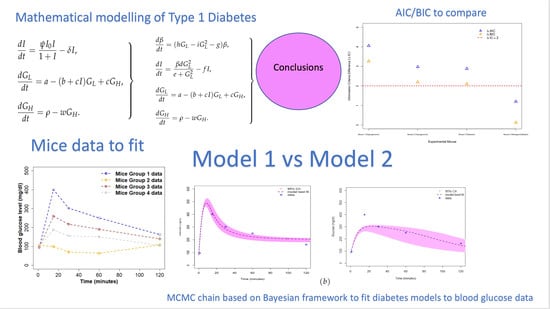
Examining Type 1 Diabetes Mathematical Models Using Experimental Data
Abstract
1. Introduction
2. Methods
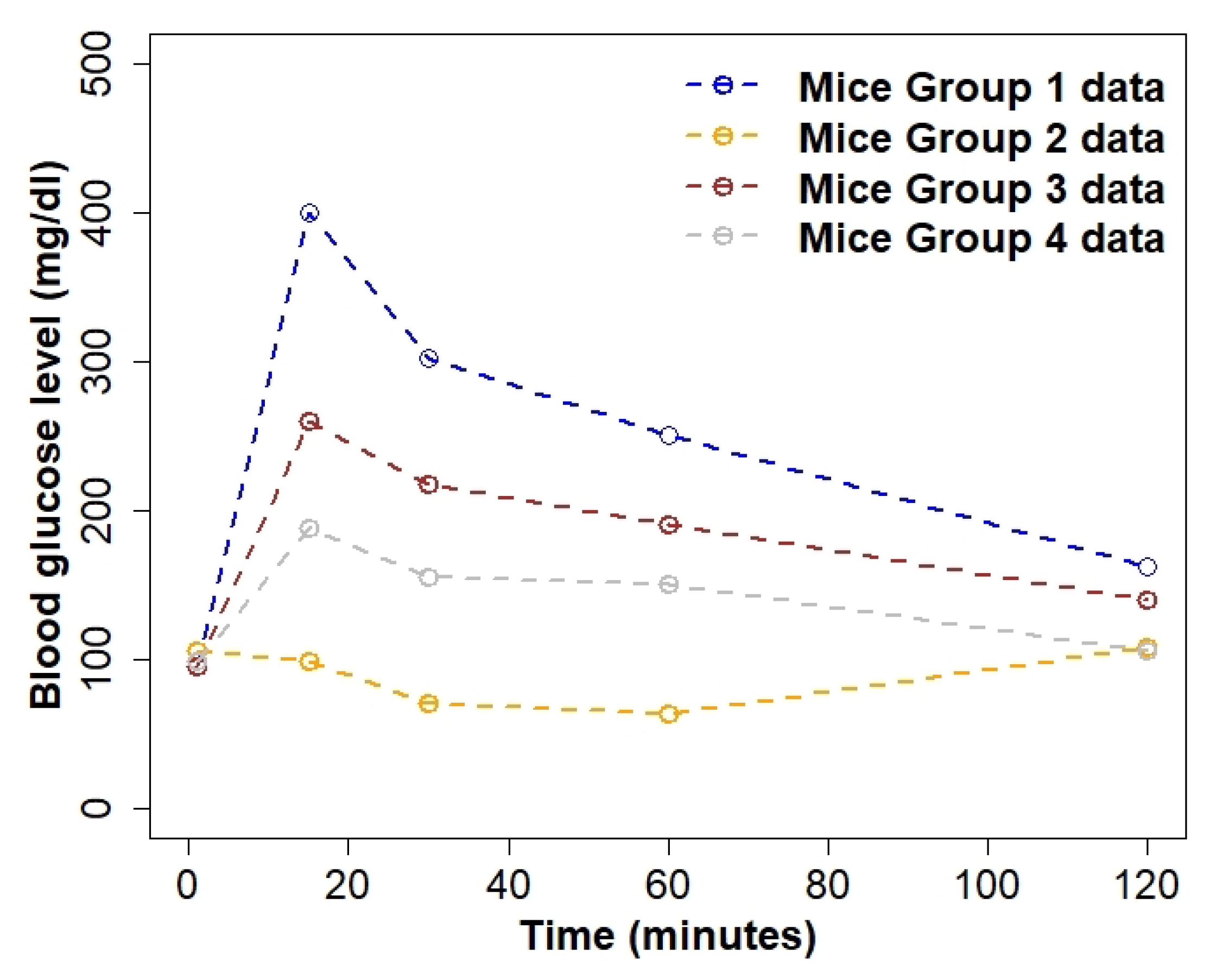
2.1. Data
2.2. Diabetes Candidate Models
2.3. Model Calibration
- Select a dataset and candidate model.
- Use functions modFit and modCost to find the best-fit parameters using least squares fit. The function modFit conducts constrained fitting of the model to data when fitting a model to data with lower and/or upper bounds. The function modCost calculates the discrepancy of a model solution with observed data. This function estimates the residuals, and the variable and model costs (sum of squared residuals), given a solution of the model and data.
