Cervical Microbiome in Women Infected with HPV16 and High-Risk HPVs
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Recruitment
2.2. Total Nucleic Acid Extraction from Cervical Specimens
2.3. The Study of the Bacterial and Fungal Communities
2.4. The Study of the Viral Community
2.5. Illumina Sequencing and Data Analysis for the Viral Community
2.6. Statistics
3. Results
3.1. Characteristics of Clinical Samples
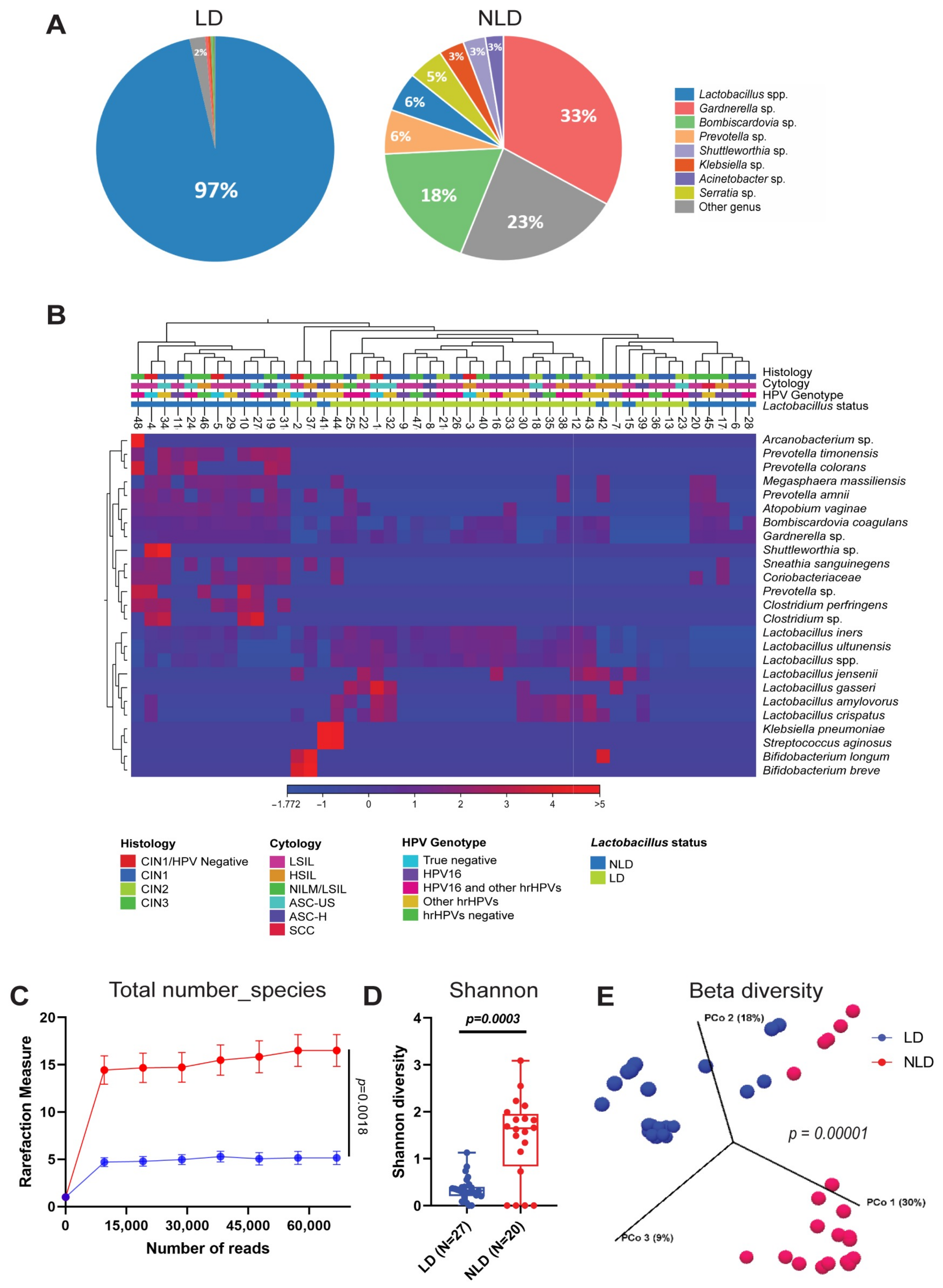
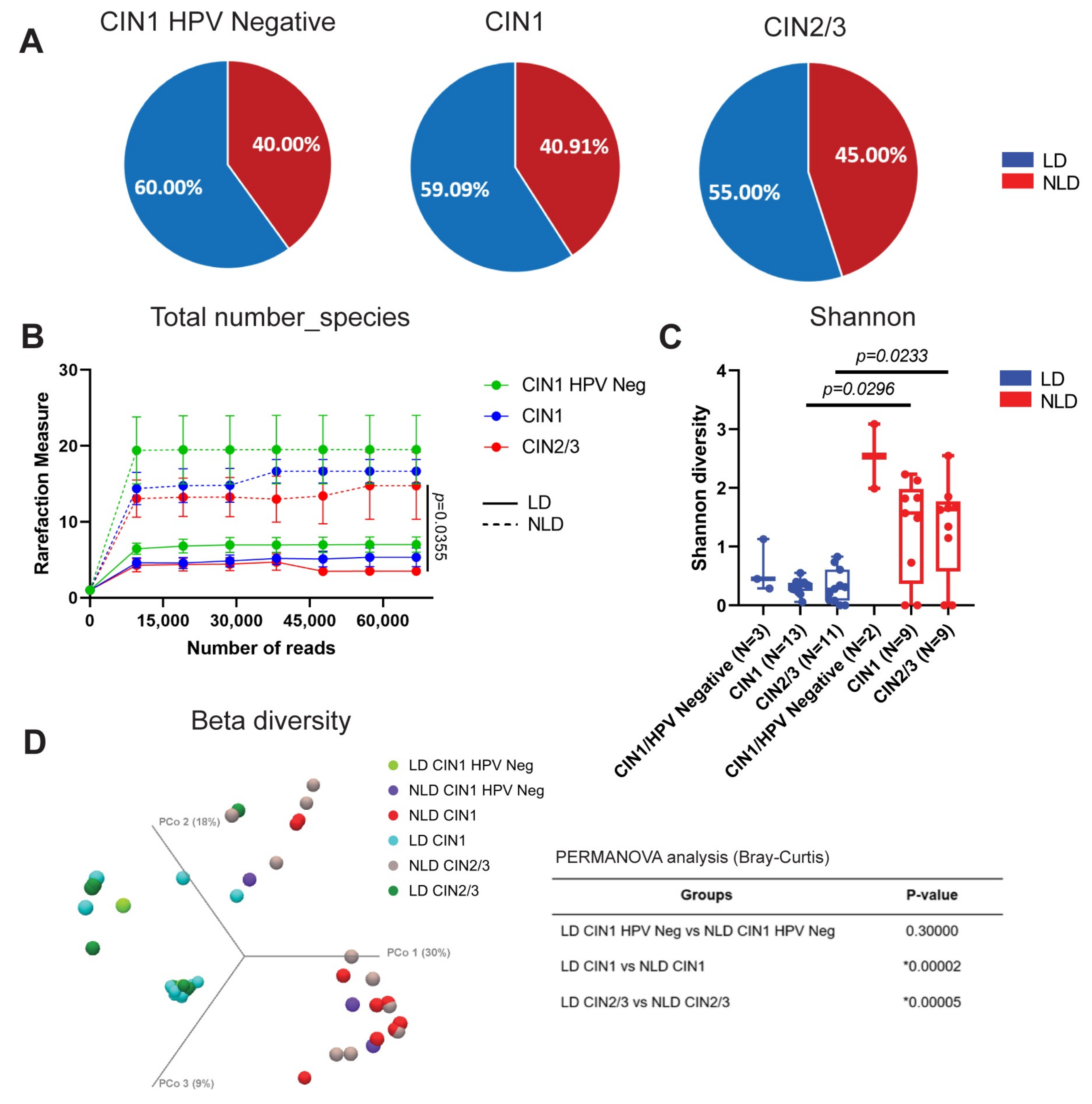
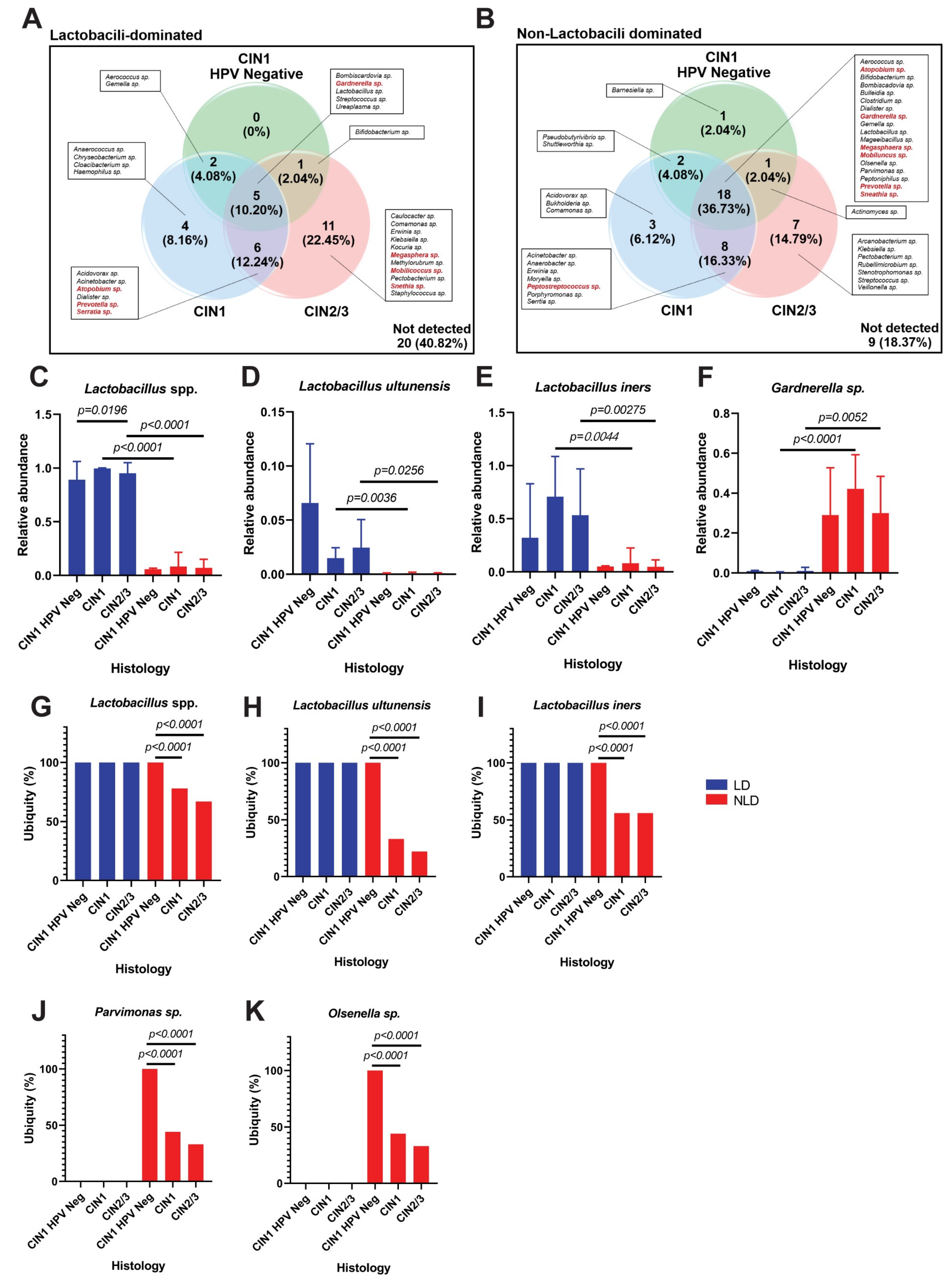
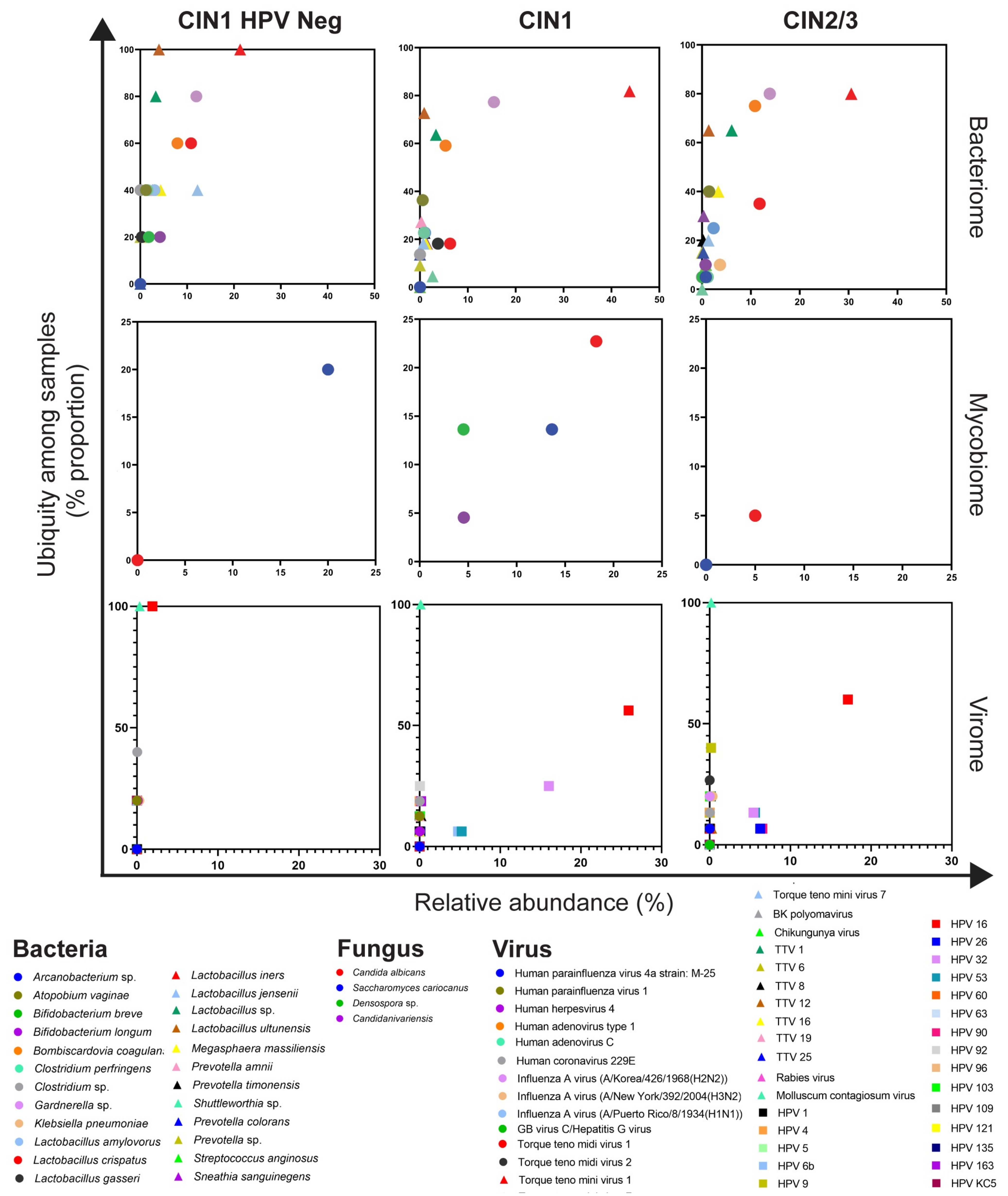
3.2. Cervical Bacterial Community in Association to Histological Characteristics and HPV Infection
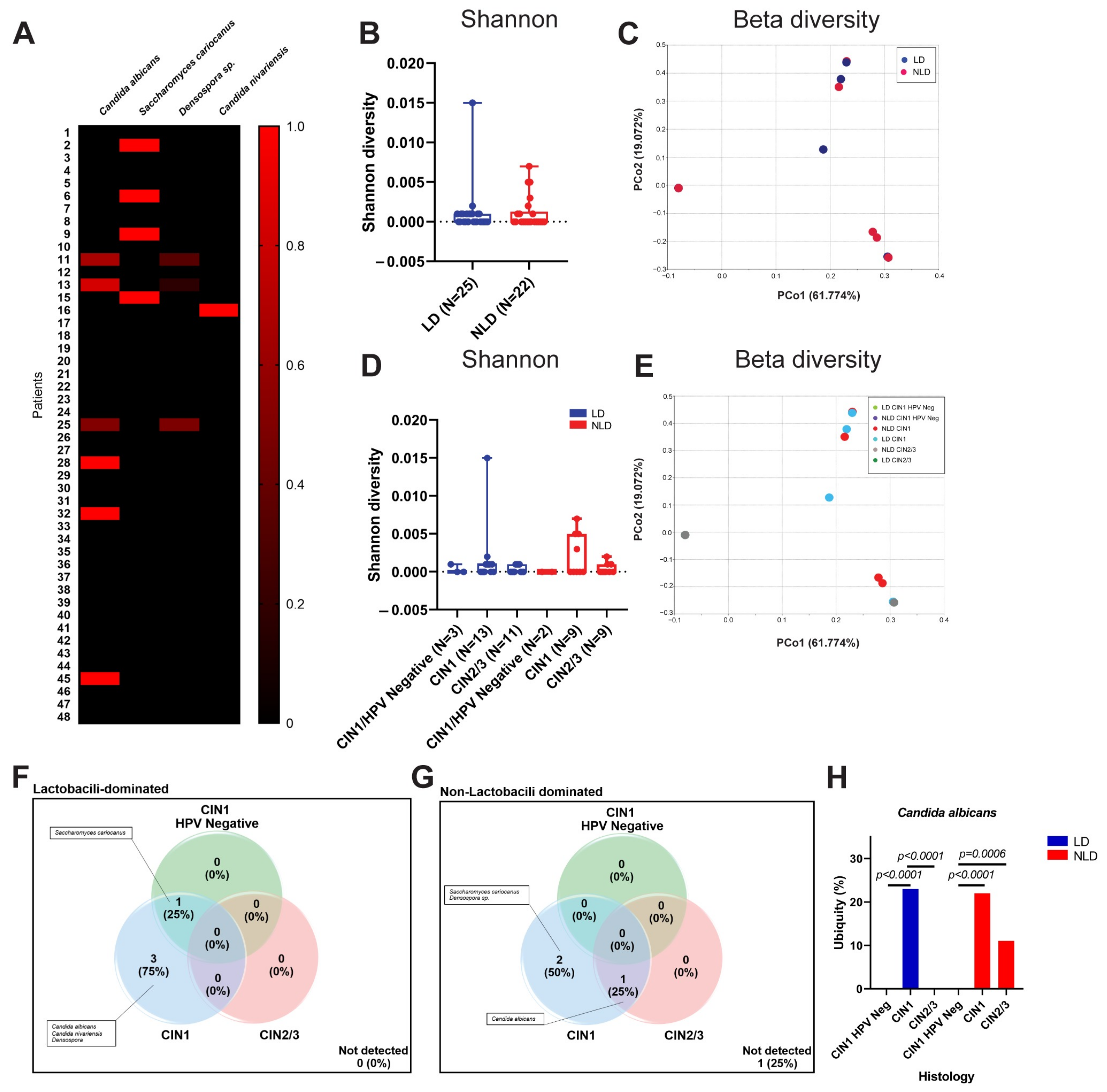
3.3. Cervical Fungal Community in Association to Histological Characteristics and HPV Infection
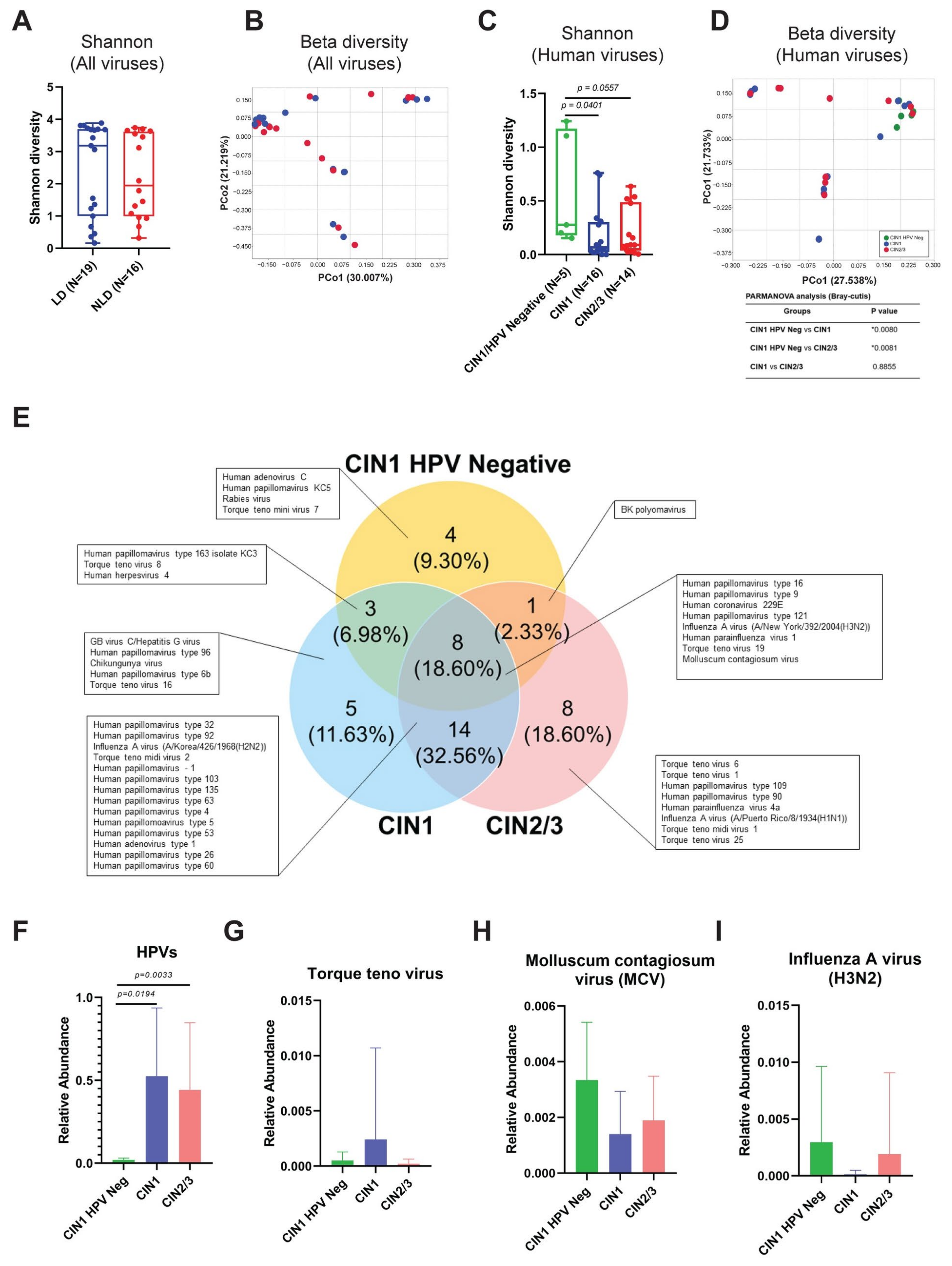
3.4. Cervical Viral Community in Association to Histological Characteristics and HPV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Forman, D.; Mathers, C.D.; Bray, F. Breast and cervical cancer in 187 countries between 1980 and 2010. Lancet 2012, 379, 1390–1391. [Google Scholar] [CrossRef]
- Saraiya, M.; Unger, E.R.; Thompson, T.D.; Lynch, C.F.; Hernandez, B.Y.; Lyu, C.W.; Steinau, M.; Watson, M.; Wilkinson, E.J.; Hopenhayn, C.; et al. US assessment of HPV types in cancers: Implications for current and 9-valent HPV vaccines. J. Natl. Cancer Inst. 2015, 107, djv086. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.M.; Franco, E.L.; Wheeler, C.M.; Moscicki, A.B.; Romanowski, B.; Roteli-Martins, C.M.; Jenkins, D.; Schuind, A.; Costa Clemens, S.A.; Dubin, G.; et al. Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: Follow-up from a randomised control trial. Lancet 2006, 367, 1247–1255. [Google Scholar] [CrossRef]
