1. Introduction
With the rapid development of the economy and technology, public demands for improving healthcare are getting stronger and how to utilize information technology to achieve auxiliary diagnosis has received increasing social attention [
1,
2,
3,
4]. Moreover, the spread of the concept of digital health [
5,
6,
7] promotes deep integration between information technology and healthcare, where a large number of machine learning (ML) models and data mining (DM) methods have been introduced into the traditional medical diagnosis pattern. To date, there are various existing studies adopting the technologies of ML and DM to predict diseases, such as predicting stable MCI patients [
8], forecasting nuanced yet significant MT errors of clinical symptoms [
9], survival risk prediction for esophageal cancer [
10], conducting breast cancer diagnosis [
11], preconception prediction for gestational diabetes mellitus [
12], predicting Alzheimer’s disease [
13], and heart disease prediction [
14,
15].
Disease prediction analysis based on ML and DM is a research trend in medical informatics. These research findings could provide a scientific and reliable diagnostic basis for medical workers and provide an effective technical support for the early intervention of related diseases. For example, Derevitskii et al. construct a hybrid predictive modelling for Thyrotoxic atrial fibrillation, which could be used as part of a decision support system for medical staff who work with thyrotoxicosis patients [
16]. Similarly, Muhammad et al. develop a machine leaning predictive model for coronary artery disease (CAD), which could be used to develop an expert system for diagnosis of CAD patients in Nigeria [
17]. Nevertheless, these aforementioned studies still contain a critical limitation, i.e., the existing proposed models mainly focus on a certain disease and mostly neglect expanding the application’s universality for predicting various other diseases, which means that these models may not obtain accurate prediction results in analyzing those other diseases. Furthermore, a model with better performance when predicting different diseases would be more significant for the medical workers and the auxiliary diagnosis process. Therefore, constructing a disease prediction model with self-adaptive ability, strong generalization ability, and strong robustness is the motivation to explore the technology-oriented pathway for auxiliary diagnosis in digital health age.
There may be large differences among the disease data from the real world, such as different attribute dimensions and different inner structures. In fact, an ideal disease prediction model which meets current medical needs should combine prediction accuracy with broad applicability. To this end, we take the kernel extreme learning machine (KELM) as the base model, which has advantages in generalization ability and robustness, and we introduce an improved swarm intelligence optimization algorithm to optimize the KELM, i.e., sparrow search algorithm (SSA) with the enhanced global searching ability (EGSSA). Finally, we design a novel disease prediction model, i.e., multi-strategies optimization-based kernel extreme learning machine (MsO-KELM). The main contributions and innovations of the MsO-KELM are highlighted as follows:
(1) To effectively predict various diseases, we utilize the EGSSA to optimize the base model by designing four novel strategies, i.e., the hunger-state foraging strategy of producers (PHFS), the parallel strategy for exploration and exploitation (EEPS), the perturbation–exploration strategy (PES), and the parameter self-adaptive strategy (PSAS). These strategies can enhance the prediction accuracy of the model and allow the model to be applied to various diseases.