- Use Gaussian likelihood to draw model parameter posteriors for a set of varied parameters (i.e., for each candidate model) assuming uniform non-informative priors.
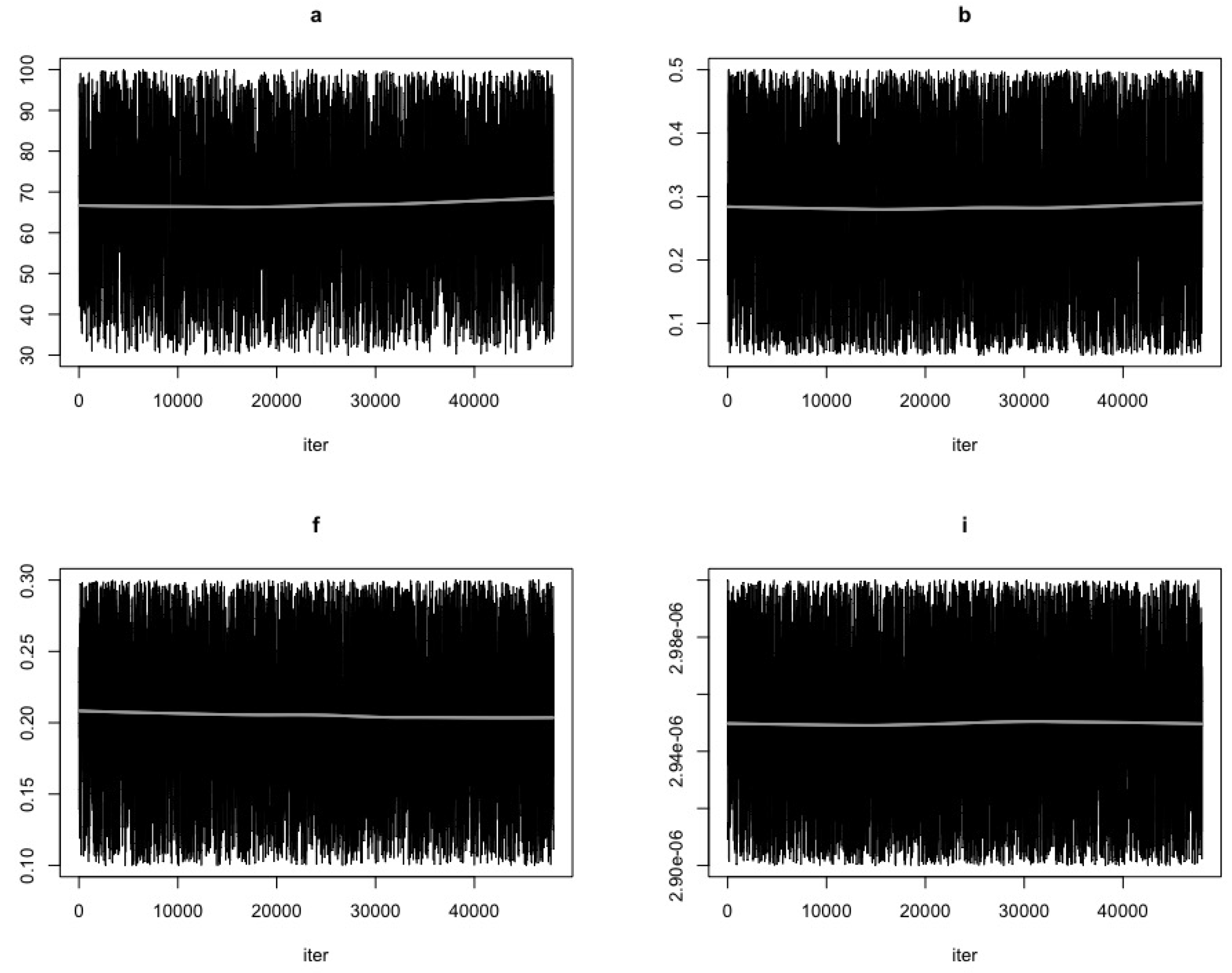
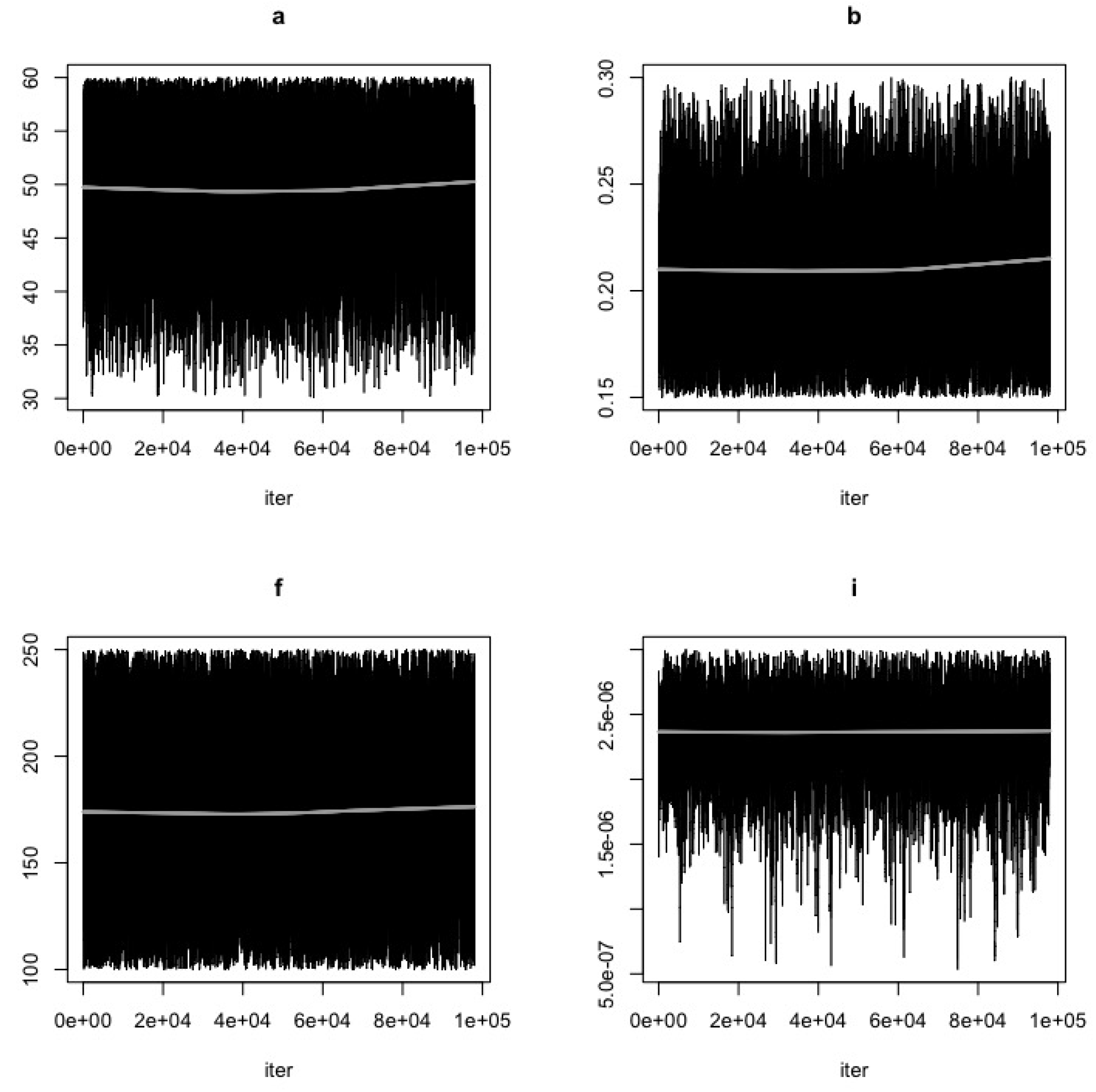
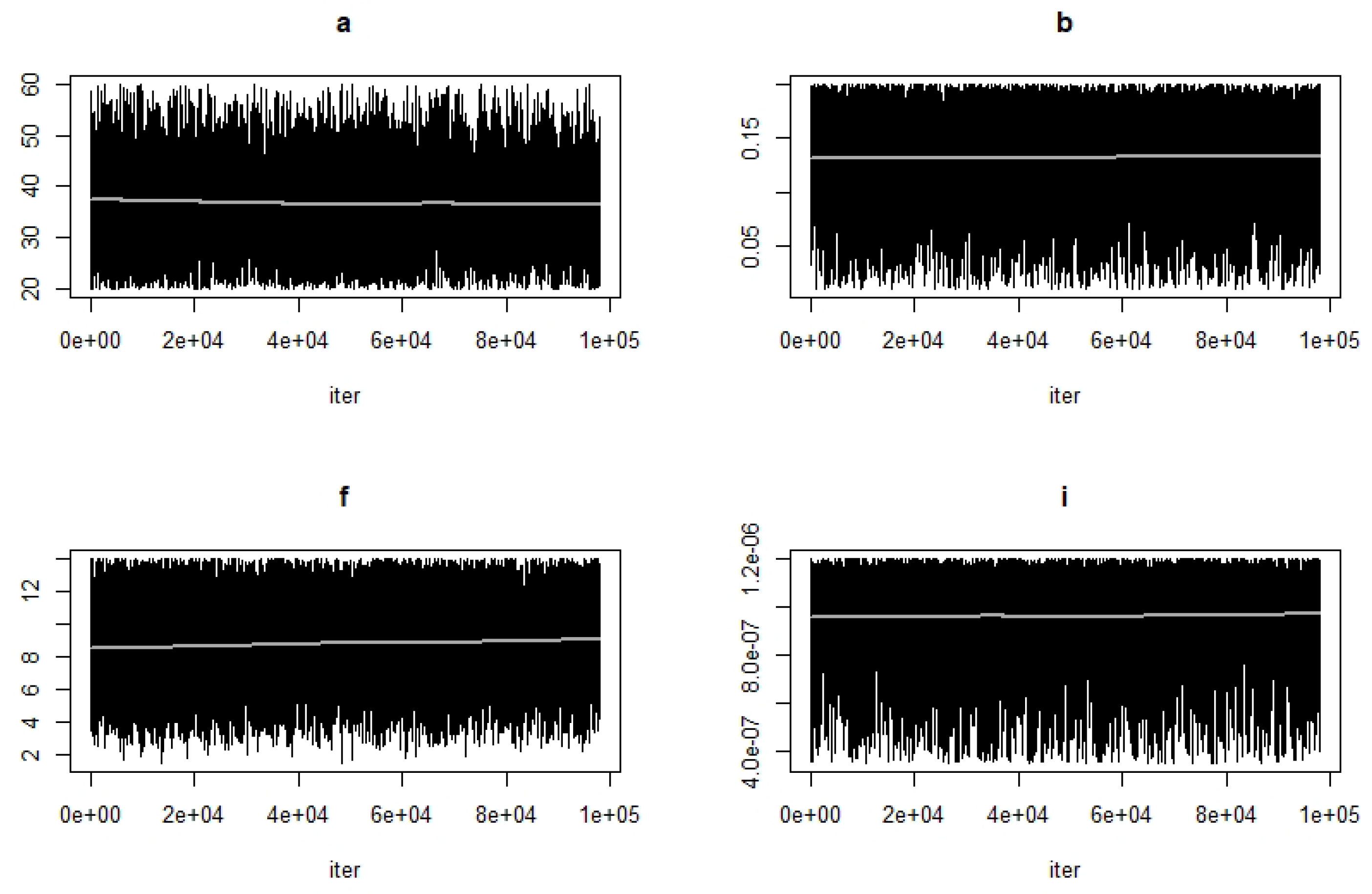
- Use the function modMCMC to perform MCMC simulations assuming Gaussian likelihood and visually examine chain convergence.
- Store the model estimates in a table.
- Compute uncertainty range for each parameter estimate and calculate AIC and BIC values.
- Plot relevant model output.
- Repeat steps 1–7 for each dataset and candidate model.
