Mutational Analysis of EGFR Mutations in Non-Small Cell Lung Carcinoma—An Indian Perspective of 212 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Population Selection and Data Collection
2.2. Tissue Preparation and DNA Extraction
2.3. Detection of EGFR Mutations
2.4. EGFR Mutation Screening
2.5. Statistical Analysis
3. Results
3.1. Demographic and Histological Characteristics of Patient Samples
3.2. EGFR Mutation Screening and Detection
3.3. Molecular Characterization of EGFR Mutations
Uncommon Mutations of EGFR
3.4. Metastatic Location and Features of EGFR Mutations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Au, J.S.K.; Thongprasert, S.; Srinivasan, S.; Tsai, C.M.; Khoa, M.T.; Heeroma, K.; Itoh, Y.; Cornelio, G.; Yang, P.C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non–small-cell lung cancer of adeno-carcinoma histology (PIONEER). J. Thorac. Oncol. 2014, 9, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Ju, M.H.; Kim, H.R.; Kim, J.B.; Kim, Y.H.; Kim, D.K.; Park, S.I. Surgical outcomes in small cell lung cancer. Korean J. Thorac. Cardiovasc. Surg. 2012, 45, 40. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod. Pathol. 2012, 25, S18–S30. [Google Scholar] [CrossRef]
- Wilcox, H.B.; Al-Zoughool, M.; Garner, M.J.; Jiang, H.; Klotz, J.B.; Krewski, D.; Nicholson, W.J.; Schoenberg, J.B.; Villeneuve, P.J.; Zielinski, J.M. Case-control study of radon and lung cancer in New Jersey. Radiat. Prot. Dosim. 2007, 128, 169–179. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Riely, G.J. New Pathologic Classification of Lung Cancer: Relevance for Clinical Practice and Clinical Trials. J. Clin. Oncol. 2013, 31, 992–1001. [Google Scholar] [CrossRef]
- Rubin, P.; Hansen, J.T. TNM Staging Atlas; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008. [Google Scholar]
- Basumallik, N.; Agarwal, M. Small Cell Lung Cancer; StatPearls Publishing: Tampa, FL, USA, 2021. [Google Scholar]
- Dela Cruz, C.S.; Tanoue, L.T.; Matthay, R.A. Lung Cancer: Epidemiology, Etiology, and Prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef]
- Tanaka, T.; Matsuoka, M.; Sutani, A.; Gemma, A.; Maemondo, M.; Inoue, A.; Okinaga, S.; Nagashima, M.; Oizumi, S.; Uematsu, K.; et al. Frequency of and variables associated with the EGFR mutation and its subtypes. Int. J. Cancer 2009, 126, 651–655. [Google Scholar] [CrossRef]
