Evaluation Study of xMAP TIER Assay on a Microsphere-Based Platform for Detecting First-Line Anti-Tuberculosis Drug Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Mycobacteria tuberculosis Strain Collection and Culture
2.2. Phenotypic Broth Microdilution Drug Susceptibility Test
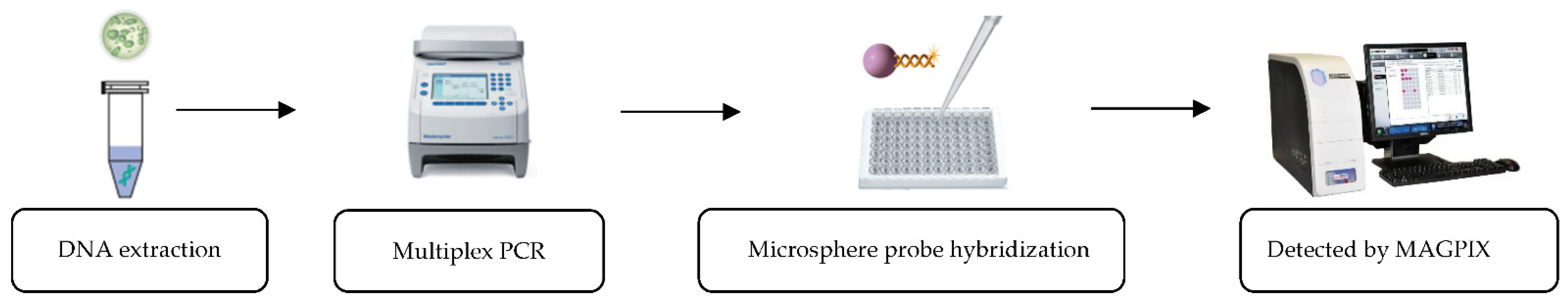
2.3. xMAP TIER Assay
2.4. Whole Genome Sequencing (WGS)
2.5. Statistical Analysis
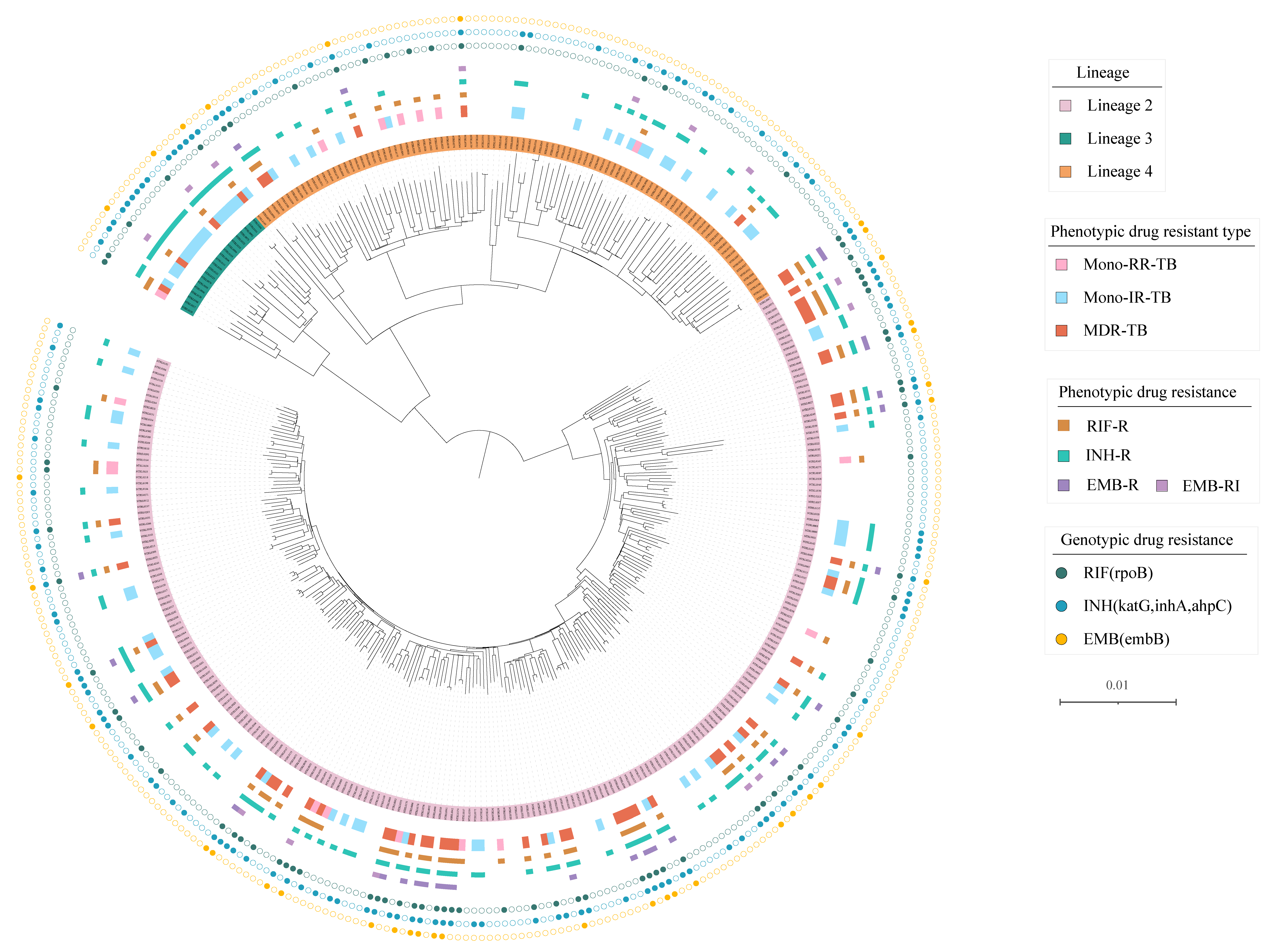
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report. 2021. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2021 (accessed on 27 October 2022).
- Zhao, Y.; Xu, S.; Wang, L.; Chin, D.P.; Wang, S.; Jiang, G.; Xia, H.; Zhou, Y.; Li, Q.; Ou, X.; et al. National survey of drug-resistant tuberculosis in China. N. Engl. J. Med. 2012, 366, 2161–2170. [Google Scholar] [CrossRef] [PubMed]
- Kapur, V.; Li, L.L.; Iordanescu, S.; Hamrick, M.R.; Wanger, A.; Kreiswirth, B.N.; Musser, J.M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 1994, 32, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Musser, J.M.; Kapur, V.; Williams, D.L.; Kreiswirth, B.N.; van Soolingen, D.; van Embden, J.D. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J. Infect. Dis. 1996, 173, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.V.; Amin, A.G.; Göksel, S.; Stager, C.E.; Dou, S.J.; El Sahly, H.; Moghazeh, S.L.; Kreiswirth, B.N.; Musser, J.M. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2000, 44, 326–336. [Google Scholar] [CrossRef]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef]