3. Results
Parameter Estimating and Uncertainty
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
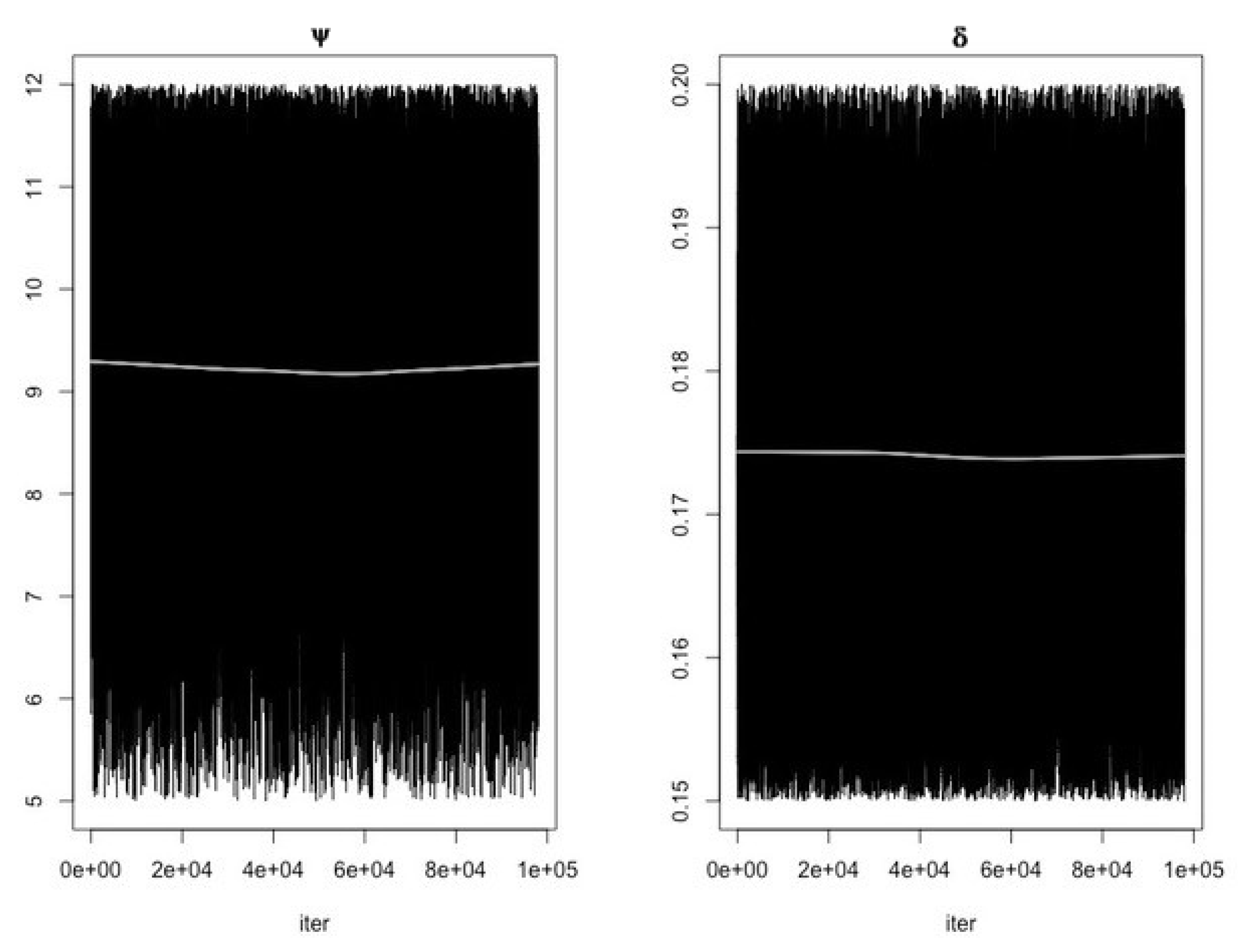
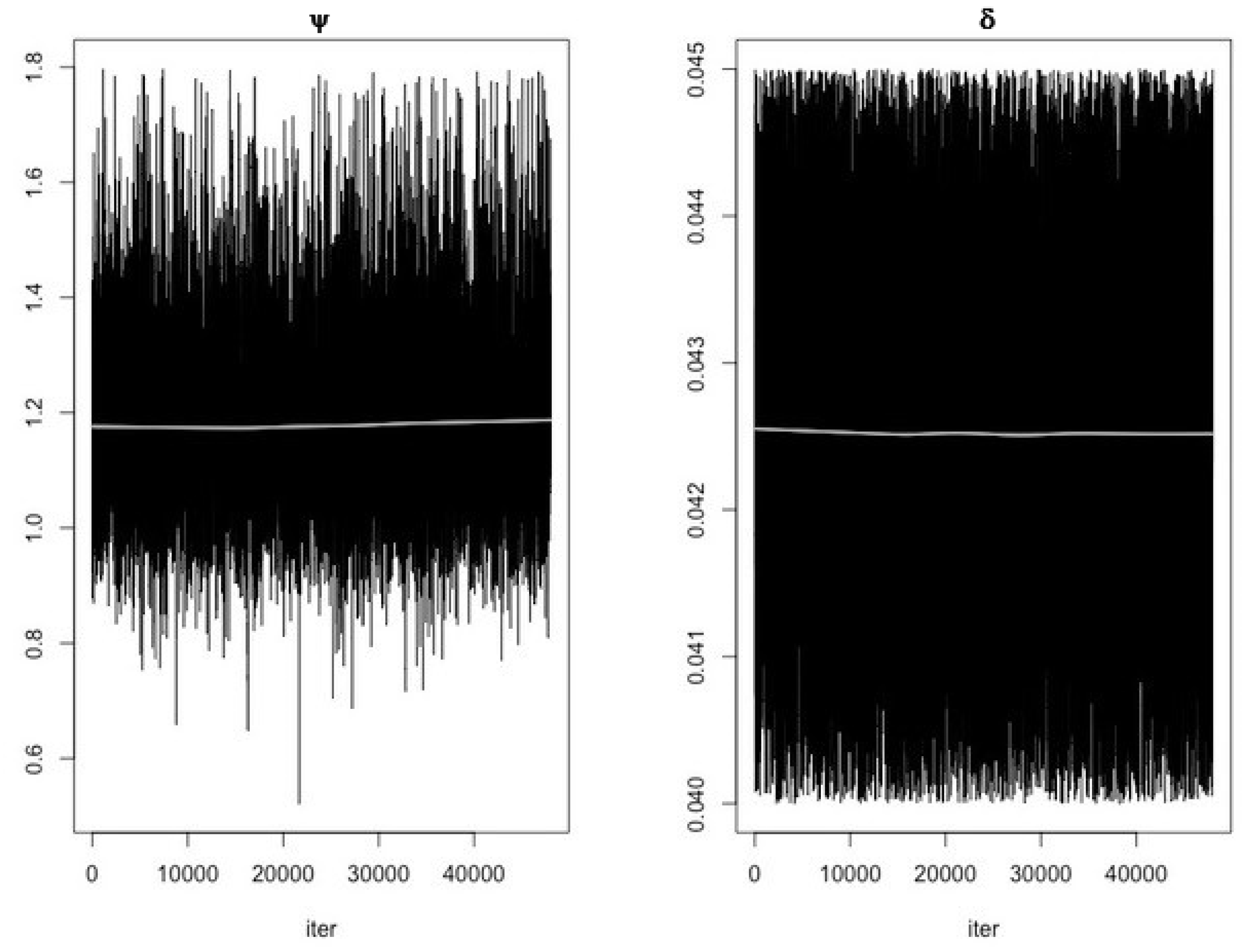
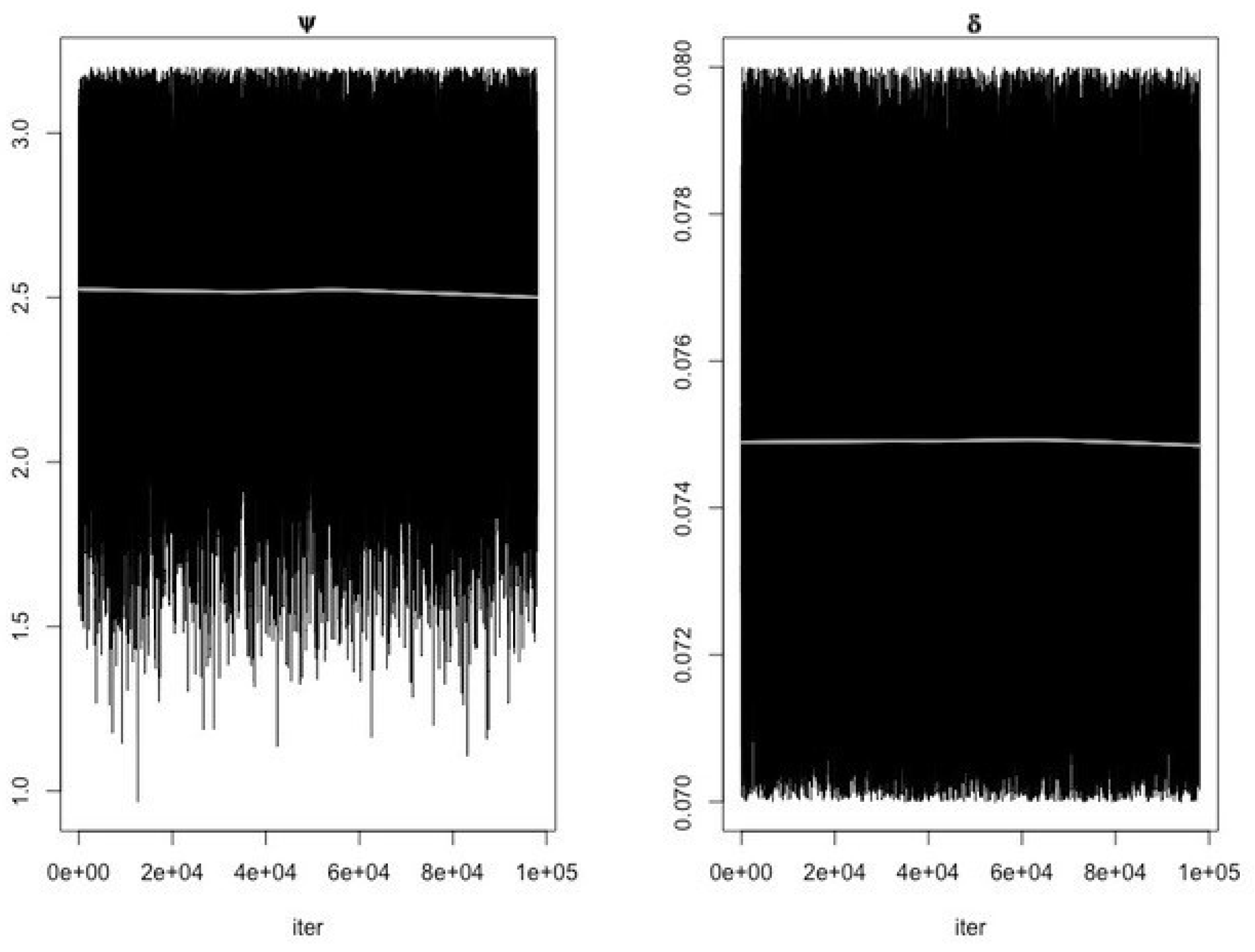
References
- International Diabetes Federation. Available online: https://www.idf.org/ (accessed on 18 August 2020).
