Influence of Local Muscle Cooling on Mitochondrial-Related Gene Expression at Rest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Skin Temperature
2.3. Arterial Hemodynamics
2.4. Intramuscular Temperature and Biopsies
2.5. mRNA Analyses
2.6. Statistical Analysis
3. Results
3.1. Temperature
3.2. Arterial Hemodynamics
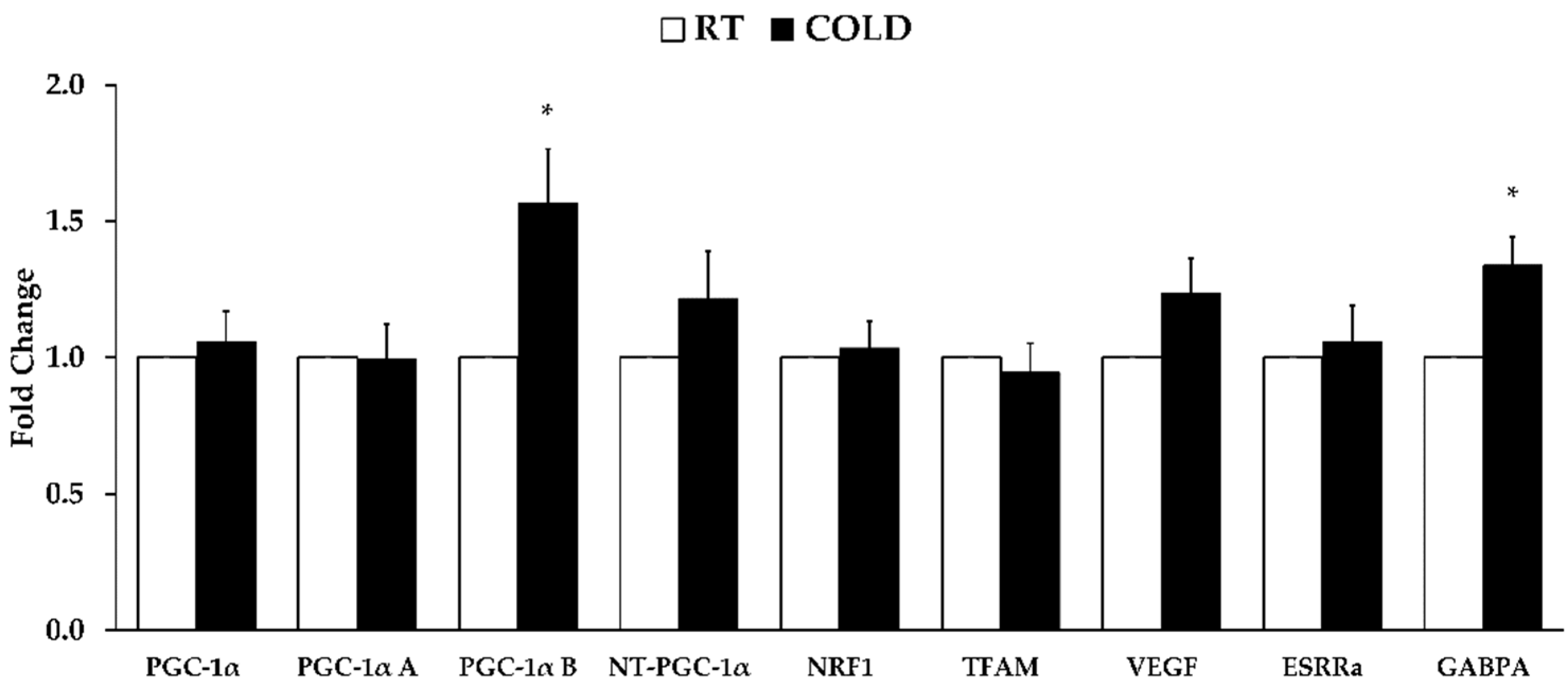
3.3. Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Derbré, F.; Gomez-Cabrera, M.C.; Nascimento, A.L.; Sanchis-Gomar, F.; Martinez-Bello, V.E.; Tresguerres, J.A.F.; Fuentes, T.; Gratas-Delamarche, A.; Monsalve, M.; Viña, J. Age Associated Low Mitochondrial Biogenesis may be Explained by Lack of Response of PGC-1α to Exercise Training. Age 2012, 34, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Bournat, J.C.; Brown, C.W. Mitochondrial dysfunction in obesity. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 446–452. [Google Scholar] [CrossRef]
- Bose, A.; Beal, M.F. Mitochondrial Dysfunction in Parkinson’s Disease. J. Neurochem. 2016, 139, 216–231. [Google Scholar] [CrossRef]
- Hoppeler, H.; Luthi, P.; Claassen, H.; Weibel, E.R.; Howald, H. The Ultrastructure of the Normal Human Skeletal Muscle. Pflugers. Archiv. 1973, 344, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Garesse, R.; Vallejo, C.G. Animal Mitochondrial Biogenesis and Function: A Regulatory Cross-Talk between Two Genomes. Gene 2001, 263, 1–16. [Google Scholar] [CrossRef]
- Wright, G.L.; Maroulakou, I.G.; Eldridge, J.; Liby, T.L.; Sridharan, V.; Tsichlis, P.N.; Muise-Helmericks, R.C. VEGF Stimulation of Mitochondrial Biogenesis: Requirement of AKT3 Kinase. FASEB J. 2008, 22, 3264–3275. [Google Scholar] [CrossRef]
- Chinsomboon, J.; Ruas, J.; Gupta, R.K.; Thom, R.; Shoag, J.; Rowe, G.C.; Sawada, N.; Raghuram, S.; Arany, Z. The Transcriptional Coactivator PGC-1α Mediates Exercise-Induced Angiogenesis in Skeletal Muscle. Proc. Natl. Acad. Sci. USA 2009, 106, 21401–21406. [Google Scholar] [CrossRef] [PubMed]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a Selective Autophagy Receptor for Mitochondrial Clearance. EMBO Rep. 2010, 11, 45–51. [Google Scholar] [CrossRef]
- Mammucari, C.; Milan, G.; Romanello, V.; Masiero, E.; Rudolf, R.; Del Piccolo, P.; Burden, S.J.; Di Lisi, R.; Sandri, C.; Zhao, J.; et al. FoxO3 Controls Autophagy in Skeletal Muscle in vivo. Cell Metab. 2007, 6, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [Green Version]