- Villa, L.L.; Costa, R.L.; Petta, C.A.; Andrade, R.P.; Ault, K.A.; Giuliano, A.R.; Wheeler, C.M.; Koutsky, L.A.; Malm, C.; Lehtinen, M.; et al. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: A randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet. Oncol. 2005, 6, 271–278. [Google Scholar] [CrossRef]
- Harper, D.M.; Franco, E.L.; Wheeler, C.; Ferris, D.G.; Jenkins, D.; Schuind, A.; Zahaf, T.; Innis, B.; Naud, P.; De Carvalho, N.S.; et al. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: A randomised controlled trial. Lancet 2004, 364, 1757–1765. [Google Scholar] [CrossRef]
- Brooks, G.F.; Carroll, K.C.; Butel, J.S.; Morse, S.A.; Mietzner, T.A. Medical Microbiology, 27th ed.; McGraw-Hill Education: New York, NY, USA, 2013. [Google Scholar]
- McBride, A.A. Oncogenic human papillomaviruses. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160273. [Google Scholar] [CrossRef]
- Ghittoni, R.; Accardi, R.; Chiocca, S.; Tommasino, M. Role of human papillomaviruses in carcinogenesis. Ecancermedicalscience 2015, 9, 526. [Google Scholar] [CrossRef]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef]
- Bauman, N.M.; Smith, R.J. Recurrent respiratory papillomatosis. Pediatr. Clin. N. Am. 1996, 43, 1385–1401. [Google Scholar] [CrossRef]
- Snoeck, R.; Wellens, W.; Desloovere, C.; Van Ranst, M.; Naesens, L.; De Clercq, E.; Feenstra, L. Treatment of severe laryngeal papillomatosis with intralesional injections of cidofovir [(S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine]. J. Med. Virol. 1998, 54, 219–225. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Monographs on the Evaluation of Carcinogenic Risks to Humans; International Agency for Research on Cancer: Lyon, France, 2004.