- Cantley, J.; Ashcroft, F.M. Q&A: Insulin secretion and type 2 diabetes: Why do β-cells fail? BMC Biol. 2015, 13, 33. [Google Scholar]
- Al Ali, H.; Daneshkhah, A.; Boutayeb, A.; Malunguza, N.J.; Mukandavire, Z. Exploring dynamical properties of a Type 1 diabetes model using mathematical and global sensitivity approaches. 2021; submitted. [Google Scholar]
- Mayo Clinic. Diabetes—Symptoms And Causes. 2020. Available online: https://www.mayoclinic.org/diseases-conditions/diabetes/symptoms-causes/syc-20371444 (accessed on 10 August 2021).
- Peric, S.; Stulnig, T.M. Diabetes and COVID-19. Wien. Klin. Wochenschr. 2020, 132, 356–361. [Google Scholar] [CrossRef]
- Singh, A.K.; Gupta, R.; Ghosh, A.; Misra, A. Diabetes in COVID-19: Prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 303–310. [Google Scholar] [CrossRef]
- Diabetes.co.uk, Beta Cells—What They Do, Role in Insulin. Available online: http://www.diabetes.co.uk/body/beta-cells.html (accessed on 10 August 2021).
- Boutayeb, W.; Lamlili, M.E.N.; Boutayeb, A.; Derouich, M. Mathematical modelling and simulation of β-cell mass, insulin and glucose dynamics: Effect of genetic predisposition to diabetes. J. Biomed. Sci. Eng. 2014, 7, 45811. [Google Scholar] [CrossRef]
- Ajmera, I.; Swat, M.; Laibe, C.; Le Novere, N.; Chelliah, V. The impact of mathematical modeling on the understanding of diabetes and related complications. CPT Pharmacometr. Syst. Pharmacol. 2013, 2, 1–14. [Google Scholar] [CrossRef]
- Mayo Clinic, Diabetes; Diagnosis and Treatment, Mayo Foundation for Medical Education and Research. Available online: https://www.mayoclinic.org/diseases-conditions/diabetes/diagnosis-treatment/drc-20371451 (accessed on 11 May 2021).
- Lombarte, M.; Lupo, M.; Brenda, L.F.; Campetelli, G.; Marilia, A.B.; Basualdo, M.; Rigalli, A. In vivo measurement of the rate constant of liver handling of glucose and glucose uptake by insulin-dependent tissues, using a mathematical model for glucose homeostasis in diabetic rats. J. Theor. Biol. 2018, 439, 205–215. [Google Scholar] [CrossRef]
- Lombarte, M.; Lupo, M.; Campetelli, G.; Basualdo, M.; Rigalli, A. Mathematical model of glucose–insulin homeostasis in healthy rats. Math. Biosci. 2013, 245, 269–277. [Google Scholar] [CrossRef]
- Al Ali, H.; Daneshkhah, A.; Boutayeb, A.; Merabet, N.; Mukandavire, Z. Using mathematical and sensitivity analysis approaches to understand glucose homeostasis model with growth hormone. 2021; submitted. [Google Scholar]
- Aradóttir, T.B.; Boiroux, D.; Bengtsson, H.; Poulsen, N.K. Modelling of fasting glucose–insulin dynamics from sparse data. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 2354–2357. [Google Scholar]
- Asadi, S.; Nekoukar, V. Adaptive fuzzy integral sliding mode control of blood glucose level in patients with type 1 diabetes: In silico studies. Math. Biosci. 2018, 305, 122–132. [Google Scholar] [CrossRef]
- Shi, X.Y.; Kuang, Y.; Makroglou, A.; Mokshagundam, S.; Li, J. Oscillatory dynamics of an intravenous glucose tolerance test model with delay interval. Chaos Interdiscip. J. Nonlinear Sci. 2017, 27, 114–324. [Google Scholar] [CrossRef] [PubMed]
- Panunzi, S.; Palumbo, P.; de Gaetano, A. A discrete single delay model for the intra-venous glucose tolerance test. Theor. Biol. Med. Model. 2007, 4, 35. [Google Scholar] [CrossRef]
- de Gaetano, A.; Arino, O. Mathematical modeling of the intravenous glucose tolerance test. J. Math. Biol. 2000, 40, 136–168. [Google Scholar] [CrossRef]
- Li, J.; Kuang, Y.; Kuang, Y.; Li, B. Analysis of IVGTT glucose–insulin interaction models with time delay. Discret. Contin. Dyn. Syst.-B 2001, 1, 103–124. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Song, X.Y. Mathematical model for diabetes mellitus with impulsive injections of glucose–insulin. Chin. Q. J. Math. 2017, 32, 118–133. [Google Scholar]
- Song, X.; Huang, M.; Li, J. Modeling impulsive insulin delivery in insulin pump with time delays. SIAM J. Appl. Math. 2014, 74, 1763–1785. [Google Scholar] [CrossRef]
- Kovatchev, B. Automated closed-loop control of diabetes: The artificial pancreas. Bioelectron. Med. 2018, 4, 14. [Google Scholar] [CrossRef]
- Gu, Y.; Wei, H.-L.; Balikhin, M.M. Nonlinear predictive model selection and model averaging using information criteria. Syst. Sci. Control Eng. 2018, 6, 319–328. [Google Scholar] [CrossRef]
- Topp, B.; Promislow, K.; Devries, G.; Miura, R.M.; Finegood, T.D. A model of β-cell mass, insulin, and glucose kinetics: Pathways to diabetes. J. Theor. Biol. 2000, 206, 605–619. [Google Scholar] [CrossRef]
- Bergman, R.N.; Toffolo, G.; Finegood, D.T.; Bowden, C.R.; Cobelli, C. Quantitative estimation of beta cell sensitivity to glucose in the intact organism: A minimal model of insulin kinetics in the dog. Diabetes 1980, 29, 979–990. [Google Scholar]
- Boutayeb, W.; Lamlili, M.E.N.; Boutayeb, A.; Derouich, M. The Impact of Obesity on Predisposed People to Type 2 Diabetes: Mathematical Model. In Bioinformatics and Biomedical Engineering, Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering 2015, Granada, Spain, 15–17 April 2015; Ortuño, F., Rojas, I., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9043, pp. 613–622. [Google Scholar]
- Bolie, V.W. Coefficients of normal blood glucose regulation. J. Appl. Physiol. 1961, 16, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Bergman, N.R.; Phillips, S.L.; Cobelli, C. Physiologic evaluation of factors controlling glucose tolerance in man: Measurement of insulin sensitivity and beta-cell glucose sensitivity from the response to intravenous glucose. J. Clin. Investig. 1981, 68, 1456–1467. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.D.; Danielle, J.L.; Daniel, B.R.; Thomas, B.V.; Stephen, A.W. A Model of β-Cell Mass, Insulin, Glucose, and Receptor Dynamics with Applications to Diabetes. 2001. Available online: https://ecommons.cornell.edu/bitstream/handle/1813/32174/BU-1579-M.pdf?sequence=1 (accessed on 10 November 2021).
- Huard, B.; Bridgewater, A.; Angelova, M. Mathematical investigation of diabetically impaired ultradian oscillations in the glucose–insulin regulation. J. Theor. Biol. 2017, 418, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Makroglou, A.; Li, J.; Kuang, Y. Mathematical models and software tools for the glucose–insulin regulatory system and diabetes: An overview. Appl. Numer. Math. 2006, 56, 559–573. [Google Scholar] [CrossRef]
- Xu, J.; Huang, G.; Guo, T.L. Bisphenol S modulates type 1 diabetes development in non-obese diabetic (NOD) mice with diet-and sex-related effects. Toxics 2019, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Schwarz, G.E. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Bowe, J.E.; Franklin, Z.J.; Hauge-Evans, A.C.; King, A.J.; Persaud, S.J.; Jones, P.M. Metabolic phenotyping guidelines: Assessing glucose homeostasis in rodent models. J. Endocrinol. 2014, 222, G13–G25. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.F.; Gong, X.; Bakh, N.A.; Carr, K.; Phillips, N.F.; Ismail-Beigi, F.; Strano, M.S. Connecting rodent and human pharmacokinetic models for the design and translation of glucose-responsive insulin. Diabetes 2020, 69, 1815–1826. [Google Scholar] [CrossRef]
- Bergman, R.N.; Ider, Y.Z.; Bowden, C.R.; Cobelli, C. Quantitative estimation of insulin sensitivity. Am. J. Physiol.-Endocrinol. Metab. 1979, 236, E677. [Google Scholar] [CrossRef]
- Bergman, R.N. Toward physiological understanding of glucose tolerance: Minimal model approach. Diabetes 1989, 38, 1512–1527. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kuang, Y. Systemically modeling the dynamics of plasma insulin in subcutaneous injection of insulin analogues for type 1 diabetes. Math. Biosci. Eng. 2009, 6, 41. [Google Scholar]
- Koutny, T. Modelling of glucose dynamics for diabetes. In Proceedings of the International Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 26–28 April 2017; Springer: Cham, Switzerland, 2017; pp. 314–332. [Google Scholar]
- Magdelaine, N.; Chaillous, L.; Guilhem, I.; Poirier, J.Y.; Krempf, M.; Moog, C.H.; Le Carpentier, E. A long-term model of the glucose—insulin dynamics of type 1 diabetes. IEEE Trans. Biomed. Eng. 2015, 62, 1546–1552. [Google Scholar] [CrossRef]
- Sanofi-Aventis. Product Monograph Apidra Insulin Glulisine. Available online: http://products.sanofi.ca/en/apidra.pdf (accessed on 25 October 2021).
