Transcriptome Analysis of Acinetobacter baumannii in Rapid Response to Subinhibitory Concentration of Minocycline
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions, and Susceptibility Testing
2.2. Total RNA Extraction and rRNA Removal
2.3. High-Throughput Sequencing cDNA Library Construction and RNA-Seq Sequencing
2.4. Transcriptome Analysis
2.5. Candidate sRNA Identification and RT-qPCR Validation
2.6. Statistical Analysis
3. Results
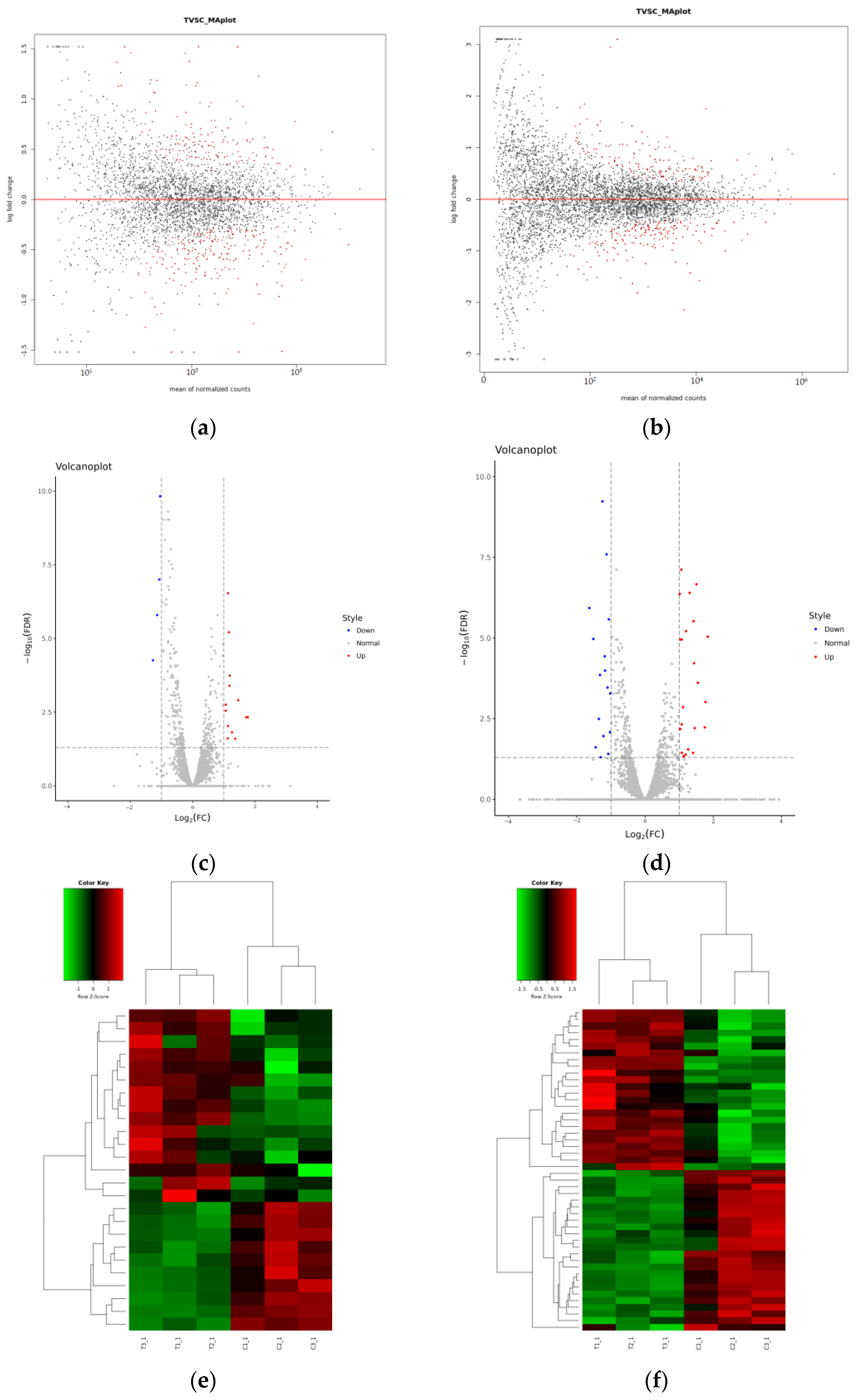
3.1. Transcriptome Analysis of A. baumannii Strain ATCC19606
3.2. Differentially Expressed mRNA and sRNA
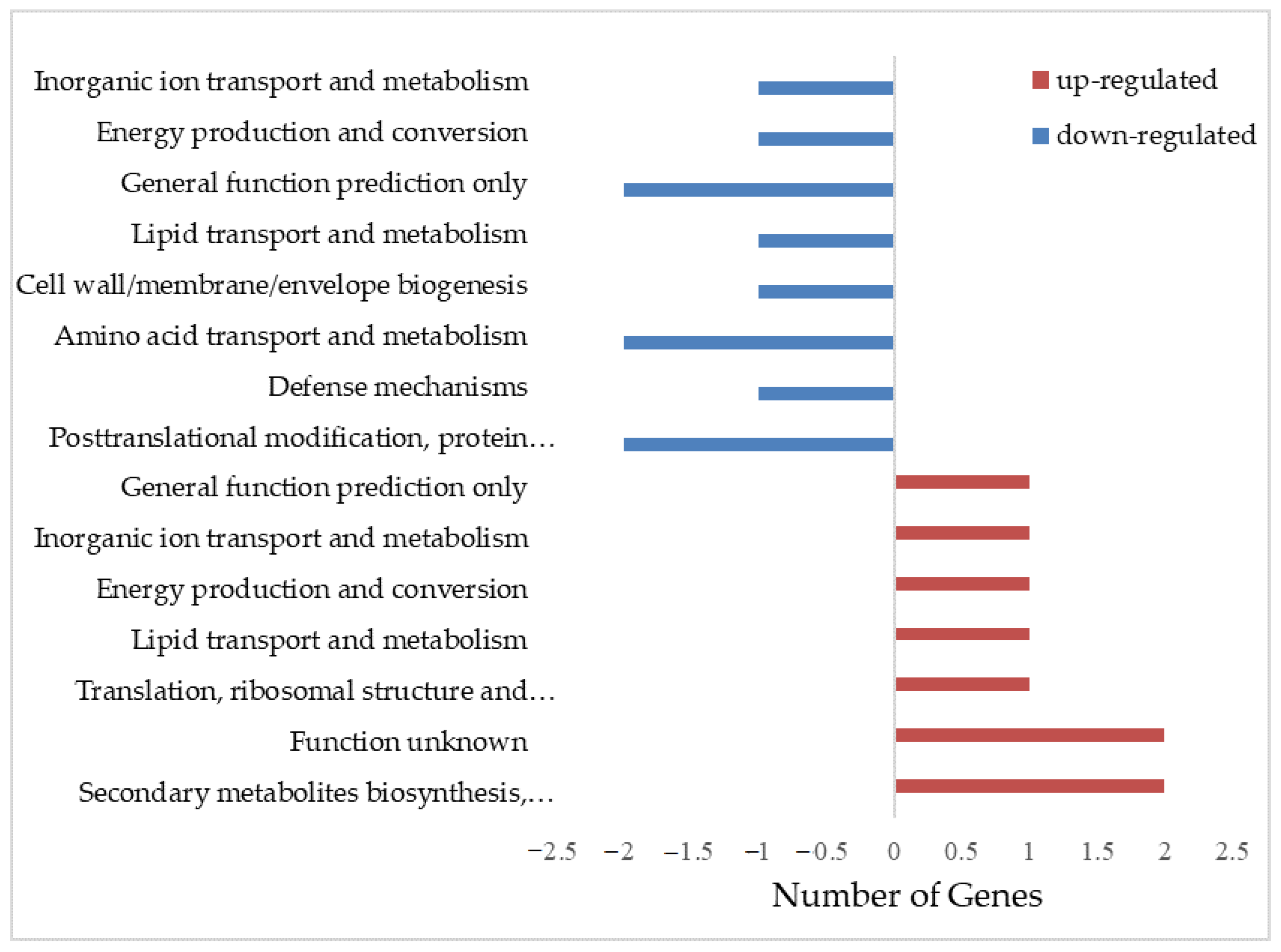
3.3. Functional Annotation of Differentially Expressed Genes
3.4. Genes Upregulated by Minocycline Treatment
3.5. Genes Downregulated by Minocycline Treatment
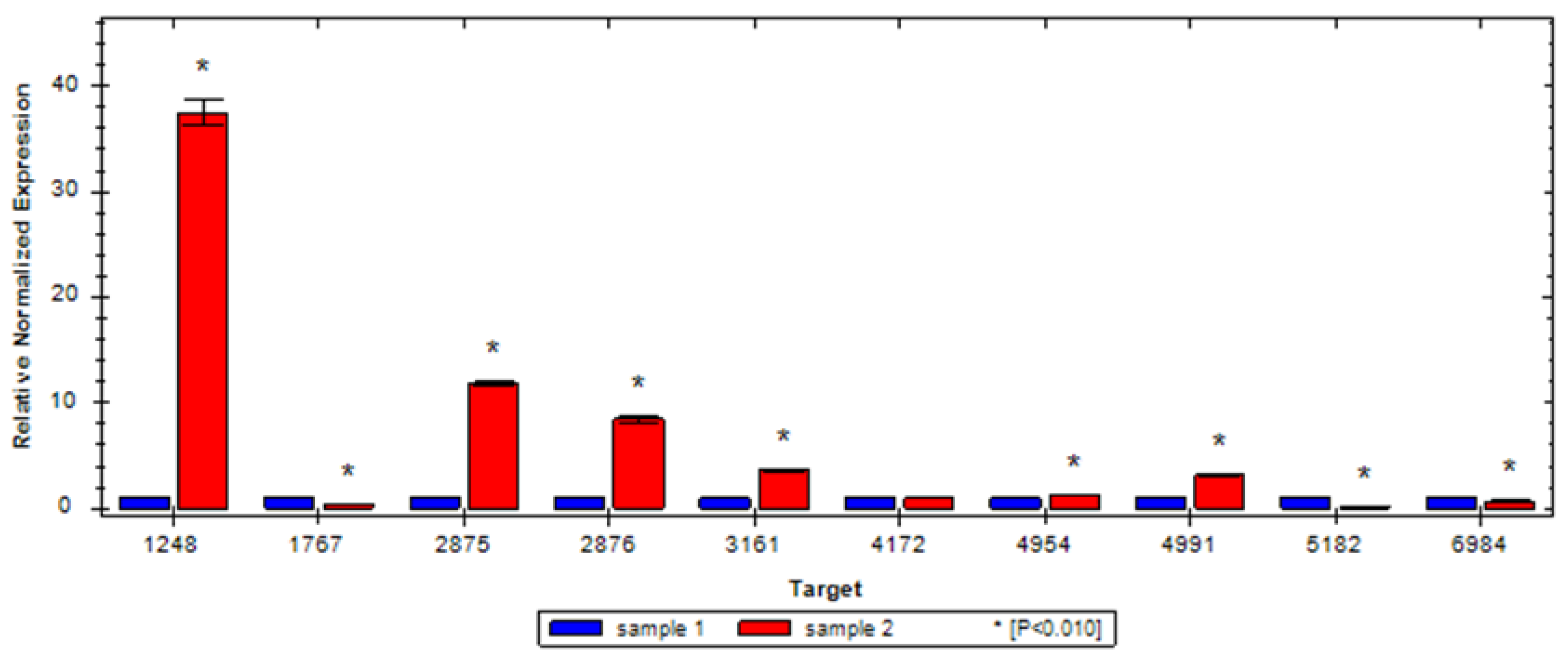
3.6. Validation of sRNA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Towner, K.J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 2009, 73, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Maboni, G.; Seguel, M.; Lorton, A.; Sanchez, S. Antimicrobial resistance patterns of Acinetobacter spp. of animal origin reveal high rate of multidrug resistance. Vet. Microbiol. 2020, 245, 108702. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Xu, H.; Jing, C.; Deng, J.; Wang, H.; Hua, C.; Chen, Y.; Chen, X.; Zhang, T.; Zhang, H.; et al. Bacterial Epidemiology and Antimicrobial Resistance Profiles in Children Reported by the ISPED Program in China, 2016 to 2020. Microbiol. Spectr. 2021, 9, e0028321. [Google Scholar] [CrossRef] [PubMed]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-Treat Resistance in Gram-negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-line Agents. Clin. Infect. Dis. 2018, 67, 1803–1814. [Google Scholar] [CrossRef]
- Kümmerer, K. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 2003, 52, 5–7. [Google Scholar] [CrossRef]
- Goff, D.A.; Kaye, K.S. Minocycline: An old drug for a new bug: Multidrug-resistant Acinetobacter baumannii. Clin. Infect. Dis. 2014, 59 (Suppl. 6), S365–S366. [Google Scholar] [CrossRef]
