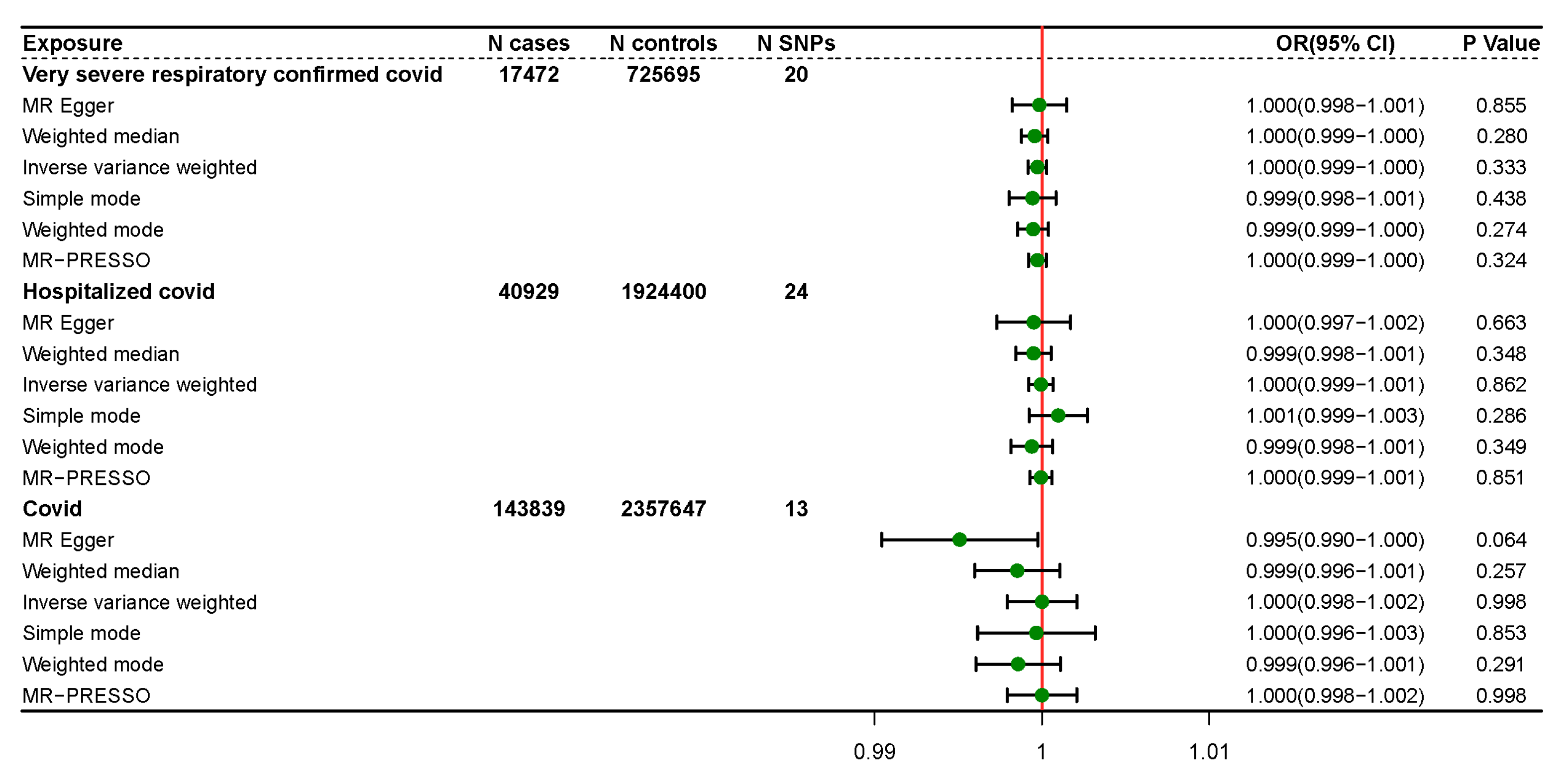
3.2. TSMR Results
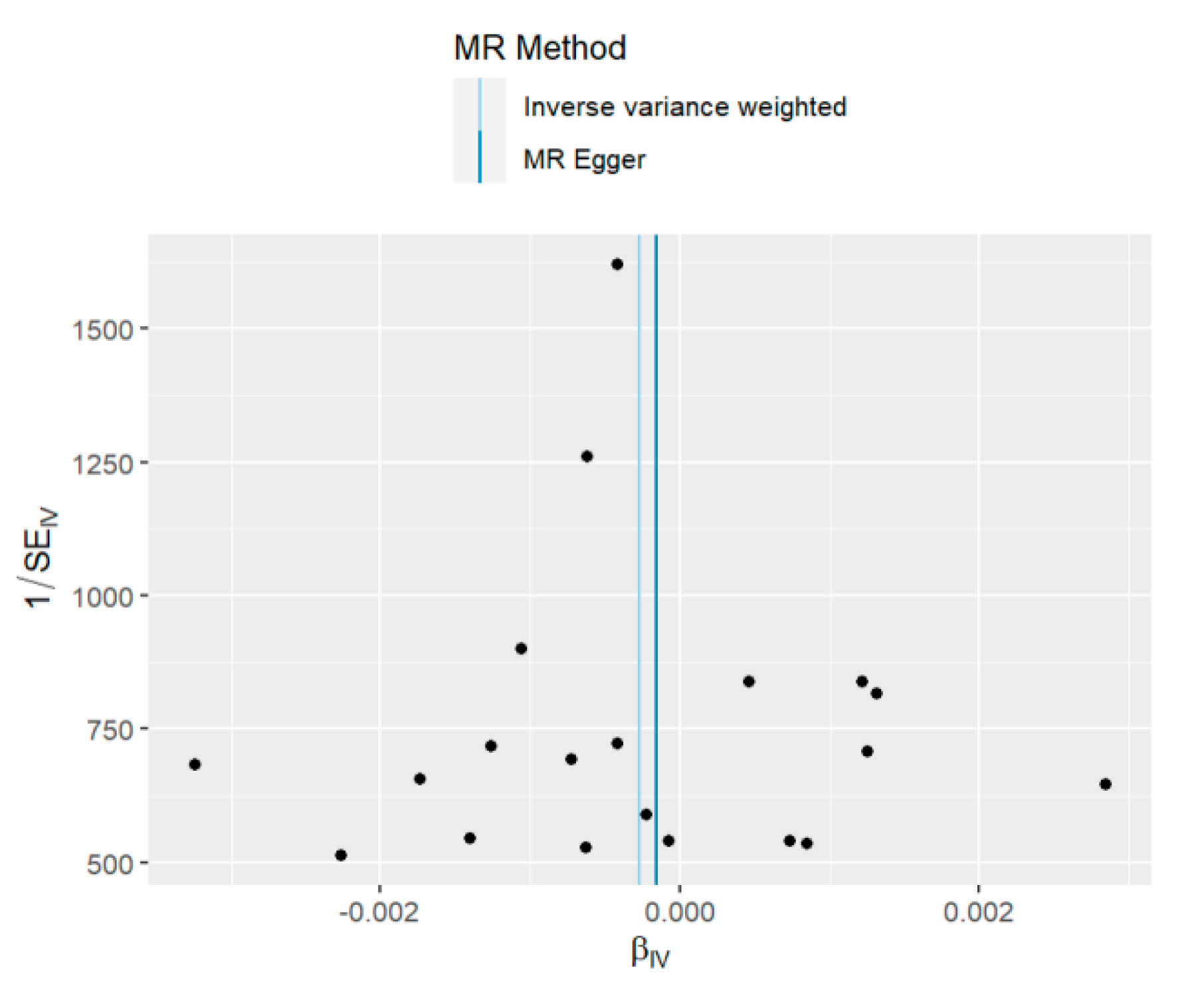
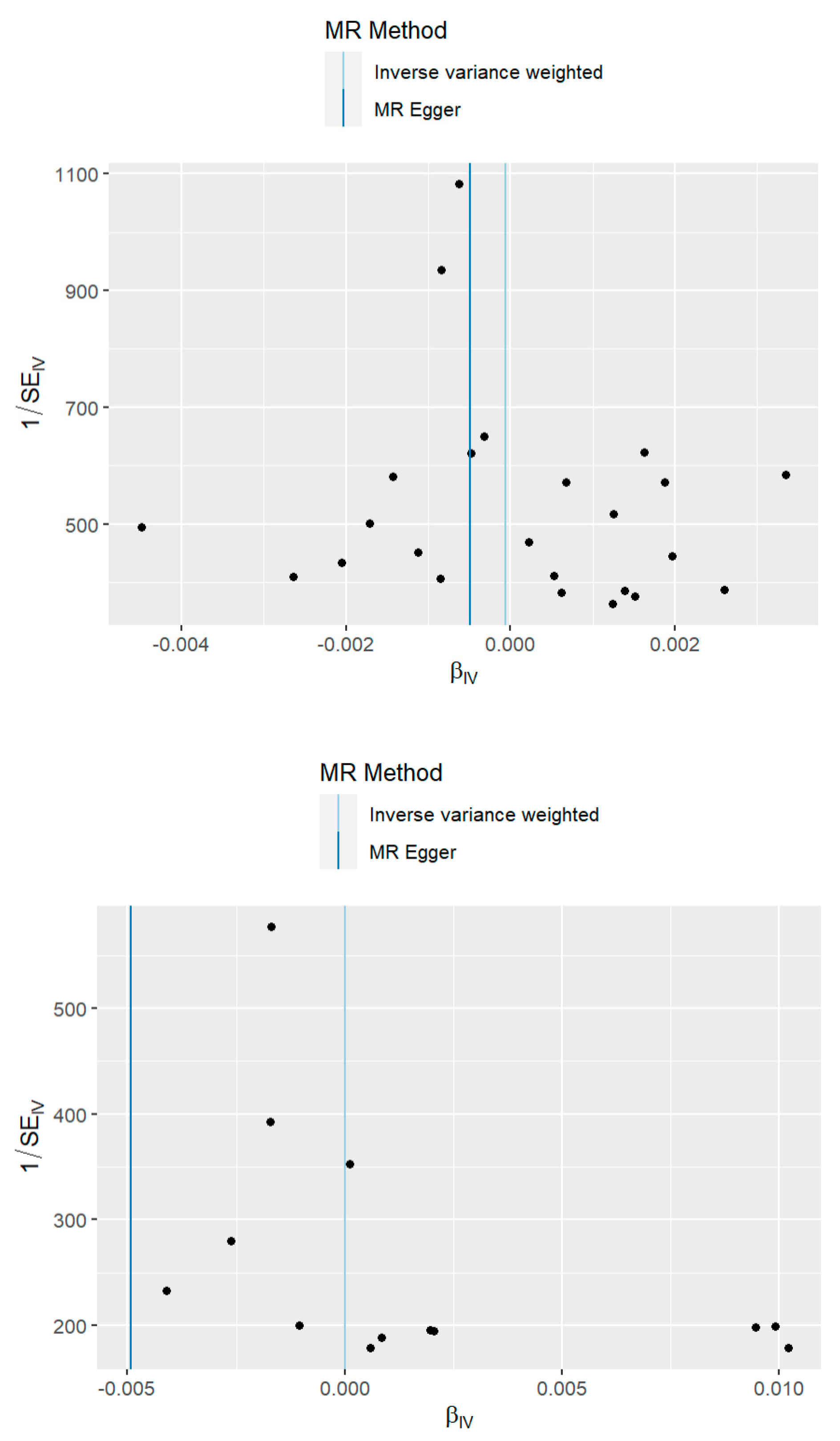
3.3. Sensitivity Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Premeaux, T.A.; Yeung, S.T.; Bukhari, Z.; Bowler, S.; Alpan, O.; Gupta, R.; Ndhlovu, L.C. Emerging Insights on Caspases in COVID-19 Pathogenesis, Sequelae, and Directed Therapies. Front. Immunol. 2022, 13, 842740. [Google Scholar] [CrossRef]
- Barak, D.; Gallo, E.; Rong, K.; Tang, K.; Du, W. Experience of the COVID-19 pandemic in Wuhan leads to a lasting increase in social distancing. Sci. Rep. 2022, 12, 18457. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 1 December 2022).
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef]
- Cao, W.; Fang, Z.; Hou, G.; Han, M.; Xu, X.; Dong, J.; Zheng, J. The psychological impact of the COVID-19 epidemic on college students in China. Psychiatry Res. 2020, 287, 112934. [Google Scholar] [CrossRef]
- Saag, M.S. Misguided Use of Hydroxychloroquine for COVID-19: The Infusion of Politics into Science. JAMA 2020, 324, 2161–2162. [Google Scholar] [CrossRef]
- Groff, D.; Sun, A.; Ssentongo, A.E.; Ba, D.M.; Parsons, N.; Poudel, G.R.; Lekoubou, A.; Oh, J.S.; Ericson, J.E.; Ssentongo, P.; et al. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw. Open 2021, 4, e2128568. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 16144. [Google Scholar] [CrossRef]
- Mazza, M.G.; De Lorenzo, R.; Conte, C.; Poletti, S.; Vai, B.; Bollettini, I.; Melloni, E.M.T.; Furlan, R.; Ciceri, F.; Rovere-Querini, P.; et al. Anxiety and depression in COVID-19 survivors: Role of inflammatory and clinical predictors. Brain Behav. Immun. 2020, 89, 594–600. [Google Scholar] [CrossRef]
- Whittaker, A.; Anson, M.; Harky, A. Neurological Manifestations of COVID-19: A systematic review and current update. Acta Neurol. Scand. 2020, 142, 14–22. [Google Scholar] [CrossRef]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’Em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. eClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef]