- Taylor, A.; Finster, J.; Mintz, D. Metabolic clearance and production rates of human growth hormone. J. Clin. Investig. 1969, 48, 2349–2358. [Google Scholar] [CrossRef][Green Version]
- Buppajarntham, S.; Junpaparp, P.; Salameh, R.; Anastasopoulou, C.; Eric, B.S. Insulin: Reference Range, Interpretation, Collection and Panels. Available online: https://emedicine.medscape.com/article/2089224-overview (accessed on 23 March 2019).
- Wisse, B.; Zieve, D. Growth Hormone Test: MedlinePlus Medical Encyclopedia, A.D.A.M. Available online: https://medlineplus.gov/ency/article/003706.htm (accessed on 23 March 2019).
- Soetaert, K.; Petzoldt, T. Inverse modelling, sensitivity and Monte Carlo analysis in R using package FME. J. Stat. Softw. 2010, 33, 1–28. [Google Scholar] [CrossRef]
- Haario, H.; Laine, M.; Mira, A.; Saksman, E. DRAM: Efficient Adaptive MCMC. Stat. Comput. 2006, 16, 339–354. [Google Scholar] [CrossRef]
- Magombedze, G.; Eda, S.; Stabel, J. Predicting the role of IL-10 in the regulation of the adaptive immune responses in Mycobacterium avium subsp. paratuberculosis infections using mathematical models. PLoS ONE 2015, 10, e0141539. [Google Scholar] [CrossRef]
- Mukandavire, Z.; Nyabadza, F.; Malunguza, N.J.; Cuadros, D.F.; Shiri, T.; Musuka, G. Quantifying early COVID-19 outbreak transmission in South Africa and exploring vaccine efficacy scenarios. PLoS ONE 2020, 15, e0236003. [Google Scholar] [CrossRef]
- Gelman, A.; Varlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis; Chapman & Hall/CRC: Boca Raton, FL, USA, 2004; Volume 2. [Google Scholar]
- Kraegen, E.W.; Chisholm, D.J. Insulin responses to varying profiles of subcutaneous insulin infusion: Kinetic modelling studies. Diabetologia 1984, 26, 208–213. [Google Scholar] [CrossRef]
- Puckett, W.R.; Lightfoot, E.N. A model for multiple subcutaneous insulin injections developed from individual diabetic patient data. Am. J. Physiol.-Endocrinol. Metab. 1995, 269, E1115–E1124. [Google Scholar] [CrossRef]
- Rossetti, P.; Pampanelli, S.; Fanelli, C.; Porcellati, F.; Costa, E.; Torlone, E.; Scionti, L.; Bolli, G.B. Intensive replacement of basal insulin in patients with type 1 diabetes given rapid-acting insulin analog at mealtime: A 3-month comparison between administration of NPH insulin four times daily and glargine insulin at dinner or bedtime. Diabetes Care 2003, 26, 1490–1496. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, S.E.; Nishida, K.E.; Sakakida, M.I.; Konno, Y.U.; Ichinose, K.E.; Uehara, M.A.; Nowak, T.A.; Shichiri, M.O. Closed-loop subcutaneous insulin infusion algorithm with a short-acting insulin analog for long-term clinical application of a wearable artificial endocrine pancreas. Front. Med. Biol. Eng. 1997, 8, 197–211. [Google Scholar] [PubMed]