- Fonseca-Moutinho, J.A. Smoking and cervical cancer. ISRN Obs. Gynecol. 2011, 2011, 847684. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Fettweis, J.M.; Brooks, J.P.; Jefferson, K.K.; Buck, G.A. The changing landscape of the vaginal microbiome. Clin. Lab. Med. 2014, 34, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Donders, G.G.; Bosmans, E.; Dekeersmaecker, A.; Vereecken, A.; Van Bulck, B.; Spitz, B. Pathogenesis of abnormal vaginal bacterial flora. Am. J. Obs. Gynecol. 2000, 182, 872–878. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef] [PubMed]
- Gajer, P.; Brotman, R.M.; Bai, G.; Sakamoto, J.; Schutte, U.M.; Zhong, X.; Koenig, S.S.; Fu, L.; Ma, Z.S.; Zhou, X.; et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012, 4, 132ra152. [Google Scholar] [CrossRef]
- Aroutcheva, A.; Gariti, D.; Simon, M.; Shott, S.; Faro, J.; Simoes, J.A.; Gurguis, A.; Faro, S. Defense factors of vaginal lactobacilli. Am. J. Obs. Gynecol. 2001, 185, 375–379. [Google Scholar] [CrossRef]
- Eschenbach, D.A.; Davick, P.R.; Williams, B.L.; Klebanoff, S.J.; Young-Smith, K.; Critchlow, C.M.; Holmes, K.K. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J. Clin. Microbiol. 1989, 27, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Witkin, S.S.; Mendes-Soares, H.; Linhares, I.M.; Jayaram, A.; Ledger, W.J.; Forney, L.J. Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: Implications for protection against upper genital tract infections. mBio 2013, 4, e00460-13. [Google Scholar] [CrossRef]
- Selle, K.; Klaenhammer, T.R. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol. Rev. 2013, 37, 915–935. [Google Scholar] [CrossRef]