- Kedor, C.; Freitag, H.; Meyer-Arndt, L.; Wittke, K.; Hanitsch, L.G.; Zoller, T.; Steinbeis, F.; Haffke, M.; Rudolf, G.; Heidecker, B.; et al. A prospective observational study of post-COVID-19 chronic fatigue syndrome following the first pandemic wave in Germany and biomarkers associated with symptom severity. Nat. Commun. 2022, 13, 5104. [Google Scholar] [CrossRef]
- Sandler, C.X.; Lloyd, A.R. Chronic fatigue syndrome: Progress and possibilities. Med. J. Aust. 2020, 212, 428–433. [Google Scholar] [CrossRef]
- Prins, J.B.; van der Meer, J.W.; Bleijenberg, G. Chronic fatigue syndrome. Lancet 2006, 367, 346–355. [Google Scholar] [CrossRef]
- Natelson, B.H.; Brunjes, D.L.; Mancini, D. Chronic Fatigue Syndrome and Cardiovascular Disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2021, 78, 1056–1067. [Google Scholar] [CrossRef]
- Lim, E.-J.; Ahn, Y.-C.; Jang, E.-S.; Lee, S.-W.; Lee, S.-H.; Son, C.-G. Systematic review and meta-analysis of the prevalence of chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME). J. Transl. Med. 2020, 18, 100. [Google Scholar] [CrossRef]
- Wong, T.L.; Weitzer, D.J. Long COVID and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)—A Systemic Review and Comparison of Clinical Presentation and Symptomatology. Medicina 2021, 57, 418. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef]
- A Lawlor, D. Commentary: Two-sample Mendelian randomization: Opportunities and challenges. Int. J. Epidemiol. 2016, 45, 908–915. [Google Scholar] [CrossRef]
- The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020, 28, 715–718. [CrossRef]
- The COVID-19 Host Genetics Initiative. Available online: https://www.covid19hg.org/ (accessed on 9 November 2022).
- Non-Cancer Illness Code, Self-Reported: Chronic Fatigue Syndrome. Available online: https://gwas.mrcieu.ac.uk/datasets/ukb-b-8961/ (accessed on 9 November 2022).