- Richards, C.S.; Bale, S.; Bellissimo, D.B.; Das, S.; Grody, W.W.; Hegde, M.R.; Lyon, E.; Ward, B.E. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008, 10, 294–300. [Google Scholar] [CrossRef]
- Wieduwilt, M.J.; Moasser, M.M. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell. Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Girard, N. Optimizing outcomes in EGFR mutation-positive NSCLC: Which tyrosine kinase inhibitor and when? Futur. Oncol. 2018, 14, 1117–1132. [Google Scholar] [CrossRef] [PubMed]
- Noronha, V.; Prabhash, K.; Thavamani, A.; Chougule, A.; Purandare, N.; Joshi, A.; Sharma, R.; Desai, S.; Jambekar, N.; Dutt, A.; et al. EGFR Mutations in Indian Lung Cancer Patients: Clinical Correlation and Outcome to EGFR Targeted Therapy. PLoS ONE 2013, 8, e61561. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhou, C. Lung cancer in never smokers—The East Asian experience. Transl. Lung Cancer Res. 2018, 7, 450–463. [Google Scholar] [CrossRef]
- Dogan, S.; Shen, R.; Ang, D.C.; Johnson, M.L.; D’Angelo, S.P.; Paik, P.K.; Brzostowski, E.B.; Riely, G.J.; Kris, M.G.; Zakowski, M.F.; et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3,026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant CancersMolecular Epidemiology of Lung Cancer Driver Mutations. Clin. Cancer Res. 2012, 18, 6169–6177. [Google Scholar] [CrossRef]
- Chougule, A.; Prabhash, K.; Noronha, V.; Joshi, A.; Thavamani, A.; Chandrani, P.; Upadhyay, P.; Utture, S.; Desai, S.; Jambhekar, N.; et al. Frequency of EGFR Mutations in 907 Lung Adenocarcioma Patients of Indian Ethnicity. PLoS ONE 2013, 8, e76164. [Google Scholar] [CrossRef]
- Melosky, B.; Kambartel, K.; Häntschel, M.; Bennetts, M.; Nickens, D.J.; Brinkmann, J.; Kayser, A.; Moran, M.; Cappuzzo, F. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol. Diagn. Ther. 2021, 26, 7–18. [Google Scholar] [CrossRef]
- Yoon, H.-Y.; Ryu, J.-S.; Sim, Y.S.; Kim, D.; Lee, S.Y.; Choi, J.; Park, S.; Ryu, Y.J.; Lee, J.H.; Chang, J.H. Clinical significance of EGFR mutation types in lung adenocarcinoma: A multi-centre Korean study. PLoS ONE 2020, 15, e0228925. [Google Scholar] [CrossRef]
- Cortes-Funes, H.; Gomez, C.; Rosell, R.; Valero, P.; Garcia-Giron, C.; Velasco, A.; Izquierdo, A.; Diz, P.; Camps, C.; Castellanos, D.; et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann. Oncol. 2005, 16, 1081–1086. [Google Scholar] [CrossRef]
- Tu, H.-Y.; Ke, E.-E.; Yang, J.-J.; Sun, Y.-L.; Yan, H.-H.; Zheng, M.-Y.; Bai, X.-Y.; Wang, Z.; Su, J.; Chen, Z.-H.; et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer 2017, 114, 96–102. [Google Scholar] [CrossRef]