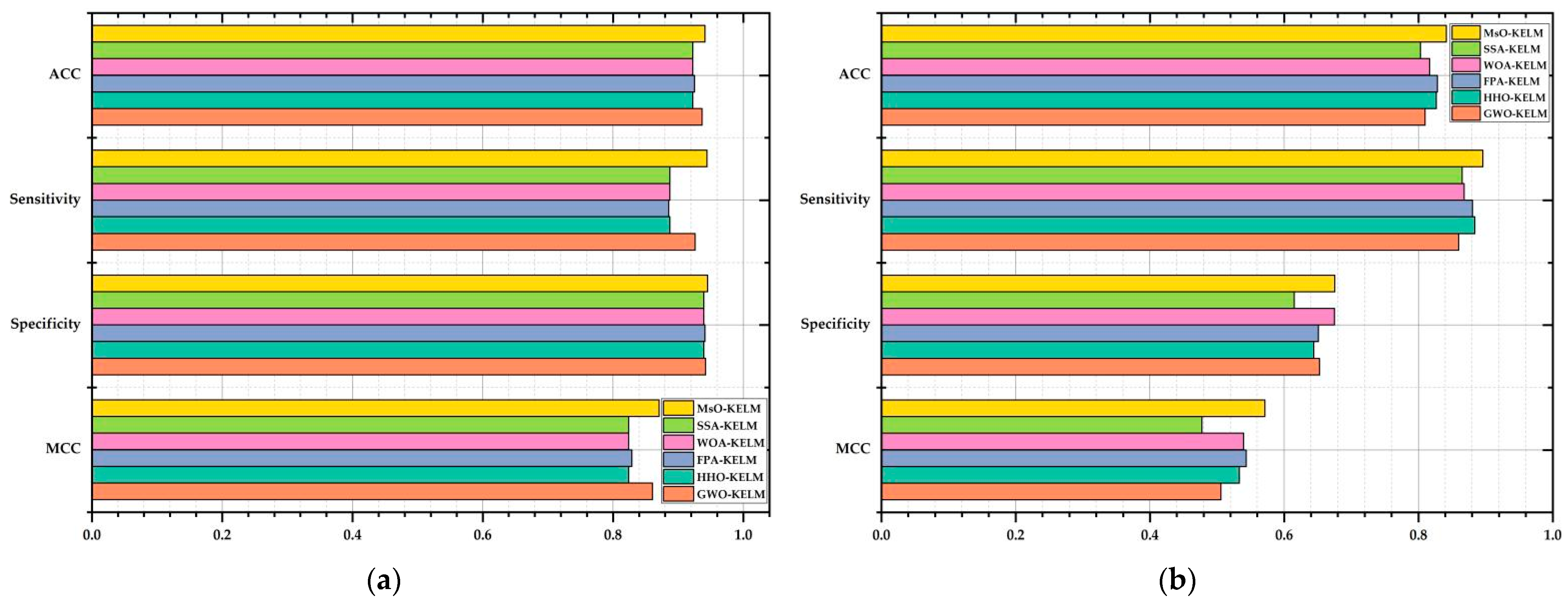
(2) To verify the prediction performance of the proposed MsO-KELM, we adopt six different disease datasets as the experimental samples, consisting of the Breast Cancer dataset (cancer), the Parkinson dataset (Parkinson’s disease), the Autistic Spectrum Disorder Screening Data for Children dataset (Autism Spectrum Disorder), the Heart Disease dataset (heart disease), the Cleveland dataset (heart disease), and the Bupa dataset (liver disease), and we evaluate the prediction performance by four different evaluation metrics, i.e., the ACC, the sensitivity, the specificity, and the MCC. Notably, the ACC is the most significant metric to evaluate the prediction accuracy.
(3) To elaborate the details of the MsO-KELM, we conduct two-stage experiments in this paper. The first experiment is mainly to prove the better optimization performance of the EGSSA, which is the basis for achieving the self-adaptive characteristic of the MsO-KELM. The second experiment is mainly to compare the MsO-KELM with other state-of-the-art prediction models.
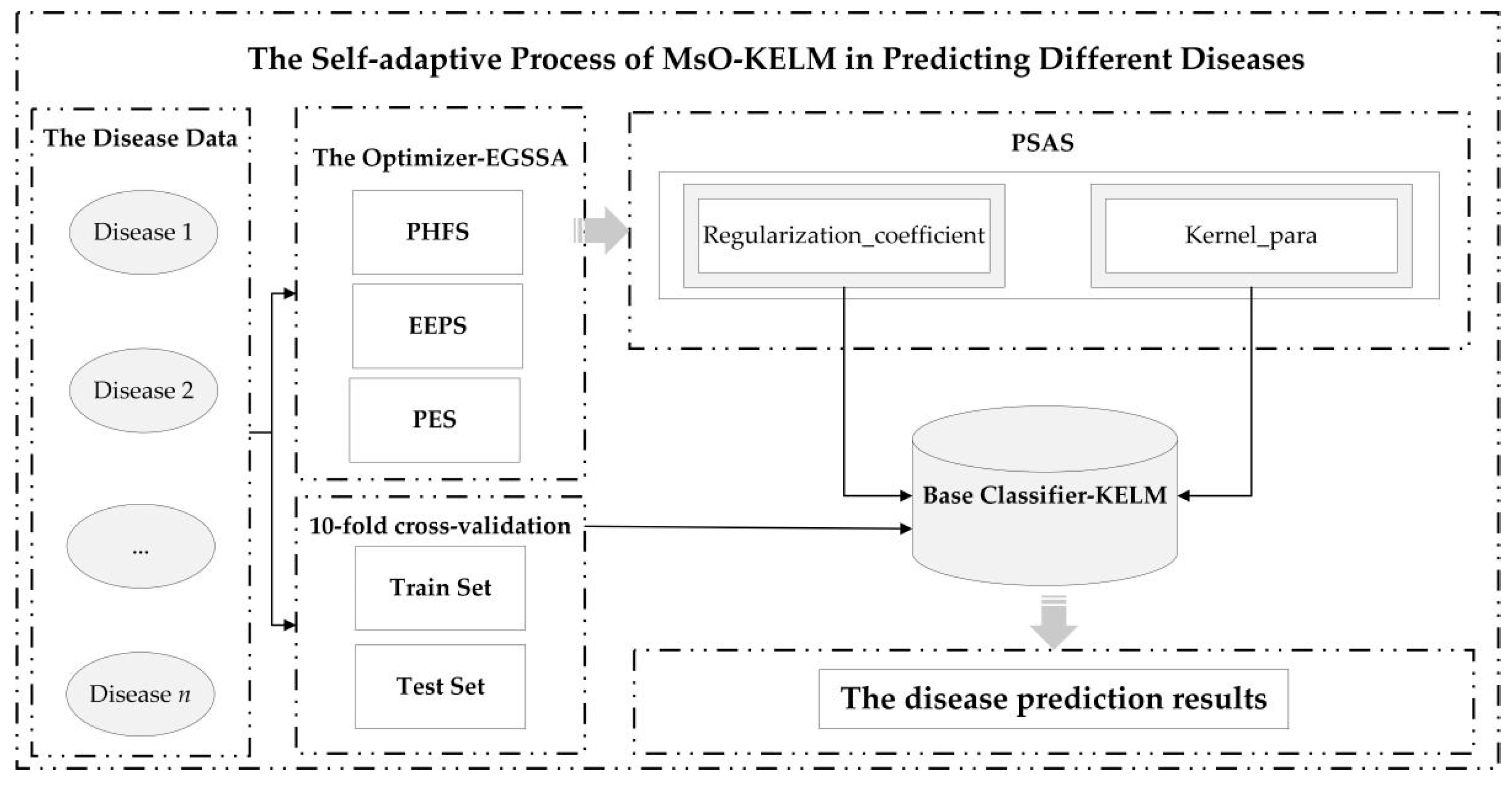
The self-adaptive prediction process of the MsO-KELM in analyzing the different diseases is shown in
Figure 1.
3. The Proposed Methodology
The core idea of the proposed methodology is to construct a self-adaptive disease prediction model with high accuracy, strong generalization ability, and strong robustness. Therefore, we select an excellent base-classifier (KELM) and design an enhanced meta-heuristic algorithm (EGSSA) as the optimizer to finally construct the self-adaptive disease prediction model, i.e., the MsO-KELM. Specifically, there are four novel strategies in the MsO-KELM, i.e., the hunger-state foraging