- Li, C.; Liu, J.; Lin, J.; Shang, H. COVID-19 and risk of neurodegenerative disorders: A Mendelian randomization study. Transl. Psychiatry 2022, 12, 283. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 2016, 40, 597–608. [Google Scholar] [CrossRef]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Bae, S.-C.; Lee, Y.H. Vitamin D level and risk of systemic lupus erythematosus and rheumatoid arthritis: A Mendelian randomization. Clin. Rheumatol. 2018, 37, 2415–2421. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, F.P.; Smith, G.D.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Huang, Y.; Hu, J.; Shao, Z. Mendelian randomization study of inflammatory bowel disease and bone mineral density. BMC Med. 2020, 18, 312. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Alwan, N.A. Track COVID-19 sickness, not just positive tests and deaths. Nature 2020, 584, 170. [Google Scholar] [CrossRef]
- Deumer, U.-S.; Varesi, A.; Floris, V.; Savioli, G.; Mantovani, E.; López-Carrasco, P.; Rosati, G.M.; Prasad, S.; Ricevuti, G. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): An Overview. J. Clin. Med. 2021, 10, 4786. [Google Scholar] [CrossRef]
- Komaroff, A.L.; Lipkin, W.I. Insights from Myalgic Encephalomyelitis/Chronic Fatigue Syndrome May Help Unravel the Pathogenesis of Post-Acute COVID-19 Syndrome. Trends Mol. Med. 2021, 27, 895–906. [Google Scholar] [CrossRef]
- Morrow, A.K.; Malone, L.A.; Kokorelis, C.; Petracek, L.S.; Eastin, E.F.; Lobner, K.L.; Neuendorff, L.; Rowe, P.C. Long-Term COVID 19 Sequelae in Adolescents: The Overlap with Orthostatic Intolerance and ME/CFS. Curr. Pediatr. Rep. 2022, 10, 31–44. [Google Scholar] [CrossRef]
- Retornaz, F.; Rebaudet, S.; Stavris, C.; Jammes, Y. Long-term neuromuscular consequences of SARS-Cov-2 and their similarities with myalgic encephalomyelitis/chronic fatigue syndrome: Results of the retrospective CoLGEM study. J. Transl. Med. 2022, 20, 429. [Google Scholar] [CrossRef]
- Mackay, A. A Paradigm for Post-Covid-19 Fatigue Syndrome Analogous to ME/CFS. Front. Neurol. 2021, 12, 701419. [Google Scholar] [CrossRef]