- Fragkou, P.C.; Poulakou, G.; Blizou, A.; Blizou, M.; Rapti, V.; Karageorgopoulos, D.E.; Koulenti, D.; Papadopoulos, A.; Matthaiou, D.K.; Tsiodras, S. The Role of Minocycline in the Treatment of Nosocomial Infections Caused by Multidrug, Extensively Drug and Pandrug Resistant Acinetobacter baumannii: A Systematic Review of Clinical Evidence. Microorganisms 2019, 7, 159. [Google Scholar] [CrossRef]
- Garrido-Mesa, N.; Zarzuelo, A.; Gálvez, J. Minocycline: Far beyond an antibiotic. Br. J. Pharm. 2013, 169, 337–352. [Google Scholar] [CrossRef]
- Ku, N.S.; Lee, S.-H.; Lim, Y.-S.; Choi, H.; Ahn, J.Y.; Jeong, S.J.; Shin, S.J.; Choi, J.Y.; Choi, Y.H.; Yeom, J.-S.; et al. In vivo efficacy of combination of colistin with fosfomycin or minocycline in a mouse model of multidrug-resistant Acinetobacter baumannii pneumonia. Sci. Rep. 2019, 9, 17127. [Google Scholar] [CrossRef]
- Asadi, A.; Abdi, M.; Kouhsari, E.; Panahi, P.; Sholeh, M.; Sadeghifard, N.; Amiriani, T.; Ahmadi, A.; Maleki, A.; Gholami, M. Minocycline, focus on mechanisms of resistance, antibacterial activity, and clinical effectiveness: Back to the future. J. Glob. Antimicrob. Resist. 2020, 22, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Flamm, R.K.; Shortridge, D.; Castanheira, M.; Sader, H.S.; Pfaller, M.A. Activity of Minocycline against U.S. Isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus Species Complex, Stenotrophomonas maltophilia, and Burkholderia cepacia Complex: Results from the SENTRY Antimicrobial Surveillance Program, 2014 to 2018. Antimicrob. Agents Chemother. 2019, 63, e01154-19. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Cnockaert, M.; Vaneechoutte, M.; Woodford, N.; Nemec, A.; Dijkshoorn, L.; Swings, J. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 2005, 156, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lu, J.; Zhao, J.; Zhang, X.; Yu, H.H.; Velkov, T.; Li, J. Complete genome sequence and genome-scale metabolic modelling of Acinetobacter baumannii type strain ATCC 19606. Int. J. Med. Microbiol. 2020, 310, 151412. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Sun, X.; Li, M.; Wang, Y.; Jiao, H.; Li, G. The Involvement of the csy1 Gene in the Antimicrobial Resistance of Acinetobacter baumannii. Front. Med. 2022, 9, 797104. [Google Scholar] [CrossRef]