- Opichka, M.; Shute, R.; Marshall, K.; Slivka, D. Effects of Exercise in a Cold Environment on Gene Expression for Mitochondrial Biogenesis and Mitophagy. Cryobiology 2019, 90, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Leick, L.; Plomgaard, P.; Grønlykke, L.; Al-Abaiji, F.; Wojtaszewski, J.F.P.; Pilegaard, H. Endurance Exercise Induces mRNA Expression of Oxidative Enzymes in Human Skeletal Muscle Late in Recovery. Scand. J. Med. Sci. Sports 2010, 20, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Slivka, D.R.; Heesch, M.W.S.; Dumke, C.L.; Cuddy, J.S.; Hailes, W.S.; Ruby, B.C. Effects of Post-Exercise Recovery in a Cold Environment on Muscle Glycogen, PGC-1 α, and Downstream Transcription Factors. Cryobiology 2013, 66, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Shute, R.J.; Marshall, K.; Opichka, M.; Schnitzler, H.; Ruby, B.C.; Slivka, D.R. Effects of 7 °C Environmental Temperature Acclimation during a 3-Week Training Period. J. Appl. Physiol. 2020, 128, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Slivka, D.R.; Dumke, C.L.; Tucker, T.J.; Cuddy, J.S.; Ruby, B.C. Human mRNA Response to Exercise and Temperature. Int. J. Sports Med. 2012, 33, 94–100. [Google Scholar] [CrossRef]
- Larson, C.; Opichka, M.; McGlynn, M.L.; Collins, C.W.; Slivka, D.R. Exercise- and Cold-Induced Human PGC-1α mRNA Isoform Specific Responses. Int. J. Environ. Res. Public Health 2020, 17, 5740. [Google Scholar] [CrossRef] [PubMed]
- Zak, R.B.; Shute, R.J.; Heesch, M.W.S.; La Salle, D.T.; Bubak, M.P.; Dinan, N.E.; Laursen, T.L.; Slivka, D.R. Impact of Hot and Cold Exposure on Human Skeletal Muscle Gene Expression. Appl. Physiol. Nutr. Metab. 2017, 42, 319–325. [Google Scholar] [CrossRef]
- Meister, B.; Collins, C.W.; McGlynn, M.L.; Slivka, D.R. Effect of Local Cold Application during Exercise on Gene Expression Related to Mitochondrial Homeostasis. Appl. Physiol. Nutr. Metab. 2021, 46, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Shute, R.J.; Heesch, M.W.S.; Laursen, T.; Slivka, D.R. Local Muscle Cooling does Not Impact Expression of Mitochondrial-Related Genes. J. Therm. Biol. 2017, 67, 35–39. [Google Scholar] [CrossRef]
- Tucker, T.J.; Slivka, D.R.; Cuddy, J.S.; Hailes, W.S.; Ruby, B.C. Effect of local cold application on glycogen recovery. J. Sports Med. Phys. Fit. 2012, 52, 158–164. [Google Scholar]