- Harrison, P.T.; Vyse, S.; Huang, P.H. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin. Cancer Biol. 2019, 61, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Vaclova, T.; Grazini, U.; Ward, L.; O’Neill, D.; Markovets, A.; Huang, X.; Chmielecki, J.; Hartmaier, R.; Thress, K.S.; Smith, P.D.; et al. Clinical impact of subclonal EGFR T790M mutations in advanced-stage EGFR-mutant non-small-cell lung cancers. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cang, S.; Liu, D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J. Hematol. Oncol. 2016, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Shi, S.; Chang, J.Y. Treatment modes for EGFR mutations in patients with brain metastases from non-small cell lung cancer: Controversy, causes, and solutions. Transl. Lung Cancer Res. 2019, 8, 524–531. [Google Scholar] [CrossRef]
- Shin, D.Y.; Kim, C.H.; Park, S.; Baek, H.; Yang, S.H. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J. Thorac. Oncol. 2014, 9, 195–199. [Google Scholar] [CrossRef]
- Bhatt, V.R.; D’Souza, S.P.; Smith, L.M.; Cushman-Vokoun, A.M.; Noronha, V.; Verma, V.; Joshi, A.; Chougule, A.; Jambhekar, N.; Kessinger, A.; et al. Epidermal Growth Factor Receptor Mutational Status and Brain Metastases in Non–Small-Cell Lung Cancer. J. Glob. Oncol. 2017, 3, 208–217. [Google Scholar] [CrossRef]
- Baek, M.Y.; Ahn, H.K.; Park, K.R.; Park, H.-S.; Kang, S.M.; Park, I.; Kim, Y.S.; Hong, J.; Sym, S.J.; Park, J.; et al. Epidermal growth factor receptor mutation and pattern of brain metastasis in patients with non-small cell lung cancer. Korean J. Intern. Med. 2018, 33, 168–175. [Google Scholar] [CrossRef]
- Kelly, W.J.; Shah, N.J.; Subramaniam, D.S. Management of Brain Metastases in Epidermal Growth Factor Receptor Mutant Non-Small-Cell Lung Cancer. Front. Oncol. 2018, 8, 208. [Google Scholar] [CrossRef]
- Sekine, A.; Kato, T.; Hagiwara, E.; Shinohara, T.; Komagata, T.; Iwasawa, T.; Satoh, H.; Tamura, K.; Kasamatsu, T.; Hayashihara, K.; et al. Metastatic brain tumors from non-small cell lung cancer with EGFR mutations: Distinguishing influence of exon 19 deletion on radiographic features. Lung Cancer 2012, 77, 64–69. [Google Scholar] [CrossRef]


| Global Population (n = 212) | EGFR Mutated Patients (n = 82; 38.67%) | EGFR Non-Mutated Patients (n = 130; 61.32%) | p Value | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Male | 121 (57%) | 37 (45.12%) | 84 (64.6%) | 0.007 |