- Wang, S.F.; Ou, X.C.; Li, Q.; Zheng, H.W.; Wang, Y.F.; Zhao, Y.L. The Abbott RealTime MTB assay and the Cepheid GeneXpert assay show comparable performance for the detection of Mycobacterium tuberculosis in sputum specimens. Int. J. Infect. Dis. 2016, 45, 78–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ou, X.; Li, Q.; Su, D.; Xia, H.; Wang, S.; Zhao, B.; Zhao, Y. A pilot study: VereMTB detection kit for rapid detection of multidrug-resistant mycobcterium tuberculosis in clinical sputum samples. PLoS ONE 2020, 15, e0228312. [Google Scholar] [CrossRef]
- Penn-Nicholson, A.; Georghiou, S.B.; Ciobanu, N.; Kazi, M.; Bhalla, M.; David, A.; Conradie, F.; Ruhwald, M.; Crudu, V.; Rodrigues, C.; et al. Detection of isoniazid, fluoroquinolone, ethionamide, amikacin, kanamycin, and capreomycin resistance by the Xpert MTB/XDR assay: A cross-sectional multicentre diagnostic accuracy study. Lancet Infect. Dis. 2022, 22, 242–249. [Google Scholar] [CrossRef]
- WHO. Consolidated Guidelines on Tuberculosis Module 3: Diagnosis—Rapid Diagnostics for Tuberculosis Detection; World Health Organization: Geneva, Switzerland, 2021. Available online: https://apps.who.int/iris/bitstream/handle/10665/342331/9789240029415-eng.pdf (accessed on 7 July 2021).