- Rossman, M.J.; Groot, H.J.; Garten, R.S.; Witman, M.A.H.; Richardson, R.S. Vascular function assessed by passive leg movement and flow-mediated dilation: Initial evidence of construct validity. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H1277–H1286. [Google Scholar] [CrossRef] [PubMed]
- Trinity, J.D.; Barrett-O’keefe, Z.; Ives, S.J.; Morgan, G.; Rossman, M.J.; Donato, A.J.; Runnels, S.; Morgan, D.E.; Gmelch, B.S.; Bledsoe, A.D.; et al. Endogenous Endothelin-1and Femoral Artery Shear Rate: Impact of Age and Implications for Atherosclerosis. J. Hypertens. 2016, 34, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Bergström, J. Muscle Electrolytes in Man Determined by Neutron Activation Analysis on Needle Biopsy Specimens. Scand. J. Clin. Lab. Investig. 1962, 14, 1–110. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2(−ΔΔ C(T)). Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Freitag, L.; Clijsen, R.; Deflorin, C.; Taube, W.; Taeymans, J.; Hohenauer, E. Intramuscular Temperature Changes in the Quadriceps Femoris Muscle after Post-Exercise Cold-Water Immersion (10 °C for 10 Min): A Systematic Review with Meta-Analysis. Front. Sport Act. Living 2021, 3, 100. [Google Scholar] [CrossRef] [PubMed]
- Tansey, E.A.; Johnson, C.D. Recent Advances in Thermoregulation. Adv. Physiol. Educ. 2015, 39, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Yen, T.C.; Zhao, K.; Kosiba, W.A.; Johnson, J.M.; Yen, T.C.; Zhao, K.; Kosiba, W.A. Sympathetic, sensory, and nonneuronal contributions to the cutaneous vasoconstrictor response to local cooling. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H1573–H1579. [Google Scholar] [CrossRef]
- Hodges, G.J.; Zhao, K.; Kosiba, W.A.; Johnson, J.M. The Involvement of Nitric Oxide in the Cutaneous Vasoconstrictor Response to Local Cooling in Humans. J. Physiol. 2006, 574, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Fiscus, K.A.; Kaminski, T.W.; Powers, M.E. Changes in Lower-Leg Blood Flow during Warm-, Cold-, and Contrast-Water Therapy. Arch. Phys. Med. Rehab. 2005, 86, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Kai, Y.; Kamei, Y.; Ezaki, O. Isoform-Specific Increases in Murine Skeletal Muscle Peroxisome Proliferator-Activated Receptor-Γ Coactivator-1α (PGC-1α) mRNA in Response to Β2-Adrenergic Receptor Activation and Exercise. Endocrinology 2008, 149, 4527–4533. [Google Scholar] [CrossRef]