strategy of producers (PHFS), the parallel strategy for exploration and exploitation (EEPS), the perturbation–exploration strategy (PES), and the parameter self-adaptive strategy (PSAS), where the EGSSA consists of the PHFS, the EEPS, and the PES. In addition, the PSAS will act on a parameter acquisition mechanism which is formed by combining the EGSSA with the KELM. In this section, the technological details of the MsO-KELM will be discussed as follows:
3.1. Foraging Strategy of Producers in Hunger-State (PHFS)
In the original SSA, the producers could be regarded as the leader roles in the sparrow populations, which are responsible for searching for food-rich positions and providing foraging directions for all scroungers, and the scroungers could follow the producers to achieve the foraging process. In that case, if the producers could expand the searching range to find a safer and more adequate position, it would provide more possibilities for scroungers to improve the foraging rate and finally enhance the global convergence performance.
However, the existing location update mechanism mainly focuses on the exploitation ability (local searching ability) of the original SSA, which may cause the producers trapping into the local optimum situation. To address this issue and enhance the exploration ability (global searching ability), we introduce the hunger games search algorithm (HGS) [
35,
36] into the original SSA to construct the PHFS for expanding the searching range of optimal position. Specifically, the PHFS is a hybrid strategy, which retains the advantage in exploitation ability of the original SSA while combining the exploration approach of the HGS algorithm with the location update mechanism of the producers.