- Hui, Z.; Liu, S.; Cui, R.; Zhou, B.; Hu, C.; Zhang, M.; Deng, Q.; Cheng, S.; Luo, Y.; Chen, H.; et al. A small molecule interacts with pMAC-derived hydroperoxide reductase and enhances the activity of aminoglycosides. J. Antibiot. 2021, 74, 324–329. [Google Scholar] [CrossRef]
- Hamidian, M.; Hall, R.M. Acinetobacter baumannii ATCC 19606 Carries GIsul2 in a Genomic Island Located in the Chromosome. Antimicrob. Agents Chemother. 2017, 61, e01991-16. [Google Scholar] [CrossRef]
- Yang, H.; Chen, G.; Hu, L.; Liu, Y.; Cheng, J.; Li, H.; Ye, Y.; Li, J. In vivo activity of daptomycin/colistin combination therapy in a Galleria mellonella model of Acinetobacter baumannii infection. Int. J. Antimicrob. Agents 2015, 45, 188–191. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Lewis, J.S., 2nd. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: Background, Organization, Functions, and Processes. J. Clin. Microbiol. 2020, 58, e01864-19. [Google Scholar] [CrossRef]
- Brady, M.F.; Jamal, Z.; Pervin, N. Acinetobacter. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Laureti, L.; Matic, I.; Gutierrez, A. Bacterial Responses and Genome Instability Induced by Subinhibitory Concentrations of Antibiotics. Antibiotics 2013, 2, 100–114. [Google Scholar] [CrossRef]
- Singh, S.; Khanna, D.; Kalra, S. Minocycline and Doxycycline: More Than Antibiotics. Curr. Mol. Pharm. 2021, 14, 1046–1065. [Google Scholar] [CrossRef] [PubMed]
- Manwar, M.R.; Shao, C.; Shi, X.; Wang, J.; Lin, Q.; Tong, Y.; Kang, Y.; Yu, J. The bacterial RNA ligase RtcB accelerates the repair process of fragmented rRNA upon releasing the antibiotic stress. Sci. China Life Sci. 2020, 63, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Englert, M.; Sheppard, K.; Aslanian, A.; Yates, J.R., 3rd; Söll, D. Archaeal 3’-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. USA 2011, 108, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Shuman, S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011, 286, 7727–7731. [Google Scholar] [CrossRef]