- Bradford, L.L.; Ravel, J. The vaginal mycobiome: A contemporary perspective on fungi in women’s health and diseases. Virulence 2017, 8, 342–351. [Google Scholar] [CrossRef]
- Yamamoto, T.; Zhou, X.; Williams, C.J.; Hochwalt, A.; Forney, L.J. Bacterial populations in the vaginas of healthy adolescent women. J. Pediatr. Adolesc. Gynecol. 2009, 22, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, J.M.; Brooks, J.P.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss, J.F.; The Vaginal Microbiome, C.; Jefferson, K.K.; Buck, G.A. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272–2282. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Gonzalez, D.; Samwel, K.; Kahesa, C.; Mwaiselage, J.; Aluthge, N.; Fernando, S.; West, J.T.; Wood, C.; Angeletti, P.C. Relationship between the Cervical Microbiome, HIV Status, and Precancerous Lesions. mBio 2019, 10, e02785-18. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Romaguera, J.; Zhao, C.; Vargas-Robles, D.; Ortiz-Morales, G.; Vazquez-Sanchez, F.; Sanchez-Vazquez, M.; de la Garza-Casillas, M.; Martinez-Ferrer, M.; White, J.R.; et al. Cervicovaginal Fungi and Bacteria Associated With Cervical Intraepithelial Neoplasia and High-Risk Human Papillomavirus Infections in a Hispanic Population. Front. Microbiol. 2018, 9, 2533. [Google Scholar] [CrossRef]
- Zarate, S.; Taboada, B.; Yocupicio-Monroy, M.; Arias, C.F. Human Virome. Arch. Med. Res. 2017, 48, 701–716. [Google Scholar] [CrossRef]
- Wylie, K.M.; Mihindukulasuriya, K.A.; Zhou, Y.; Sodergren, E.; Storch, G.A.; Weinstock, G.M. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 2014, 12, 71. [Google Scholar] [CrossRef]