In the original SSA, the location update process of producers is shown as Equation (4), and the key calculation function affecting the convergence efficiency is shown in Equation (11):
where the
α and
itermax are the parameters set manually. It is clearly shown that Equation (11) has a descending trend and will eventually converge to 0, which means the producers can easily repeat the searching behavior at a certain position with the number of individuals increasing, and eventually fall into a local optimum. By contrast, the HGS has a significant advantage in exploration ability, and the hungry feature function affecting the convergence efficiency is shown in Equation (12):
where the
par_maunal is a parameter set manually. It is clearly shown that Equation (12) is not a single ascending or descending trend, but the function trend is affected by the parameter value. When the inputting value is less than the
par_maunal value, the function could have an ascending trend with the inputting value increasing. Moreover, when the inputting value is larger than the
par_maunal value, the function could have a descending trend with the inputting value increasing. Therefore, introducing the hungry feature of HGS into the searching behavior of producers could expand the searching range of optimal position, and ultimately enhance the exploration ability of SSA.
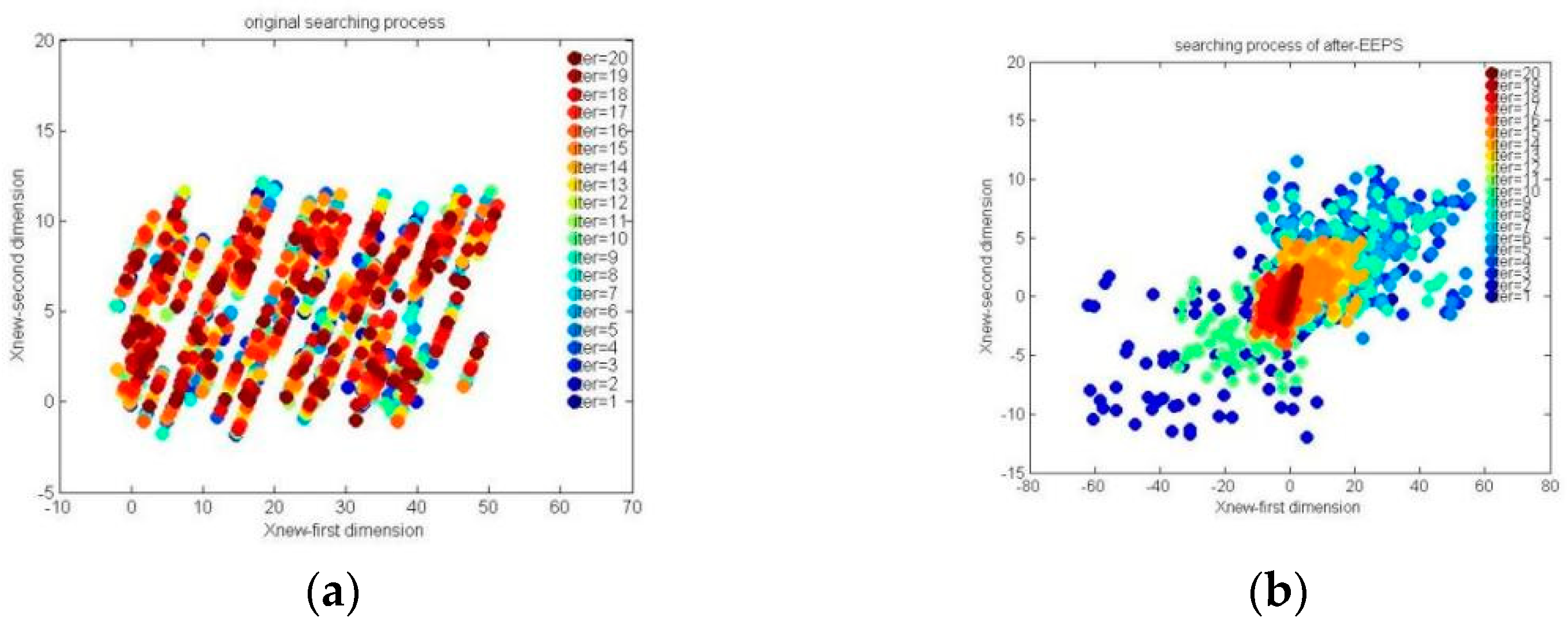
The searching processes based on Equations (11) and (12) are shown in
Figure 2.