| Female | 91 (43%) | 45 (54.88%) | 46 (35.38%) | 0.007 |
| Male (≤60) | 42 (34.71%) | 12 (32.4%) | 30 (35.7%) | 0.726 |
| Male (>60) | 79 (65.29%) | 25 (67.5%) | 54 (64.3%) | 0.726 |
| Female (≤60) | 41 (45%) | 20 (44.44%) | 21 (45.6%) | 0.907 |
| Female (>60) | 50 (54.94%) | 25 (55.5%) | 25 (54.3%) | 0.907 |
| Histological characteristics | ||||
| Adenocarcinoma | 173 (82.4%) | 71 (86.58%) | 102 (77.7%) | 0.379 |
| Squamous cell carcinoma | 17 (7.9%) | 5 (6.09%) | 12 (9.09%) | 0.379 |
| Adenosquamous carcinoma | 14 (6.54%) | 3 (3.65%) | 11 (8.33%) | 0.379 |
| Sarcomatoid carcinoma | 3 (1.4%) | 2 (2.43%) | 1 (0.75%) | 0.379 |
| Large cell carcinoma | 3 (1.4%) | 1 (1.21%) | 2 (1.51%) | 0.379 |
| Histology Type | Number of Samples | Exon |
|---|---|---|
| Adenocarcinoma | 71 | 18, 19, 20, 21 |
| Squamous cell carcinoma | 5 | 18, 19, 21 |
| Adenosquamous carcinoma | 3 | 18, 20, 21 |
| Large cell carcinoma | 1 | 19 |
| Sarcomatoid carcinoma | 3 | 18, 21 |
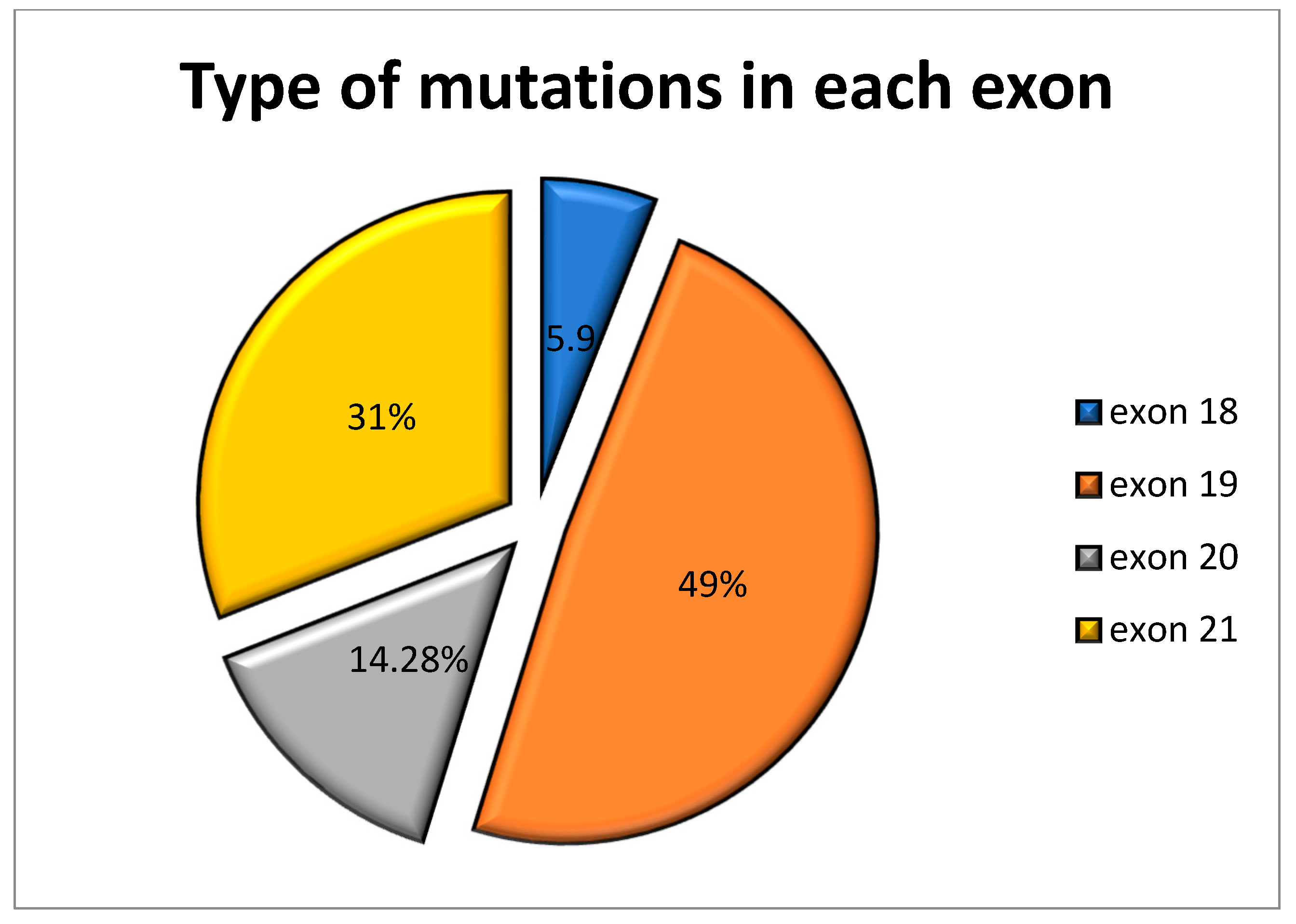
| Exon No. | Number of Mutations | cDNA Change | Amino Acid Change | Type of Mutation | Metastasis |
|---|---|---|---|---|---|
| 18 | 1 | c.2156G>C | Gly719Ala | Substitution | No |
| 18 | 1 | c.2155G>A | Gly719Ser | Substitution | No |
| 18 | 1 | c.2134T>C | Phe712Leu | Substitution | No |
| 18 | 1 | c.2155G>T | Gly719Cys | Substitution | No |
| 19 | 7 | c.2235_2249del | Glu746_Ala750 | Deletion | No |
| 19 | 7 | c.2236_2250del | Glu746_Ala750 | Deletion | No |
| 19 | 1 | c.2236_2252del | E746_T751 | Deletion | No |
| 19 | 2 | c.2239_2248delinsC | Leu747_Ala750>Pro | Deletion and Insertion | No |
| 19 | 1 | c.2240_2257del | Leu747_Pro753 | Deletion | Yes |
| 19 | 1 | c.2240_2267delinsGCCAA | Leu747_Ala755 | Deletion and Insertion | Yes |
| 19 | 1 | c.2258 C>T | Pro753Leu | Substitution | No |
| 19 | 1 | c.2240_2254del | Leu747_Thr751 | Deletion | Yes |
| 20 | 4 | c.2303G>T | S768I | Substitution | Yes |
| 20 | 1 | c.2290_2291insTCCGGGAAGCCT | Ala763_Tyr764insPheArgGluAla | Substitution | Yes |
| 20 | 1 | c.2314C>T | Pro772Ser | Insertion | No |
| 20 | 1 | c.2362 C>T | Leu788Phe | Substitution | No |
| 20 | 2 | c.2369C>T | T790M | Substitution | No |
| 21 | 23 | c.2573T>G | L858R | Substitution | No/Yes |
| 21 | 2 | c.2582T>A | L861Q | Substitution | Yes |
| Frequency of EGFR Negative Mutation (n = 50) | Frequency of EGFR Positive Mutation (n = 28) | Total (n = 78) | ||||