- Siqueira, J.D.; Curty, G.; Xutao, D.; Hofer, C.B.; Machado, E.S.; Seuanez, H.N.; Soares, M.A.; Delwart, E.; Soares, E.A. Composite Analysis of the Virome and Bacteriome of HIV/HPV Co-Infected Women Reveals Proxies for Immunodeficiency. Viruses 2019, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, H.; Chen, H.; Li, Z.; Ding, W.; Wang, J.; Yin, Y.; Jin, L.; Sun, S.; Jing, C.; et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBioMedicine 2021, 73, 103639. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.P.; Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000, 38, 3623–3630. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Koljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Briese, T.; Kapoor, A.; Mishra, N.; Jain, K.; Kumar, A.; Jabado, O.J.; Lipkin, W.I. Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis. mBio 2015, 6, e01491-15. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Nie, K.; Zhang, C.; Zhang, Y.; Wang, J.; Niu, P.; Ma, X. VIP: An integrated pipeline for metagenomics of virus identification and discovery. Sci. Rep. 2016, 6, 23774. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Sirichoat, A.; Buppasiri, P.; Engchanil, C.; Namwat, W.; Faksri, K.; Sankuntaw, N.; Pasomsub, E.; Chantratita, W.; Lulitanond, V. Characterization of vaginal microbiota in Thai women. PeerJ 2018, 6, e5977. [Google Scholar] [CrossRef]
- Koedooder, R.; Mackens, S.; Budding, A.; Fares, D.; Blockeel, C.; Laven, J.; Schoenmakers, S. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum. Reprod. Update 2019, 25, 298–325. [Google Scholar] [CrossRef] [PubMed]
- Mastromarino, P.; Di Pietro, M.; Schiavoni, G.; Nardis, C.; Gentile, M.; Sessa, R. Effects of vaginal lactobacilli in Chlamydia trachomatis infection. Int. J. Med. Microbiol. 2014, 304, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Breshears, L.M.; Edwards, V.L.; Ravel, J.; Peterson, M.L. Lactobacillus crispatus inhibits growth of Gardnerella vaginalis and Neisseria gonorrhoeae on a porcine vaginal mucosa model. BMC Microbiol. 2015, 15, 276. [Google Scholar] [CrossRef]