According to
Figure 2, we could find that the original location update method can easily search in a local space (like the red region in
Figure 2a), but the hungry roles of HGS could search in a relatively global region (like
Figure 2b). Therefore, the PHFS could be described as follows:
where the meaning of
is similar to that of
in Equation (4), the
indicates the location of last iteration, the
rand is a random number, and the
o is an adjustment parameter which is set to 2 in this paper.
3.2. Parallel Strategy for Exploration and Exploitation (EEPS)
As shown in Equation (4), we could find that the location update process of producers consists of two different stages. The PHFS acts on stage 1 (R < ST), and the EEPS described below is going to act on stage 2 (R ≥ ST). According to the principle of SSA, the producers would move to a safer place when perceiving danger coming. In the new location update process, the producers would search globally with a normally distributed random manner and eventually converge to the optimal position. Nevertheless, there are still some limitations in this moving process, i.e., (i) the existing searching range could continue to be expanded; and (ii) the single global searching process could affect the convergence accuracy. Therefore, we design the EEPS to balance the exploration process and exploitation process in this paper. In fact, the balancing effect of EEPS is able to dynamically adjust the position searching approach of the producers, i.e., by promoting the producers exploring the whole space with a global searching approach for expanding the searching range of the potential optimal position in the early stage, while exploiting the current area with a local searching approach when a certain area is close to the optimal position.
Similarly, inspired by the literature [
37,
38] related to HGS, we introduce the pattern of food-approaching into the location update process of producers in stage 2, and construct the balance factor shown in Equation (14):
where the meaning of
para is a random number in the range of (0, 1), the
δ indicates a control parameter (it is set to 2), the
iter indicates the current number of iterations, and the
iter_max indicates the maximum number of iterations.
Subsequently, the EEPS, which combines the balance factor with the original location update mechanism, is shown in Equation (15):
where the meaning of the parameters are similar to Equations (4), (6) and (14).
The comparison results of the original searching process and the searching process based on after-EEPS are shown in
Figure 3a,b, respectively.
In
Figure 3, the points in the same color are the positions of all producers in one iteration, while different colors indicate different iterations.
Figure 3a shows that the original location update mechanism of producers in stage 2 could achieve a global searching process to some extent, but it lacks the local exploitation behavior and the global searching range is not large enough, which can finally affect the convergence accuracy. As a comparison,
Figure 3b shows that the EEPS not only expands the global searching range, but retains the exploitation ability, which can be clearly seen in the color changing process of points.
3.3. Perturbation–Exploration Strategy (PES)
Notably, the PHFS and EEPS only enhance the exploration ability of producer populations, which means that there remains the possibility of trapping into local optimum for the whole sparrow populations. Therefore, we introduce the Cauchy distribution operator [
39,
40,
41] to construct the PES to further expand the searching range and avoid the local optimum situation in late iterations.
In the PES, we adopt the Cauchy distribution operator to obtain a variant of the current optimal individual. Moreover, we compare the fitness of the current optimal individual with that of the variant to save the better solution. In this paper, the probability density function of Cauchy distribution operator is shown as follows:
where the
a is a parameter (in this paper, the
a is equal to 1), and the
k indicates a variable, the value range of which is from negative infinity to positive infinity. Based on the Cauchy distribution operator, we could obtain the perturbation–exploration process as follows:
where the
r is a Cauchy distribution random variable generating function.
3.4. Parameter Self-Adaptive Strategy (PSAS)
As we all know, obtaining the appropriate parameter values (k and c) is most significant for the original KELM. However, the existing method of parameter taking is to adopt grid searching, which not only increases the computational cost but also affects the final results of parameter obtaining. To achieve a self-adaptive process of these two parameters, we propose the PSAS via combining the EGSSA with the KELM.
Specifically, there are four significant stages in the PSAS, and the technical details are as follows: (i) achieving the location initialization process of sparrow populations (this paper adopts the random generation method of the original SSA); (ii) the core parameters (
k and
c) could be automatically obtained by adopting the EGSSA; (iii) to eliminate the randomness of these two obtained parameters, this paper utilizes 10-fold cross-validation [
18] to re-obtain the optimal parameter values; and (iv) the re-obtained optimal parameter values are introduced into the original KELM, and finally the MsO-KELM is formed to perform the prediction performance on six different disease datasets based on 10-fold cross-validation.