- Temmel, H.; Müller, C.; Sauert, M.; Vesper, O.; Reiss, A.; Popow, J.; Martinez, J.; Moll, I. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 2017, 45, 4708–4721. [Google Scholar]
- Lin, C.; Su, S.; Ho, K.; Hsu, Y.; Lee, K. Bactericidal effect of sulbactam against Acinetobacter baumannii ATCC 19606 studied by 2D-DIGE and mass spectrometry. Int. J. Antimicrob. Agents 2014, 44, 38–46. [Google Scholar] [CrossRef]
- Hua, X.; Chen, Q.; Li, X.; Yu, Y. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int. J. Antimicrob. Agents 2014, 44, 337–344. [Google Scholar] [CrossRef]
- Sivinski, J.; Ambrose, A.J.; Panfilenko, I.; Zerio, C.J.; Machulis, J.M.; Mollasalehi, N.; Kaneko, L.K.; Stevens, M.; Ray, A.M.; Park, Y.; et al. Functional Differences between E. coli and ESKAPE Pathogen GroES/GroEL. mBio 2021, 12, e02167-20. [Google Scholar] [CrossRef]
- Stevens, M.; Abdeen, S.; Salim, N.; Ray, A.M.; Washburn, A.; Chitre, S.; Sivinski, J.; Park, Y.; Hoang, Q.Q.; Chapman, E.; et al. HSP60/10 chaperonin systems are inhibited by a variety of approved drugs, natural products, and known bioactive molecules. Bioorg. Med. Chem. Lett. 2019, 29, 1106–1112. [Google Scholar] [CrossRef]
- Abdeen, S.; Salim, N.; Mammadova, N.; Summers, C.M.; Frankson, R.; Ambrose, A.J.; Anderson, G.G.; Schultz, P.G.; Horwich, A.L.; Chapman, E.; et al. GroEL/ES inhibitors as potential antibiotics. Bioorg. Med. Chem. Lett. 2016, 26, 3127–3134. [Google Scholar] [CrossRef]
- Stevens, M.; Howe, C.; Ray, A.M.; Washburn, A.; Chitre, S.; Sivinski, J.; Park, Y.; Hoang, Q.Q.; Chapman, E.; Johnson, S.M. Analogs of nitrofuran antibiotics are potent GroEL/ES inhibitor pro-drugs. Bioorg. Med. Chem. 2020, 28, 115710. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, K.; Gandra, R.F.; Wisniewski, E.S.; Osaku, C.A.; Kadowaki, M.K.; Felipach-Neto, V.; Haus, L.F.A.; Simão, R.C.G. DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J. Med. Microbiol. 2010, 59 Pt 9, 1061–1068. [Google Scholar] [CrossRef]