- Yin, F.; Chan, J.F.; Zhu, Q.; Fu, R.; Chen, J.H.; Choi, G.K.; Tee, K.M.; Li, L.; Qian, S.; Yam, W.C.; et al. Development and in-use evaluation of a novel Luminex MicroPlex microsphere-based (TRIOL) assay for simultaneous identification of Mycobacterium tuberculosis and detection of first-line and second-line anti-tuberculous drug resistance in China. J. Clin. Pathol. 2017, 70, 342–349. [Google Scholar] [CrossRef]
- Liu, C.F.; Song, Y.M.; He, P.; Liu, D.X.; He, W.C.; Li, Y.M.; Zhao, Y.L. Evaluation of Multidrug Resistant Loop-mediated Isothermal Amplification Assay for Detecting the Drug Resistance of Mycobacterium tuberculosis. Biomed. Environ. Sci. 2021, 34, 616–622. [Google Scholar] [CrossRef]
- Liu, D.; Huang, F.; Zhang, G.; He, W.; Ou, X.; He, P.; Zhao, B.; Zhu, B.; Liu, F.; Li, Z.; et al. Whole-genome sequencing for surveillance of tuberculosis drug resistance and determination of resistance level in China. Clin. Microbiol. Infect. 2022, 28, 731.e9–731.e15. [Google Scholar] [CrossRef]
- He, W.; Tan, Y.; Liu, C.; Wang, Y.; He, P.; Song, Z.; Liu, D.; Zheng, H.; Ma, A.; Zhao, B.; et al. Drug-Resistant Characteristics, Genetic Diversity, and Transmission Dynamics of Rifampicin-Resistant Mycobacterium tuberculosis in Hunan, China, Revealed by Whole-Genome Sequencing. Microbiol. Spectr. 2022, 10, e0154321. [Google Scholar] [CrossRef] [PubMed]
- Catalogue of Mutations in Mycobacterium Tuberculosis Complex and Their Association with Drug Resistance; World Health Organization: Geneva, Switzerland, 2021.
- Walker, T.M.; Miotto, P.; Köser, C.U.; Fowler, P.W.; Knaggs, J.; Iqbal, Z.; Hunt, M.; Chindelevitch, L.; Farhat, M.; Cirillo, D.M.; et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: A genotypic analysis. Lancet Microbe 2022, 3, e265–e273. [Google Scholar] [CrossRef] [PubMed]
- Kik, S.V.; Qin, Z.Z.; Pai, M. Optimal Diagnosis: How Early and Improved Diagnosis Can Help Prevent TB Transmission, Clinical Insights: Tuberculosis Prevention; Future Medicine Ltd.: London, UK, 2014; pp. 7–32. [Google Scholar]
- Dunbar, S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 2006, 363, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M.; Grindle, K.; Pappas, T.; Marshall, D.J.; Moser, M.J.; Beaty, E.L.; Shult, P.A.; Prudent, J.R.; Gern, J.E. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 2007, 45, 2626–2634. [Google Scholar] [CrossRef]
- High-Priority Target Product Profiles for New Tuberculosis Diagnostics: Report of a Consensus Meeting; World Health Organization: Geneva, Switzerland, 2014. Available online: https://www.who.int/publications/i/item/WHO-HTM-TB-2014.18 (accessed on 5 August 2014).
- Van Ingen, J.; Aarnoutse, R.; de Vries, G.; Boeree, M.J.; van Soolingen, D. Low-level rifampicin-resistant Mycobacterium tuberculosis strains raise a new therapeutic challenge. Int. J. Tuberc. Lung Dis. 2011, 15, 990–992. [Google Scholar] [CrossRef]
- Van Deun, A.; Aung, K.J.; Hossain, A.; de Rijk, P.; Gumusboga, M.; Rigouts, L.; de Jong, B.C. Disputed rpoB mutations can frequently cause important rifampicin resistance among new tuberculosis patients. Int. J. Tuberc. Lung Dis. 2015, 19, 185–190. [Google Scholar] [CrossRef]
- Xia, H.; Song, Y.; Zheng, Y.; Wang, S.; Zhao, B.; He, W.; Liu, D.; Ou, X.; Zhou, Y.; Zhao, Y. Detection of Mycobacterium tuberculosis Rifampicin Resistance Conferred by Borderline rpoB Mutations: Xpert MTB/RIF is Superior to Phenotypic Drug Susceptibility Testing. Infect. Drug Resist. 2022, 15, 1345–1352. [Google Scholar] [CrossRef]
- Seifert, M.; Catanzaro, D.; Catanzaro, A.; Rodwell, T.C. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: A systematic review. PLoS ONE 2015, 10, e0119628. [Google Scholar] [CrossRef]