- Arnett, S.; Alleva, L.; Korossy-Horwood, R.; Clark, I. Chronic fatigue syndrome—A neuroimmunological model. Med. Hypotheses 2011, 77, 77–83. [Google Scholar] [CrossRef] [PubMed]

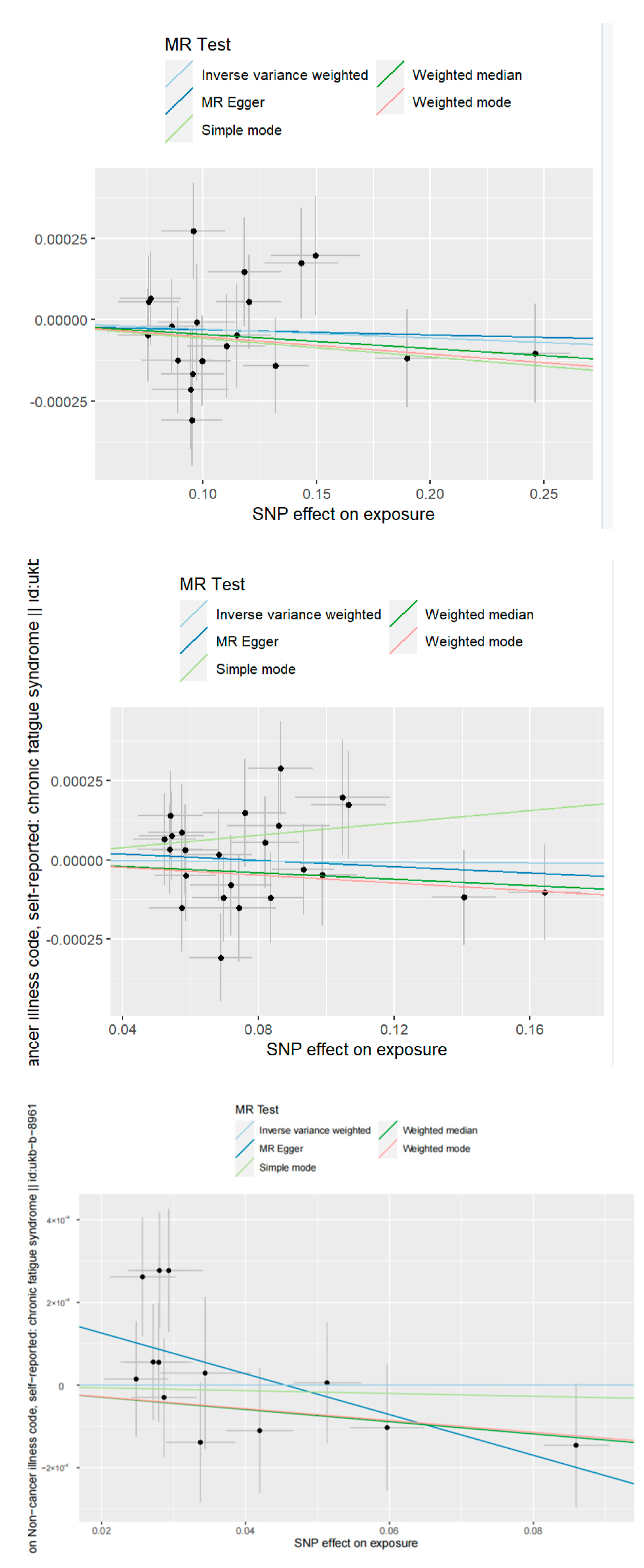
| SNP | Chromosome | Position | β.exposure | SE.exposure | pval.exposure | β.outcome | SE.outcome | pval.outcome | F-Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| Severity | ||||||||||
| rs10850097 | 12 | 113361117 | 0.09552 | 0.01399 | 8.59 × 10−12 | −0.0001661 | 0.000145037 | 0.25 | 2907.13 | |
| rs1120591 | 8 | 61537523 | 0.07600 | 0.01334 | 1.22 × 10−8 | −0.0000482 | 0.000143872 | 0.74 | 2093.94 | |
| rs1123573 | 2 | 60707588 | −0.12036 | 0.01438 | 5.64 × 10−17 | −0.0000551 | 0.000143234 | 0.70 | 4881.36 | |
| rs1128175 | 6 | 31150435 | −0.11827 | 0.01601 | 1.50 × 10−13 | −0.0001474 | 0.000166854 | 0.38 | 4038.83 | |
| rs11614702 | 12 | 133058157 | 0.09967 | 0.01306 | 2.29 × 10−14 | −0.0001258 | 0.000138689 | 0.36 | 3704.18 | |
| rs117169628 | 16 | 89264460 | 0.14967 | 0.01965 | 2.60 × 10−14 | 0.0001962 | 0.000183068 | 0.28 | 3629.32 | |
| rs12585036 | 13 | 113535741 | 0.14326 | 0.01609 | 5.28 × 10−19 | 0.0001735 | 0.000170713 | 0.31 | 5210.72 | |
| rs12610495 | 19 | 4717672 | 0.24613 | 0.01530 | 3.28 × 10−58 | −0.0001029 | 0.000151855 | 0.50 | 19,886.87 | |
| rs12614007 | 2 | 57316503 | 0.08889 | 0.01604 | 2.99 × 10−8 | −0.0001245 | 0.000162446 | 0.44 | 2191.49 | |
| rs2070788 | 21 | 42841988 | −0.07615 | 0.01336 | 1.19 × 10−8 | −0.0000560 | 0.000140635 | 0.69 | 2102.92 | |
| rs2897075 | 7 | 99630342 | 0.07714 | 0.01334 | 7.30 × 10−9 | 0.0000651 | 0.000143991 | 0.65 | 2090.65 | |
| rs2924480 | 11 | 34529831 | −0.13204 | 0.01445 | 6.52 × 10−10 | 0.0001407 | 0.000146439 | 0.34 | 5875.94 | |
| rs2983793 | 10 | 81445802 | 0.08640 | 0.01421 | 1.18 × 10−9 | −0.0000194 | 0.000146350 | 0.89 | 2729.57 | |
| rs34712979 | 4 | 106819053 | −0.11042 | 0.01712 | 1.13 × 10−10 | 0.0000806 | 0.000159248 | 0.61 | 3370.15 | |