- Schildkraut, J.A.; Coolen, J.P.M.; Burbaud, S.; Sangen, J.J.N.; Kwint, M.P.; Floto, R.A.; Op den Camp, H.J.M.; Te Brake, L.H.M.; Wertheim, H.F.L.; Neveling, K.; et al. RNA Sequencing Elucidates Drug-Specific Mechanisms of Antibiotic Tolerance and Resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2022, 66, e0150921. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Chu, Z.; Lu, J.; Li, D.; Wang, Y.; Yang, S.; Zhang, Y. Heterologous Expression of Chaperones from Hyperthermophilic Archaea Inhibits Aminoglycoside-Induced Protein Misfolding in Escherichia coli. Biochemistry 2017, 82, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Schneiders, T. Tigecycline challenge triggers sRNA production in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2012, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Hämmerle, H.; Večerek, B.; Resch, A.; Bläsi, U. Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature. RNA Biol. 2013, 10, 1834–1841. [Google Scholar] [CrossRef][Green Version]
- Sharma, R.; Arya, S.; Patil, S.D.; Sharma, A.; Jain, P.K.; Navani, N.K.; Pathania, R. Identification of novel regulatory small RNAs in Acinetobacter baumannii. PLoS ONE 2014, 9, e93833. [Google Scholar] [CrossRef]
- Álvarez-Fraga, L.; Rumbo-Feal, S.; Pérez, A.; Gómez, M.J.; Gayoso, C.; Vallejo, J.A.; Ohneck, E.J.; Valle, J.; Actis, L.A.; Beceiro, A.; et al. Global assessment of small RNAs reveals a non-coding transcript involved in biofilm formation and attachment in Acinetobacter baumannii ATCC 17978. PLoS ONE 2017, 12, e0182084. [Google Scholar] [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar]
- Kröger, C.; MacKenzie, K.D.; Alshabib, E.Y.; Kirzinger, M.W.B.; Suchan, D.M.; Chao, T.C.; Akulova, V.; Miranda-CasoLuengo, A.A.; Monzon, V.A.; Conway, T.; et al. The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res. 2018, 46, 9684–9698. [Google Scholar] [CrossRef]
- Eyraud, A.; Tattevin, P.; Chabelskaya, S.; Felden, B. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 2014, 42, 4892–4905. [Google Scholar] [CrossRef]
- Mediati, D.G.; Wu, S.; Wu, W.; Tree, J.J. Networks of Resistance: Small RNA Control of Antibiotic Resistance. Trends Genet. 2021, 37, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hoe, C.H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Schu, D.J.; Zhang, A.; Gottesman, S.; Storz, G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. Embo J. 2015, 34, 2557–2573. [Google Scholar] [CrossRef]