- Mukhopadhyay, A.; De Gaetano, A.; Arino, O. Modeling the intra-venous glucose tolerance test: A global study for a single-distributed-delay model. Discret. Contin. Dyn. Syst.-B 2004, 4, 407. [Google Scholar] [CrossRef]
- Nucci, G.; Cobelli, C. Models of subcutaneous insulin kinetics. A critical review. Comput. Methods Program. Biomed. 2000, 62, 249–257. [Google Scholar] [CrossRef]
- Tarin, C.; Teufel, E.; PicÛ, J.; Bondia, J.; Pfleiderer, H.J. Comprehensive pharmacokinetic model of insulin glargine and other insulin formulations. IEEE Trans. Biomed. Eng. 2005, 52, 1994–2005. [Google Scholar] [CrossRef] [PubMed]
- Wilinska, M.E.; Chassin, L.J.; Schaller, H.C.; Schaupp, L.; Pieber, T.R.; Hovorka, R. Insulin kinetics in type-1 diabetes: Continuous and bolus delivery of rapid acting insulin. IEEE Trans. Biomed. Eng. 2004, 52, 3–12. [Google Scholar] [CrossRef]
- Li, J.; Johnson, J.D. Mathematical models of subcutaneous injection of insulin analogues: A mini-review. Discret. Contin. Dyn. Syst. Ser. B 2009, 12, 401. [Google Scholar] [CrossRef]
- MediLexicon International. 10 Signs of Uncontrolled Diabetes—Medical News Today. Available online: https://www.medicalnewstoday.com/articles/317465#High-blood-glucose-readings (accessed on 11 August 2021).
- Gingras, V.; Taleb, N.; Roy-Fleming, A.; Legault, L.; Rabasa-Lhoret, R. The challenges of achieving postprandial glucose control using closed-loop systems in patients with type 1 diabetes. Diabetes Obes. Metab. 2018, 20, 245–256. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R.; Huyvaert, K.P. AIC model selection and multimodel inference in behavioral ecology: Some background, observations, and comparisons. Behav. Ecol. Sociobiol. 2011, 65, 23–35. [Google Scholar] [CrossRef]
- Hadj-Abo, A.; Enge, S.; Rose, J.; Kunte, H.; Fleischhauer, M. Individual differences in impulsivity and need for cognition as potential risk or resilience factors of diabetes self-management and glycemic control. PLoS ONE 2020, 15, e0227995. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, A.; Buck, C.E.; Daneshkhah, A.; Eiser, J.R.; Garthwaite, P.H.; Jenkinson, D.J.; Oakley, J.E.; Rakow, T. Uncertain Judgements: Eliciting Experts’ Probabilities; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Daneshkhah, A.; Oakley, J.E. Eliciting multivariate probability distributions. Rethink. Risk Meas. Rep. 2010, 1, 23. [Google Scholar]