| rs35617599 | 19 | 50874794 | 0.09594 | 0.01398 | 6.77 × 10−12 | 0.0002727 | 0.000148087 | 0.07 | 2960.59 | |
| rs550057 | 9 | 136146597 | 0.11508 | 0.01501 | 1.73 × 10−14 | −0.0000480 | 0.000159173 | 0.76 | 3956.96 | |
| rs646327 | 19 | 49209851 | −0.09538 | 0.01338 | 1.00 × 10−12 | 0.0003092 | 0.000139088 | 0.03 | 3310.72 | |
| rs7528403 | 1 | 65382792 | −0.09740 | 0.01686 | 7.58 × 10−9 | 0.0000079 | 0.000179455 | 0.96 | 2719.92 | |
| rs7664615 | 4 | 25448493 | −0.09472 | 0.01697 | 2.40 × 10−8 | 0.0002145 | 0.000184190 | 0.24 | 2142.10 | |
| rs9636867 | 21 | 34609944 | 0.18972 | 0.01387 | 1.32 × 10−42 | −0.0001185 | 0.000150267 | 0.43 | 12,505.91 | |
| Hospitalization | ||||||||||
| rs10774679 | 12 | 113374748 | 0.08358 | 0.00950 | 1.36 × 10−18 | −0.0001198 | 0.000143988 | 0.41 | 6191.24 | |
| rs11208552 | 1 | 65412830 | −0.05736 | 0.00989 | 6.70 × 10−9 | −0.0000870 | 0.000152264 | 0.57 | 3036.07 | |
| rs1123573 | 2 | 60707588 | −0.08188 | 0.01016 | 7.45 × 10−16 | −0.0000551 | 0.000143234 | 0.70 | 6086.70 | |
| rs117169628 | 16 | 89264460 | 0.10477 | 0.01391 | 5.09 × 10−14 | 0.0001962 | 0.000183068 | 0.28 | 4917.59 | |
| rs11790730 | 9 | 33425871 | 0.07587 | 0.01218 | 4.68 × 10−10 | 0.0001500 | 0.000170557 | 0.38 | 3468.55 | |
| rs12151726 | 2 | 198273591 | 0.05732 | 0.00975 | 4.11 × 10−9 | −0.0001513 | 0.000139912 | 0.28 | 3094.13 | |
| rs12585036 | 13 | 113535741 | 0.10643 | 0.01108 | 7.75 × 10−22 | 0.0001735 | 0.000170713 | 0.31 | 7364.79 | |
| rs12610495 | 19 | 4717672 | 0.16432 | 0.01075 | 9.89 × 10−53 | −0.0001029 | 0.000151855 | 0.50 | 22,020.48 | |
| rs17412601 | 3 | 101499275 | −0.06815 | 0.00978 | 3.24 × 10−12 | −0.0000153 | 0.000145432 | 0.92 | 4097.22 | |
| rs2068205 | 6 | 33058583 | 0.05430 | 0.00965 | 1.83 × 10−8 | 0.0000754 | 0.000140680 | 0.59 | 2706.14 | |
| rs2897075 | 7 | 99630342 | 0.05229 | 0.00929 | 1.82 × 10−8 | 0.0000651 | 0.000143991 | 0.65 | 2494.14 | |
| rs34712979 | 4 | 106819053 | −0.07190 | 0.01214 | 3.21 × 10−9 | 0.0000806 | 0.000159248 | 0.61 | 3597.65 | |