- Howden, B.P.; Beaume, M.; Harrison, P.F.; Hernandez, D.; Schrenzel, J.; Seemann, T.; Francois, P.; Stinear, T.P. Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob. Agents Chemother. 2013, 57, 3864–3874. [Google Scholar] [CrossRef] [PubMed]
| AccID | Gene Name | log2FC | Style | Description | Molecular Function |
|---|---|---|---|---|---|
| A6739_RS00130 | - | 1.261057061 | up | putative methionine/alanine importer small subunit | - |
| A6739_RS02970 | rtcB | 1.375621078 | up | RtcB family protein | GTP binding, metal ion binding, RNA ligase |
| A6739_RS03845 | groS | −1.587812857 | down | molecular chaperone GroES | ATP binding, ATP-dependent protein folding chaperone, isomerase, unfolded protein binding |
| A6739_RS03850 | groL | −1.236322333 | down | chaperonin GroL | ATP binding, ATP-dependent protein folding chaperone, isomerase, unfolded protein binding |
| entE | entE | 1.133073343 | up | “2,3-dihydroxybenzoate-AMP ligase” | (2,3-dihydroxybenzoyl) adenylate synthase, ATP binding, ligase |
| A6739_RS05090 | blaADC | −1.511731996 | down | class C beta-lactamase ADC-158 | beta-lactamase |
| A6739_RS05460 | - | 1.190591988 | up | hypothetical protein | - |
| A6739_RS05735 | mmsA | −1.046110868 | down | methylmalonate-semialdehyde dehydrogenase (CoA acylating) | malonate-semialdehyde dehydrogenase (acetylating), methylmalonate-semialdehyde dehydrogenase (acylating) |
| A6739_RS06410 | - | 1.065451376 | up | hypothetical protein | - |
| A6739_RS06520 | - | 1.183871644 | up | TonB-dependent siderophore receptor | siderophore uptake transmembrane transporter, signaling receptor |
| A6739_RS06535 | - | 3.500019666 | up | ABC transporter ATP-binding protein | ABC-type transporter, ATP binding, ATPase-coupled transmembrane transporter |
| A6739_RS07175 | - | 1.715529743 | up | hypothetical protein | - |
| A6739_RS07555 | amdA | 1.461758079 | up | amidase | amidase, indoleacetamide hydrolase |
| A6739_RS07560 | - | 1.364873266 | up | acyl-CoA dehydrogenase | oxidoreductase |