- Borgdorff, H.; Gautam, R.; Armstrong, S.D.; Xia, D.; Ndayisaba, G.F.; van Teijlingen, N.H.; Geijtenbeek, T.B.; Wastling, J.M.; van de Wijgert, J.H. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal. Immunol. 2016, 9, 621–633. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef]
- Wakabayashi, R.; Nakahama, Y.; Nguyen, V.; Espinoza, J.L. The Host-Microbe Interplay in Human Papillomavirus-Induced Carcinogenesis. Microorganisms 2019, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef]
- Dong, B.; Huang, Y.; Cai, H.; Chen, Y.; Li, Y.; Zou, H.; Lin, W.; Xue, H.; Feng, A.; Zhao, H.; et al. Prevotella as the hub of the cervicovaginal microbiota affects the occurrence of persistent human papillomavirus infection and cervical lesions in women of childbearing age via host NF-κB/C-myc. J. Med. Virol. 2022, 94, 5519–5534. [Google Scholar] [CrossRef] [PubMed]
- Santella, B.; Schettino, M.T.; Franci, G.; De Franciscis, P.; Colacurci, N.; Schiattarella, A.; Galdiero, M. Microbiota and HPV: The role of viral infection on vaginal microbiota. J. Med. Virol. 2022, 94, 4478–4484. [Google Scholar] [CrossRef]
- Petrova, M.I.; Reid, G.; Vaneechoutte, M.; Lebeer, S. Lactobacillus iners: Friend or Foe? Trends Microbiol. 2017, 25, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Klebanoff, S.J.; Hillier, S.L.; Eschenbach, D.A.; Waltersdorph, A.M. Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J. Infect. Dis. 1991, 164, 94–100. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Marchesi, J.R.; Lee, Y.S.; Bennett, P.R.; Kyrgiou, M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: What do we know and where are we going next? Microbiome 2016, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Boattini, M.; Bianco, G.; Cavallo, R.; Costa, C. Parvimonas micra bacteremia following endoscopic retrograde cholangiopancreatography: A new route of infection. Anaerobe 2018, 54, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, A.; Kokubu, E.; Warita, T.; Ishihara, K. Synergistic biofilm formation by Parvimonas micra and Fusobacterium nucleatum. Anaerobe 2020, 62, 102100. [Google Scholar] [CrossRef]
- Petrova, M.I.; van den Broek, M.; Balzarini, J.; Vanderleyden, J.; Lebeer, S. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol. Rev. 2013, 37, 762–792. [Google Scholar] [CrossRef]
- Sobel, J.D.; Vazquez, J.; Lynch, M.; Meriwether, C.; Zervos, M.J. Vaginitis due to Saccharomyces cerevisiae: Epidemiology, clinical aspects, and therapy. Clin. Infect. Dis. 1993, 16, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Acheson, N.H. Fundamentals of Molecular Virology, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Vafadar, S.; Shahdoust, M.; Kalirad, A.; Zakeri, P.; Sadeghi, M. Competitive exclusion during co-infection as a strategy to prevent the spread of a virus: A computational perspective. PLoS ONE 2021, 16, e0247200. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology, 9th ed.; Elsevier: Alpharetta, GA, USA, 2018. [Google Scholar]
- Kaelin, E.A.; Skidmore, P.T.; Łaniewski, P.; Holland, L.A.; Chase, D.M.; Herbst-Kralovetz, M.M.; Lim, E.S. Cervicovaginal DNA Virome Alterations Are Associated with Genital Inflammation and Microbiota Composition. mSystems 2022, 7, e0006422. [Google Scholar] [CrossRef]