- Daneshkhah, A.; Hosseinian-Far, A.; Sedighi, T.; Farsi, M. Prior Elicitation and Evaluation of Imprecise Judgements for Bayesian Analysis of System Reliability, Strategic Engineering for Cloud Computing and Big Data Analytics; Springer: Berlin/Heidelberg, Germany, 2017; pp. 63–79. [Google Scholar]
- Wasserman, L. Recent methodological advances in robust Bayesian inference. Bayesian Stat. 1992, 4, 483–502. [Google Scholar]
- Gustafson, P.; Wasserman, L. Local sensitivity diagnostics for Bayesian inference. Ann. Stat. 1995, 23, 2153–2167. [Google Scholar] [CrossRef]
- Smith, J.Q.; Daneshkhah, A. On the robustness of Bayesian networks to learning from non-conjugate sampling. Int. J. Approx. Reason. 2010, 51, 558–572. [Google Scholar] [CrossRef]
- Sedighi, T.; Hosseinian-Far, A.; Daneshkhah, A. Measuring local sensitivity in Bayesian inference using a new class of metrics. Commun. Stat.-Theory Methods 2021, 1–17. [Google Scholar] [CrossRef]
- Daneshkhah, A.; Bedford, T. Probabilistic sensitivity analysis of system availability using Gaussian processes. Reliab. Eng. Syst. Saf. 2013, 112, 82–93. [Google Scholar] [CrossRef]
- Hoyos, J.D.; Villa-Tamayo, M.F.; Builes-Montaño, C.E.; Ramirez-Rincón, A.; Godoy, J.L.; Garcia-Tirado, J.; Rivadeneira, P.S. Identifiability of Control-Oriented Glucose-Insulin Linear Models: Review and Analysis. IEEE Access 2021, 9, 69173–69188. [Google Scholar] [CrossRef]
- Staal, O.M.; Fougner, A.L.; Sælid, S.; Stavdahl, Ø. Glucose-insulin metabolism model reduction and parameter selection using sensitivity analysis. In Proceedings of the American Control Conference (ACC), Philadelphia, PA, USA, 10–12 July 2019; pp. 4104–4111. [Google Scholar]
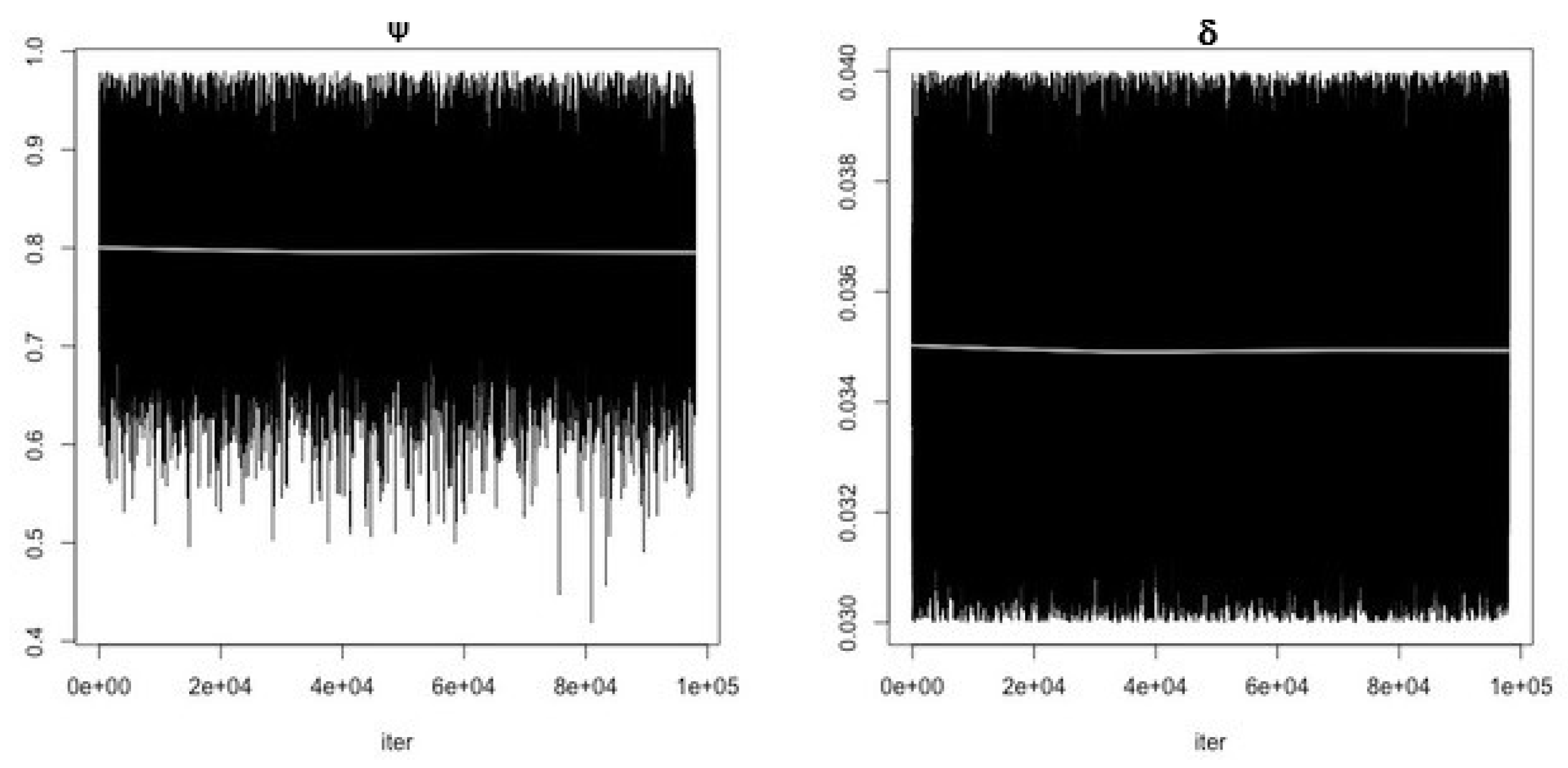
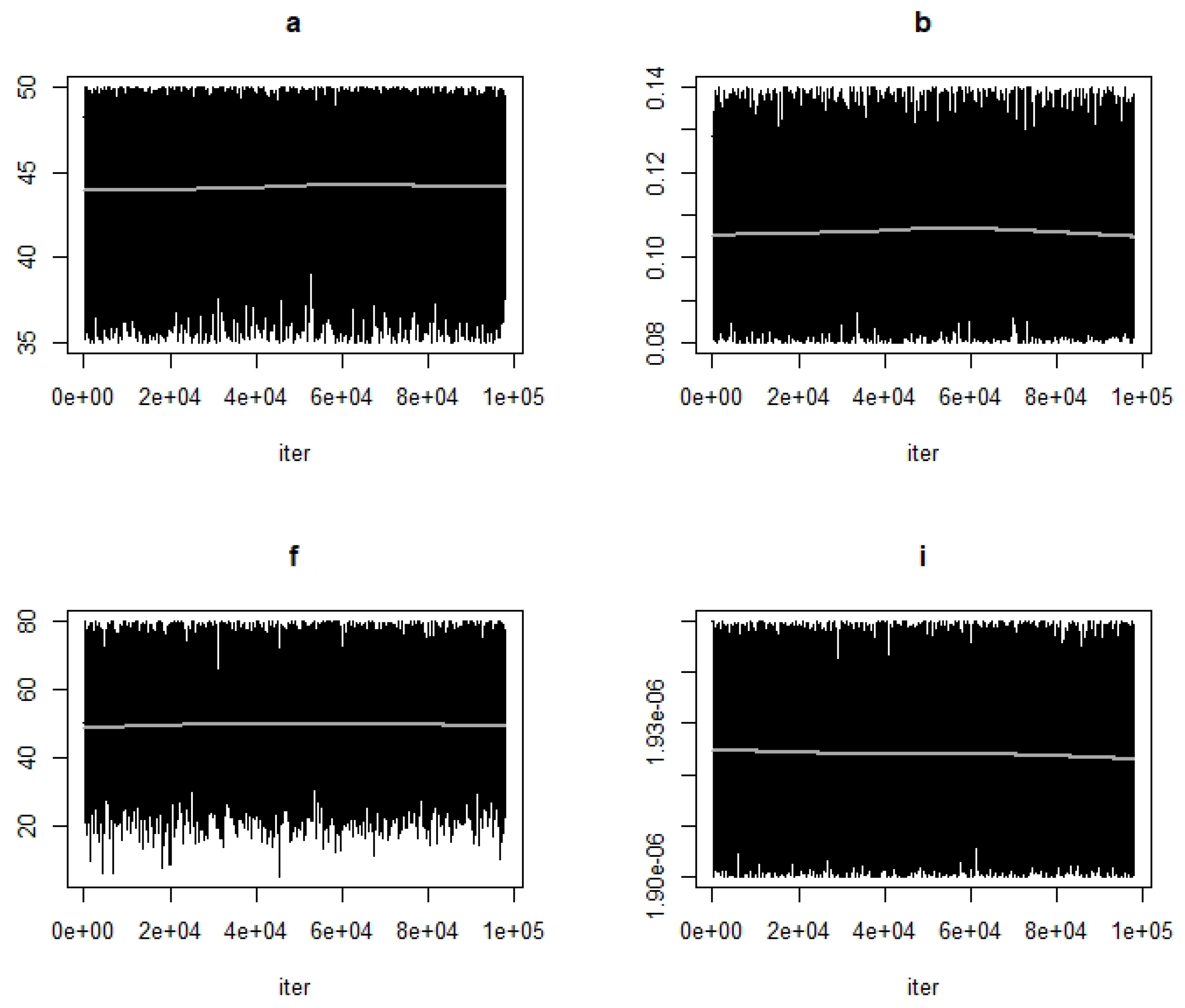
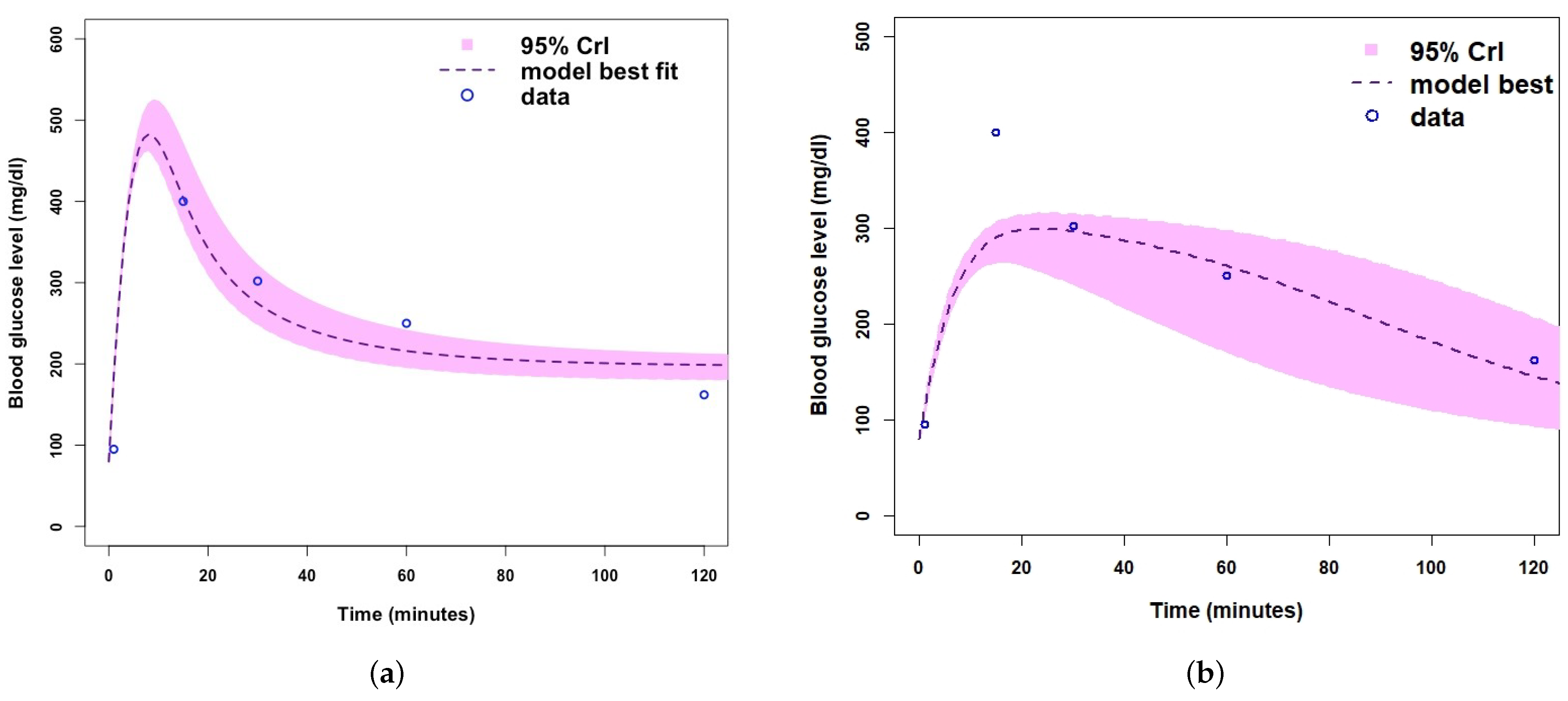
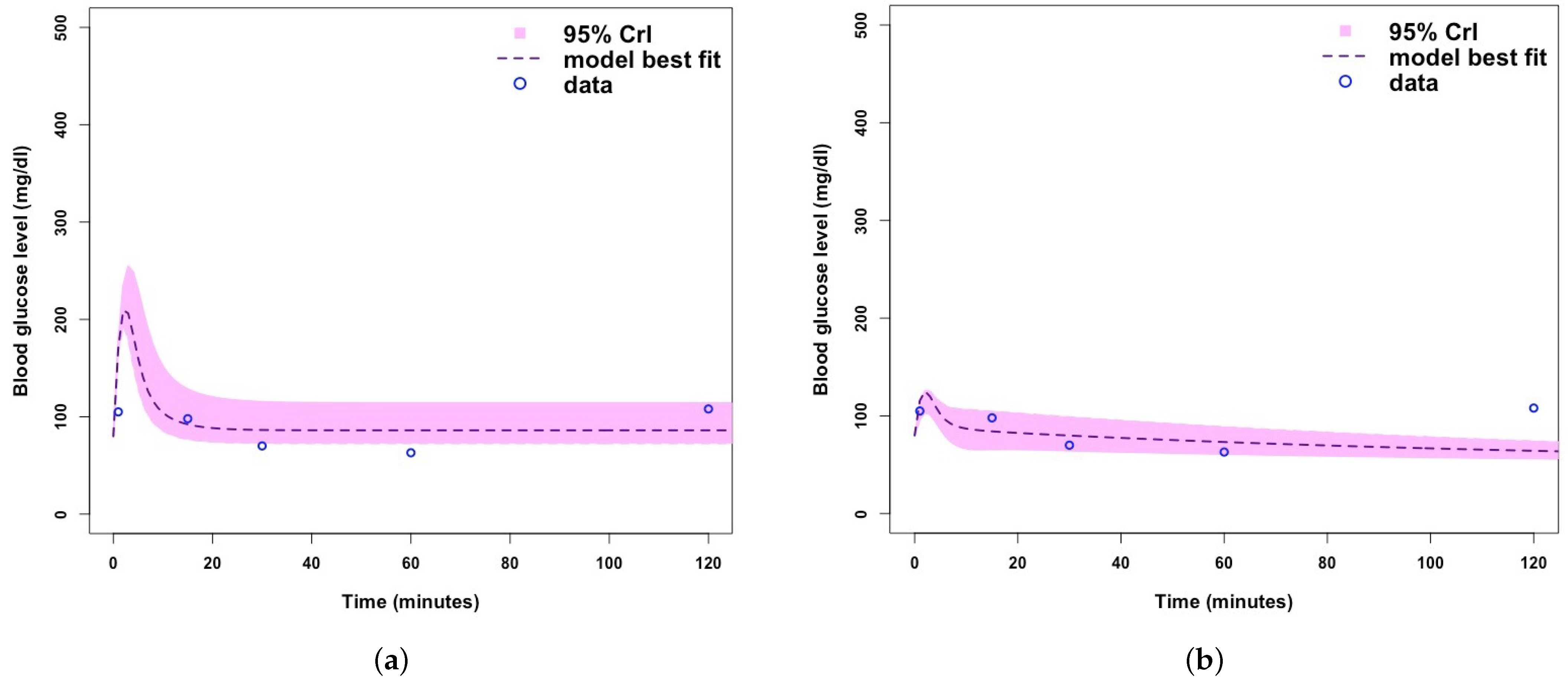
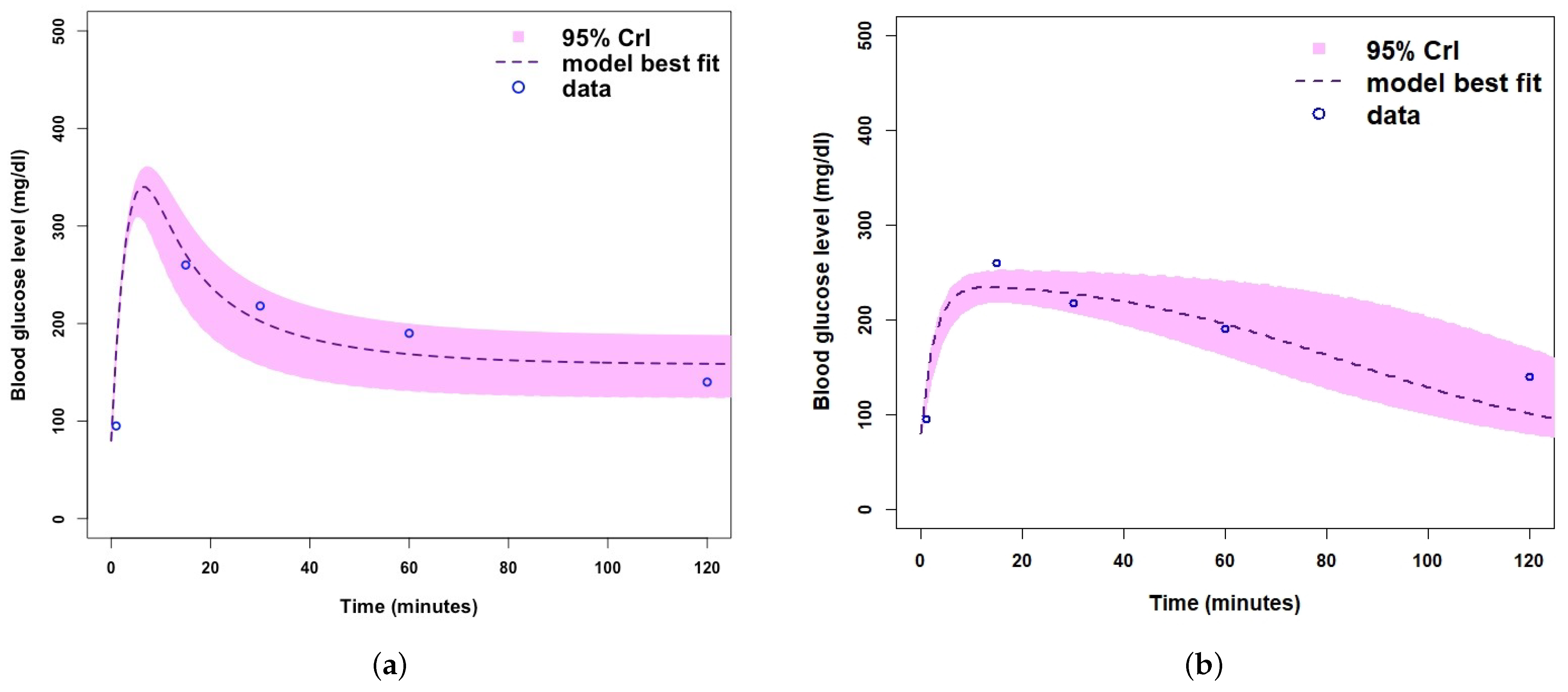
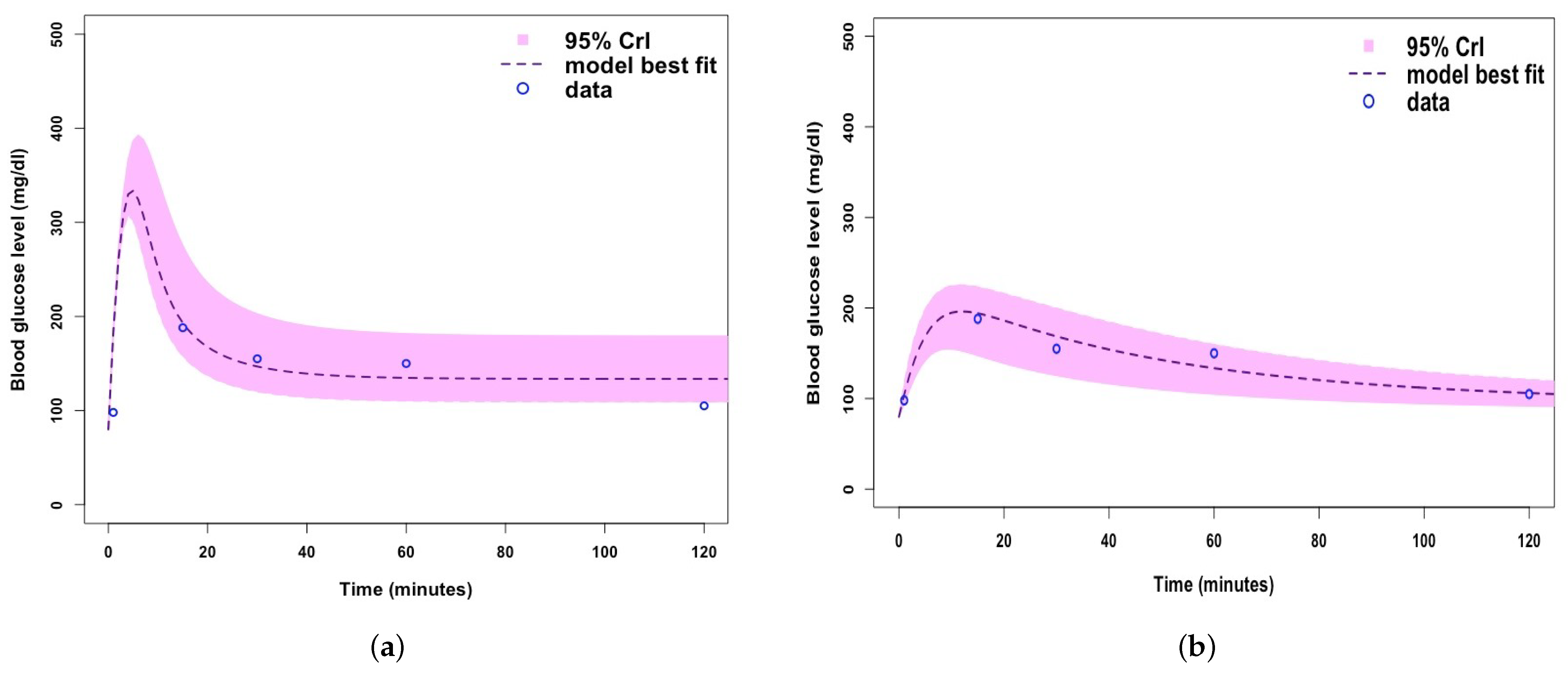
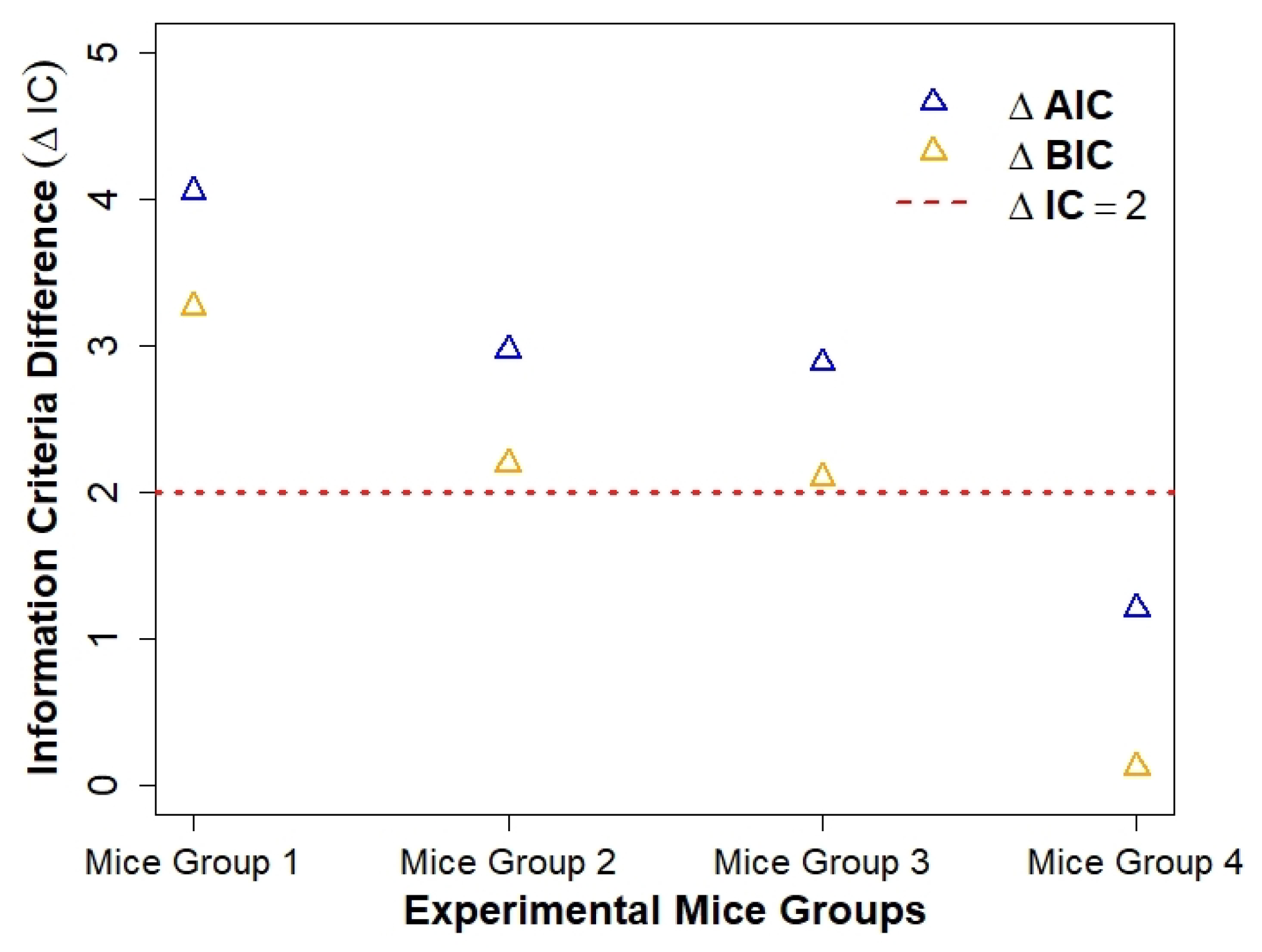
| Parameter/Variable Definition | Symbol | Baseline Value [Range] | Unit | Reference |
|---|---|---|---|---|
| Biological parameters | ||||
| Glucose production rate by liver | a | fitted * | mg/dL min | |
| Glucose clearance rate independent of insulin | b | fitted * | min | |
| Insulin induced glucose uptake rate | c | 0.85 [0.1–1] | mL/m IU min | Assumed |
| -cell maximum insulin secretory rate | d | 43.2 [40–100] | m IU/mL min mg | Assumed |
| Sigmoidal inflection point | e | 20,000 [20,000–50,000] | mg/dL | Assumed |
| Whole body insulin clearance rate | f | fitted * | min | |
| -cell natural death rate | g | 0.03 [0.03–1] | min | Assumed |
| Determines -cell glucose tolerance range factor | h | –1] | dL/mg min | Assumed |