- Maningi, N.E.; Malinga, L.A.; Antiabong, J.F.; Lekalakala, R.M.; Mbelle, N.M. Comparison of line probe assay to BACTEC MGIT 960 system for susceptibility testing of first and second-line anti-tuberculosis drugs in a referral laboratory in South Africa. BMC Infect. Dis. 2017, 17, 795. [Google Scholar] [CrossRef]
- Chandak, R.J.; Malhotra, B.; Bhargava, S.; Goel, S.K.; Verma, D.; Tiwari, J. Evaluation of MTBDRsl for detecting resistance in Mycobacterium tuberculosis to second-line drugs. Int. J. Tuberc. Lung Dis. 2019, 23, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
| Drug | Gene | Wild Type | Mutation |
|---|---|---|---|
| INH | |||
| katG | 315AGC | 315ACC | |
| inhA | -777C | -777T | |
| -770T | -770C | ||
| RIF | |||
| rpoB | 430CTG | 430CCG | |
| rpoB | 432CAA | 432AAA or 432CAA | |
| rpoB | 435GAC | 435GTC or 435GGC | |
| rpoB | 445CAC | 445TAC or 445GAC | |
| rpoB | 450TCG | 450TTG | |
| rpoB | 452CTG | 152CCG | |
| EMB | embB | 306ATG | 306GTG, 306CTG, 306ATA, 306ATT or 306ATC |
| Drugs | Phenotypically DST Resistant | Phenotypically DST Susceptible | Sensitivity (95%CI) | Specificity (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|
| xMAP Resistant | xMAP Susceptible | Total | xMAP Resistant | xMAP Susceptible | Total | |||
| Rifampicin | 74 | 4 a | 78 | 3 b | 272 | 275 | 94.9 (87.4–98.6) | 98.9 (96.9–99.8) |
| Isoniazid | 123 | 15 c | 138 | 0 | 215 | 215 | 89.1 (82.7–93.8) | 100.0 (98.3–100.0) |
| Ethambutol | 23 | 5 d | 28 | 1 e | 310 | 311 | 82.1 (63.1–93.9) | 99.7 (98.2–100.0) |
| Drugs | Genotypically Resistant by WGS | Genotypically Susceptible by WGS | Sensitivity (95%CI) | Specificity (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|
| xMAP Resistant | xMAP Susceptible | Total | xMAP Resistant | xMAP Susceptible | Total | |||
| Rifampicin | 76 | 4 | 80 | 1 | 272 | 273 | 95.0 (87.7–98.6) | 99.6 (98.0–100.0) |
| Isoniazid | 123 | 4 | 127 | 0 | 226 | 226 | 96.9 (93.8–99.9) | 100.0 (100.0–100.0) |
| Ethambutol | 31 | 5 | 36 | 0 | 317 | 317 | 86.1 (70.5–95.3) | 100.0 (98.8–100.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, X.; Zhang, Z.; Zhao, B.; Song, Z.; Wang, S.; He, W.; Pei, S.; Liu, D.; Xing, R.; Xia, H.; et al. Evaluation Study of xMAP TIER Assay on a Microsphere-Based Platform for Detecting First-Line Anti-Tuberculosis Drug Resistance. Int. J. Environ. Res. Public Health 2022, 19, 17068. https://doi.org/10.3390/ijerph192417068
Ou X, Zhang Z, Zhao B, Song Z, Wang S, He W, Pei S, Liu D, Xing R, Xia H, et al. Evaluation Study of xMAP TIER Assay on a Microsphere-Based Platform for Detecting First-Line Anti-Tuberculosis Drug Resistance. International Journal of Environmental Research and Public Health. 2022; 19(24):17068. https://doi.org/10.3390/ijerph192417068
Chicago/Turabian StyleOu, Xichao, Zhiguo Zhang, Bing Zhao, Zexuan Song, Shengfen Wang, Wencong He, Shaojun Pei, Dongxin Liu, Ruida Xing, Hui Xia, and et al. 2022. "Evaluation Study of xMAP TIER Assay on a Microsphere-Based Platform for Detecting First-Line Anti-Tuberculosis Drug Resistance" International Journal of Environmental Research and Public Health 19, no. 24: 17068. https://doi.org/10.3390/ijerph192417068
APA StyleOu, X., Zhang, Z., Zhao, B., Song, Z., Wang, S., He, W., Pei, S., Liu, D., Xing, R., Xia, H., & Zhao, Y. (2022). Evaluation Study of xMAP TIER Assay on a Microsphere-Based Platform for Detecting First-Line Anti-Tuberculosis Drug Resistance. International Journal of Environmental Research and Public Health, 19(24), 17068. https://doi.org/10.3390/ijerph192417068