In summary, the novel MsO-KELM model utilizes the four major strategies, i.e., PHFS, EEPS, PES, and PSAS, to achieve improved prediction performance, and the specific details of the MsO-KELM are shown in Algorithm 1.
| Algorithm 1: MsO-KELM model |
![Ijerph 19 12509 i001]() |
5. Discussion
Combining information technology with medical information to realize the auxiliary diagnosis is a research trend in the digital health age; therefore, researchers have conducted a large number of prediction studies on common diseases by utilizing some machine learning models. However, the existing studies have weak generalization performance, which limits their further application for other diseases. In other words, these findings may obtain better prediction results in solving a certain disease but may obtain worse prediction results in solving other, different diseases. The reason why these findings have weak generalization performance is that the researchers mainly emphasize result-orientation for a certain disease, i.e., they focus on training with single disease data to obtain an effective prediction model which is suitable for the current disease. In fact, a prediction model with better generalization performance could help medical workers to diagnose various diseases. Therefore, exploring a prediction model with strong generalization ability has important theoretical and practical significance in the current digital health age.
5.1. Theoretical Significance
This paper is an attempt to explore a technology-oriented pathway for auxiliary diagnosis. On the one hand, our investigation adopts different disease data as the experimental objects, and aims to expand the application scope of the final findings by mining the internal characteristics of these different disease data, which could provide a novel research idea for researchers to eliminate the application limitations of the existing studies, and could provide effective theoretical guidance for researchers to realize comprehensive auxiliary diagnosis in facing various diseases.
On the other hand, utilizing an enhanced meta-heuristic algorithm to optimize the operating mechanism and inner structure of the original classifier is an efficient method to improve the performance of a model. The proposed MsO-KELM not only enhances the generalization ability and rapid learning ability of the original KELM classifier, but also realizes the self-adaptive process in predicting different diseases by introducing the EGSSA optimizer. The results of the evaluation metrics show that the MsO-KELM has significant advantages among all compared models in predicting the six diseases. Specifically, the MsO-KELM can obtain the best ACC value when predicting each disease, such that the ACC value is 94.124% in predicting breast cancer, the ACC value is 91.079% in predicting Autistic Spectrum Disorder, and the ACC value is 84.167% in predicting Parkinson’s disease. These ACC values could reflect the effectiveness of the MsO-KELM in disease prediction. Compared with some specific disease models [
11,
50], there may be a slight decrease in prediction accuracy of the MsO-KELM. However, according to the No Free Lunch (NFL) theory [
51], although the MsO-KELM has slightly decreased accuracy in predicting a certain disease, it could predict more different diseases, which enhances the medical application value of the model.
5.2. Practical Significance
On the one hand, for the six different real-world diseases selected in this study, our investigation could assist doctors to diagnose or screen patients with related diseases, guide patients to prevent the diseases in a targeted manner, reduce the risk level of these diseases, and finally improve the survival quality of the patients.
On the other hand, there may be a consistent one-to-one match between the existing prediction models and their prediction targets, which means they will increase the economic cost and time cost for medical departments when analyzing different diseases. Therefore, a universal prediction model with strong generalization ability and robustness could predict more different diseases so as to improve the overall work efficiency of the medical workers.
5.3. Limitations
This study completes the construction of a self-adaptive prediction model in solving different diseases, but some limitations related to the MsO-KELM should be noted.
(i) In terms of model, although this paper presents four entirely novel optimization strategies to further optimize the prediction performance of the model, the performance improvement is accompanied by an increase in computational complexity, which is inevitable and could be comprehended according to the NFL theory. Therefore, we will further enhance the prediction performance of the MsO-KELM by redesigning a novel parameter optimization mechanism and optimizing the fundamental structure of the prediction model to reduce the computational complexity in the future studies.
(ii) In terms of data, on the one hand, the disease data analyzed in this paper are publicly available datasets, where each disease dataset has a limited sample size and some datasets even have specific geographical characteristics; on the other hand, the number of disease categories covered by the selected datasets are not enough. Therefore, we will increase the category of disease data and expand the sample size of disease data. In addition, we will also strive for collecting global disease data to eliminate the influence of the geographical factors.