| A6739_RS07580 | - | 1.127894768 | up | aromatic-ring-hydroxylating dioxygenase subunit beta | dioxygenase |
| A6739_RS08375 | azr | −1.071338077 | down | FMN-dependent NADH-azoreductase | oxidoreductase |
| A6739_RS08405 | - | −1.139044967 | down | TIGR01244 family phosphatase | hydrolase, oxidoreductase |
| A6739_RS08410 | - | −1.273424919 | down | MBL fold metallo-hydrolase | beta-lactamase, protein dimerization, zinc ion binding |
| A6739_RS09455 | - | −1.043166606 | down | amino acid ABC transporter permease | transmembrane transporter |
| A6739_RS10785 | - | 1.769148258 | up | hypothetical protein | - |
| A6739_RS11485 | plaP | −1.747668925 | down | APC family permease | transmembrane transporter |
| A6739_RS11675 | - | 1.162127898 | up | hypothetical protein | - |
| A6739_RS12210 | nlpE | 1.518804309 | down | copper resistance protein NlpE | - |
| A6739_RS14015 | pntB | 1.06376377 | up | NAD(P) transhydrogenase subunit alpha | NAD(P)+ transhydrogenase, NAD(P) binding, protein dimerization |
| A6739_RS15535 | - | 1.132093844 | up | hypothetical protein | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Ma, X. Transcriptome Analysis of Acinetobacter baumannii in Rapid Response to Subinhibitory Concentration of Minocycline. Int. J. Environ. Res. Public Health 2022, 19, 16095. https://doi.org/10.3390/ijerph192316095
Gao L, Ma X. Transcriptome Analysis of Acinetobacter baumannii in Rapid Response to Subinhibitory Concentration of Minocycline. International Journal of Environmental Research and Public Health. 2022; 19(23):16095. https://doi.org/10.3390/ijerph192316095
Chicago/Turabian StyleGao, Lili, and Xiaochun Ma. 2022. "Transcriptome Analysis of Acinetobacter baumannii in Rapid Response to Subinhibitory Concentration of Minocycline" International Journal of Environmental Research and Public Health 19, no. 23: 16095. https://doi.org/10.3390/ijerph192316095
APA StyleGao, L., & Ma, X. (2022). Transcriptome Analysis of Acinetobacter baumannii in Rapid Response to Subinhibitory Concentration of Minocycline. International Journal of Environmental Research and Public Health, 19(23), 16095. https://doi.org/10.3390/ijerph192316095