| rs383510 | 21 | 42858367 | −0.05397 | 0.00946 | 1.14 × 10−8 | −0.0001404 | 0.000139530 | 0.31 | 2836.26 | |
| rs4403445 | 8 | 61432007 | 0.05845 | 0.00903 | 9.82 × 10−11 | −0.0000500 | 0.000143867 | 0.73 | 3229.73 | |
| rs5023077 | 12 | 133141973 | −0.06962 | 0.00911 | 2.11 × 10−14 | 0.0001197 | 0.000138878 | 0.39 | 4766.32 | |
| rs550057 | 9 | 136146597 | 0.09880 | 0.01030 | 8.74 × 10−22 | −0.0000480 | 0.000159173 | 0.76 | 7169.36 | |
| rs55938136 | 17 | 43798360 | −0.08588 | 0.01524 | 1.73 × 10−8 | −0.0001077 | 0.000166166 | 0.52 | 4029.68 | |
| rs638294 | 19 | 50863023 | 0.08651 | 0.00945 | 5.67 × 10−20 | 0.0002897 | 0.000148130 | 0.05 | 6525.99 | |
| rs646327 | 19 | 49209851 | −0.06890 | 0.00933 | 1.54 × 10−13 | 0.0003092 | 0.000139088 | 0.026 | 4664.76 | |
| rs717624 | 7 | 22894487 | −0.05379 | 0.00947 | 1.34 × 10−8 | −0.0000330 | 0.000140778 | 0.81 | 2678.99 | |
| rs7515509 | 1 | 77949123 | 0.05838 | 0.00966 | 1.52 × 10−9 | 0.0000306 | 0.000142052 | 0.83 | 3101.91 | |
| rs7671107 | 4 | 25449225 | −0.07412 | 0.01099 | 1.51 × 10−11 | 0.0001520 | 0.000170930 | 0.37 | 4359.11 | |
| rs7949972 | 11 | 34502042 | −0.09310 | 0.00919 | 3.94 × 10−24 | 0.0000298 | 0.000143289 | 0.84 | 7757.94 | |
| rs9636867 | 21 | 34609944 | 0.14045 | 0.00940 | 1.83 × 10−50 | −0.0001185 | 0.000150267 | 0.43 | 17,671.68 | |
| COVID | ||||||||||
| rs10774675 | 12 | 113361237 | 0.03373 | 0.00476 | 1.38 × 10−12 | −0.0001386 | 0.000144861 | 0.34 | 1206.40 | |
| rs1123573 | 2 | 60707588 | −0.02794 | 0.00465 | 1.83 × 10−9 | −0.0000551 | 0.000143234 | 0.70 | 903.94 | |
| rs12610495 | 19 | 4717672 | 0.05967 | 0.00509 | 1.02 × 10−30 | −0.0001029 | 0.000151855 | 0.50 | 3665.96 | |
| rs12972221 | 19 | 50879140 | 0.02932 | 0.00467 | 3.30 × 10−10 | 0.0002772 | 0.000148189 | 0.06 | 921.76 | |
| rs17860169 | 21 | 34613301 | 0.04196 | 0.00461 | 8.90 × 10−20 | −0.0001103 | 0.000150124 | 0.46 | 1984.44 | |
| rs2260685 | 3 | 195497743 | 0.02716 | 0.00455 | 2.33 × 10−9 | 0.0000559 | 0.000139376 | 0.69 | 918.04 | |