| Determines -cell glucose tolerance range factor | i | fitted * | dL/mg min | |
| Growth hormone production rate by somatotropic cells | 15.06 [5–30] | mIU/mL min | [43] | |
| Growth hormone clearance rate by the liver | w | 1958.40 [2000–4000] | min | [43] |
| Insulin absorption rate | fitted * | mIU/mL min | ||
| Insulin clearance rate | fitted * | min | ||
| Insulin Bolus | 5 [5–30] | mIU/mL | [3] | |
| Model response variables | Initial conditions | |||
| -cells | 800 | mg | [8] | |
| Insulin | I | 20 | mIU/mL | [44] |
| Glucose | 80 | mg/dL | [10] | |
| Growth Hormone | 30 | mIU/mL | [45] |
| Parameter | Mice Group 1 | Mice Group 2 | Mice Group 3 | Mice Group 4 |
|---|---|---|---|---|
| * | 0.38 (0.3–0.4) | 1.6 (1.5–2) | 0.42 (0.4–0.45) | 0.79 (0.7–0.8) |
| 0.82 (0.63–0.96) | 8.11 (5.67–11.84) | 1.21 (0.93–1.60) | 2.44 (1.65–3.16) |
| Parameter | Mice Group 1 | Mice Group 2 | Mice Group 3 | Mice Group 4 |
|---|---|---|---|---|
| a | 45.28 (36.02–49.72]) | 37.29 (34.45–97.24) | 86.11 (51.79–97.05) | 30.75 (21.21–51.56) |
| b | 0.13 (0.08–0.14) | 0.11 (0.06–0.49) | 0.37 (0.20–0.41) | 0.12 (0.03–0.2) |
| f | 72.19 (22.26–76.49) | 0.28 (0.11–0.3) | 102.81 (24.87–243.27) | 10.05 (3.77–13.76) |
| * | 1.91 (1.9–1.95) | 2.91 (2.9–3) | 2.276 (2.275–2.278) | 1.18 (0.51–1.19) |
| Mouse | ||||||
|---|---|---|---|---|---|---|
| Mouse 1 | 11.73 | 10.95 | 15.78 | 14.21 | 4.05 | 3.26 |
| Mouse 2 | 11.10 | 10.32 | 14.07 | 12.51 | 2.97 | 2.19 |
| Mouse 3 | 11.33 | 10.55 | 14.21 | 12.65 | 2.88 | 2.10 |
| Mouse 4 | 11.18 | 10.70 | 12.38 | 10.82 | 1.20 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Ali, H.; Daneshkhah, A.; Boutayeb, A.; Mukandavire, Z. Examining Type 1 Diabetes Mathematical Models Using Experimental Data. Int. J. Environ. Res. Public Health 2022, 19, 737. https://doi.org/10.3390/ijerph19020737
Al Ali H, Daneshkhah A, Boutayeb A, Mukandavire Z. Examining Type 1 Diabetes Mathematical Models Using Experimental Data. International Journal of Environmental Research and Public Health. 2022; 19(2):737. https://doi.org/10.3390/ijerph19020737
Chicago/Turabian StyleAl Ali, Hannah, Alireza Daneshkhah, Abdesslam Boutayeb, and Zindoga Mukandavire. 2022. "Examining Type 1 Diabetes Mathematical Models Using Experimental Data" International Journal of Environmental Research and Public Health 19, no. 2: 737. https://doi.org/10.3390/ijerph19020737
APA StyleAl Ali, H., Daneshkhah, A., Boutayeb, A., & Mukandavire, Z. (2022). Examining Type 1 Diabetes Mathematical Models Using Experimental Data. International Journal of Environmental Research and Public Health, 19(2), 737. https://doi.org/10.3390/ijerph19020737