|---|---|---|---|---|---|---|
| Total | Percentage (%) | Total | Percentage (%) | Total | Percentage (%) | |
| Lymph node | 23 | 46 | 12 | 42.9 | 35 | 44.9 |
| Multiple sites | 10 | 20 | 5 | 17.9 | 15 | 19.2 |
| Pleural fluid | 7 | 14 | 2 | 7.1 | 9 | 11.5 |
| Brain | 2 | 4 | 6 | 21.4 | 8 | 10.3 |
| Bone | 3 | 6 | 2 | 7.1 | 5 | 6.4 |
| Spinal | 2 | 4 | - | - | 2 | 2.6 |
| Femur | - | - | 1 | 3.6 | 1 | 1.3 |
| Liver | 1 | 2 | - | - | 1 | 1.3 |
| Lung | 1 | 2 | - | - | 1 | 1.3 |
| Pericardial fluid | 1 | 2 | - | - | 1 | 1.3 |
| Exon No. | cDNA Change | Amino Acid Change | Type of Mutation | Number of Samples |
|---|---|---|---|---|
| 19 | c.2235_2249del | Glu746_Ala750 | deletion | 1 |
| 19 | c.2236_2250del | Glu746_Ala750 | deletion | 1 |
| 19 | c.2239_2248delinsC | Leu747_Ala750>Pro | deletion and insertion | 1 |
| 21 | c.2573T>G | L858R | substitution | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaler, A.K.; Patel, K.; Patil, H.; Tiwarekar, Y.; Kulkarni, B.; Hastak, M.; Athikari, N.; Rane, S.; Nikam, A.; Umarji, S.; et al. Mutational Analysis of EGFR Mutations in Non-Small Cell Lung Carcinoma—An Indian Perspective of 212 Patients. Int. J. Environ. Res. Public Health 2023, 20, 758. https://doi.org/10.3390/ijerph20010758
Kaler AK, Patel K, Patil H, Tiwarekar Y, Kulkarni B, Hastak M, Athikari N, Rane S, Nikam A, Umarji S, et al. Mutational Analysis of EGFR Mutations in Non-Small Cell Lung Carcinoma—An Indian Perspective of 212 Patients. International Journal of Environmental Research and Public Health. 2023; 20(1):758. https://doi.org/10.3390/ijerph20010758
Chicago/Turabian StyleKaler, Amrit Kaur, Khushi Patel, Harshali Patil, Yash Tiwarekar, Bijal Kulkarni, Meenal Hastak, Nivetha Athikari, Samrudhi Rane, Ankita Nikam, Smita Umarji, and et al. 2023. "Mutational Analysis of EGFR Mutations in Non-Small Cell Lung Carcinoma—An Indian Perspective of 212 Patients" International Journal of Environmental Research and Public Health 20, no. 1: 758. https://doi.org/10.3390/ijerph20010758
APA StyleKaler, A. K., Patel, K., Patil, H., Tiwarekar, Y., Kulkarni, B., Hastak, M., Athikari, N., Rane, S., Nikam, A., Umarji, S., Shaikh, I., Goyle, S., & Mistry, R. (2023). Mutational Analysis of EGFR Mutations in Non-Small Cell Lung Carcinoma—An Indian Perspective of 212 Patients. International Journal of Environmental Research and Public Health, 20(1), 758. https://doi.org/10.3390/ijerph20010758