| rs2290859 | 3 | 101525625 | −0.05132 | 0.00473 | 1.79 × 10−27 | −0.0000054 | 0.000145542 | 0.97 | 2961.04 | |
| rs505922 | 9 | 136149229 | 0.08592 | 0.00449 | 1.05 × 10−81 | −0.0001457 | 0.000148905 | 0.33 | 8474.34 | |
| rs721917 | 10 | 81706324 | 0.02802 | 0.00437 | 1.48 × 10−10 | 0.0002773 | 0.000140657 | 0.05 | 963.27 | |
| rs78295726 | 19 | 10426512 | 0.03438 | 0.00629 | 4.60 × 10−8 | 0.0000290 | 0.000182971 | 0.87 | 727.61 | |
| rs7949972 | 11 | 34502042 | −0.02867 | 0.00450 | 1.87 × 10−10 | 0.0000298 | 0.000143289 | 0.84 | 937.44 | |
| rs914615 | 1 | 155175892 | −0.02480 | 0.00437 | 1.38 × 10−8 | −0.0000145 | 0.000138969 | 0.92 | 761.27 | |
| rs9916158 | 17 | 38182229 | 0.02569 | 0.00449 | 1.10 × 10−8 | 0.0002619 | 0.000143728 | 0.07 | 763.84 |
| Exposure | Method | SNP | β | SE | p-Value | OR | OR (Lower 95% CI) | OR (Upper 95% CI) |
|---|---|---|---|---|---|---|---|---|
| Severity | MR Egger | 20 | −0.00015446 | 0.000832451 | 0.85487 | 0.99985 | 0.99822 | 1.00148 |
| Weighted median | 20 | −0.000436505 | 0.000403932 | 0.27986 | 0.99956 | 0.99877 | 1.00036 | |
| IVW | 20 | −0.000273796 | 0.000282557 | 0.33255 | 0.99973 | 0.99917 | 1.00028 | |
| Simple mode | 20 | −0.00056838 | 0.000716877 | 0.43765 | 0.99943 | 0.99803 | 1.00084 | |
| Weighted mode | 20 | −0.00052857 | 0.000468997 | 0.27377 | 0.99947 | 0.99855 | 1.00039 | |
| MR-PRESSO | 20 | −0.000273796 | 0.0002706 | 0.32436 | 0.99972 | 0.99919 | 1.00025 | |
| Hospitalization | MR Egger | 24 | −0.000495398 | 0.001123173 | 0.66347 | 0.99950 | 0.99731 | 1.00171 |
| Weighted median | 24 | −0.000506027 | 0.000538839 | 0.34768 | 0.99949 | 0.99844 | 1.00055 | |
| IVW | 24 | −6.43 × 10−5 | 0.000368448 | 0.86153 | 0.99994 | 0.99921 | 1.00066 | |
| Simple mode | 24 | 0.000972112 | 0.000889543 | 0.28579 | 1.00097 | 0.99923 | 1.00272 | |
| Weighted mode | 24 | −0.00060844 | 0.000636798 | 0.34928 | 0.99939 | 0.99815 | 1.00064 | |
| MR-PRESSO | 24 | −6.38 × 10−5 | 0.0003366 | 0.85125 | 0.99993 | 0.99928 | 1.00059 | |