- Lolomadze, E.A.; Rebrikov, D.V. Constant companion: Clinical and developmental aspects of torque teno virus infections. Arch. Virol. 2020, 165, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Morselli, S.; Foschi, C.; Laghi, L.; Zagonari, S.; Patuelli, G.; Camboni, T.; Ceccarani, C.; Consolandi, C.; Djusse, M.E.; Pedna, M.F.; et al. Torquetenovirus in pregnancy: Correlation with vaginal microbiome, metabolome and pro-inflammatory cytokines. Front. Microbiol. 2022, 13, 998849. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasivimolrattana, T.; Chantratita, W.; Sensorn, I.; Chaiwongkot, A.; Oranratanaphan, S.; Bhattarakosol, P.; Bhattarakosol, P. Cervical Microbiome in Women Infected with HPV16 and High-Risk HPVs. Int. J. Environ. Res. Public Health 2022, 19, 14716. https://doi.org/10.3390/ijerph192214716
Sasivimolrattana T, Chantratita W, Sensorn I, Chaiwongkot A, Oranratanaphan S, Bhattarakosol P, Bhattarakosol P. Cervical Microbiome in Women Infected with HPV16 and High-Risk HPVs. International Journal of Environmental Research and Public Health. 2022; 19(22):14716. https://doi.org/10.3390/ijerph192214716
Chicago/Turabian StyleSasivimolrattana, Thanayod, Wasun Chantratita, Insee Sensorn, Arkom Chaiwongkot, Shina Oranratanaphan, Pattarasinee Bhattarakosol, and Parvapan Bhattarakosol. 2022. "Cervical Microbiome in Women Infected with HPV16 and High-Risk HPVs" International Journal of Environmental Research and Public Health 19, no. 22: 14716. https://doi.org/10.3390/ijerph192214716
APA StyleSasivimolrattana, T., Chantratita, W., Sensorn, I., Chaiwongkot, A., Oranratanaphan, S., Bhattarakosol, P., & Bhattarakosol, P. (2022). Cervical Microbiome in Women Infected with HPV16 and High-Risk HPVs. International Journal of Environmental Research and Public Health, 19(22), 14716. https://doi.org/10.3390/ijerph192214716