| Susceptibility | MR Egger | 13 | −0.004929688 | 0.002395122 | 0.06406 | 0.99508 | 0.99042 | 0.99976 |
| Weighted median | 13 | −0.001478204 | 0.001303262 | 0.25670 | 0.99852 | 0.99598 | 1.00108 | |
| IVW | 13 | 2.44 × 10−6 | 0.001058907 | 0.99817 | 1.00000 | 0.99793 | 1.00208 | |
| Simple mode | 13 | −0.000339753 | 0.001793678 | 0.85293 | 0.99966 | 0.99615 | 1.00318 | |
| Weighted mode | 13 | −0.001426332 | 0.001291833 | 0.29119 | 0.99857 | 0.99605 | 1.00111 | |
| MR-PRESSO | 13 | 2.44 × 10−6 | 1.0010589 | 0.99820 | 1.00000 | 0.99792 | 1.00208 |
| Methods | Severity | Hospitalization | Susceptibility |
|---|---|---|---|
| MR Egger-Intercept Pleiotropy Test p-value | 0.88 | 0.69 | 0.05 |
| MR Egger Cochran’s Q Test p-value | 0.50 | 0.64 | 0.63 |
| IVW Cochran’s Q Test p-value | 0.56 | 0.69 | 0.30 |
| MR-PRESSO Global Test p-value | 0.59 | 0.69 | 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Cao, Y.; Wu, L. No Causal Effects Detected in COVID-19 and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Two Sample Mendelian Randomization Study. Int. J. Environ. Res. Public Health 2023, 20, 2437. https://doi.org/10.3390/ijerph20032437
Xu W, Cao Y, Wu L. No Causal Effects Detected in COVID-19 and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Two Sample Mendelian Randomization Study. International Journal of Environmental Research and Public Health. 2023; 20(3):2437. https://doi.org/10.3390/ijerph20032437
Chicago/Turabian StyleXu, Wangzi, Yu Cao, and Lin Wu. 2023. "No Causal Effects Detected in COVID-19 and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Two Sample Mendelian Randomization Study" International Journal of Environmental Research and Public Health 20, no. 3: 2437. https://doi.org/10.3390/ijerph20032437
APA StyleXu, W., Cao, Y., & Wu, L. (2023). No Causal Effects Detected in COVID-19 and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Two Sample Mendelian Randomization Study. International Journal of Environmental Research and Public Health, 20(3), 2437. https://doi.org/10.3390/ijerph20032437