- Miura, S.; Kawanaka, K.; Kai, Y.; Tamura, M.; Goto, M.; Shiuchi, T.; Minokoshi, Y.; Ezaki, O. An Increase in Murine Skeletal Muscle Peroxisome Proliferator-Activated Receptor-Γ Coactivator-1α (PGC-1α) mRNA in Response to Exercise is Mediated by Β-Adrenergic Receptor Activation. Endocrinology 2007, 148, 3441–3448. [Google Scholar] [CrossRef] [Green Version]
- Cannon, B.; Needergaard, J. Brown Adipose Tissue: Function and Physiological Significance. Physiol. Rev. 2003, 84, 277–359. [Google Scholar] [CrossRef]
- Janský, L. Humoral Thermogenesis and its Role in Maintaining Energy Balance. Physiol. Rev. 1995, 75, 237–259. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Transcriptional Paradigms in Mammalian Mitochondrial Biogenesis and Function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar] [CrossRef]
- Baldelli, S.; Aquilano, K.; Ciriolo, M.R. Punctum on Two Different Transcription Factors Regulated by PGC-1α: Nuclear Factor Erythroid-Derived 2-Like 2 and Nuclear Respiratory Factor 2. Biochim. Biophys. Acta 2013, 1830, 4137–4146. [Google Scholar] [CrossRef]
- Blesa, J.R.; Hernandez-Yago, J. Distinct Functional Contributions of 2 GABP-NRF-2 Recognition Sites within the Context of the Human TOMM70 Promoter. Biochem. Cell Biol. 2006, 84, 813–822. [Google Scholar] [CrossRef]

| Primer 1 | Primer 2 | Probe | |
|---|---|---|---|
| Reference | |||
| B-Actin | CCTTGCACATGCCGGAG | ACAGAGCCTCGCCTTTG | TCATCCATGGTGAGCTGGCGG |
| B2M | ACCTCCATGATGCTGCTTAC | GGACTGGTCTTTCTATCTCTTGT | CCTGCCGTGTGAACCATGTGACT |
| GAPDH | TGTAGTTGAGGTCAATGAAGGG | ACATCGCTCAGACACCATG | AAGGTCGGAGTCAACGGATTTGGTC |
| RPS18 | GTCAATGTCTGCTTTCCTCAAC | GTTCCAGCATATTTTGCGAGT | TCTTCGGCCCACACCCTTAATGG |
| Biogenesis | |||
| PGC-1α | AGCCTCTTTGCCCAGATCTT | GGCAATCCGTCTTCATCCAC | AGCTTTCTGGGTGGACTCAAGTGG |
| PGC-1α A | ATGGAGTGACATCGAGTGTGCT | GAGTCCACCCAGAAAGCTGT | AAGACCAGCCTCTTTGCCCAGATC |
| PGC-1α B | CTATGGATTCAATTTTGAAATGTGC | CTGATTGGTCACTGCACCAC | AAGACCAGCCTCTTTGCCCAGATC |
| NT-PGC-1α | TCACACCAAACCCACAGAGA | CTGGAAGATATGGCACAT | AAAGAAGTCCCACACACAGTCGCA |
| VEGF | GCGCTGATAGACATCCATGA | CCATGAACTTTCTGCTGTCTTG | TGCTCTACCTCCACCATGCCAAG |
| TFAM | GCCAAGACAGATGAAAACCAC | TGGGAAGGTCTGGAGCA | CGCTCCCCCTTCAGTTTTGTGTATTT |
| ESRRα | TCTCCGCTTGGTGATCTCA | CTATGGTGTGGCATCCTGTG | TGGTCCTCTTGAAGAAGGCTTTGCA |
| NRF1 | GTCATCTCACCTCCCTGTAAC | GATGCTTCAGAATTGCCAACC | ATGGAGAGGTGGAACAAAATTGGGC |
| GABPA | TGGCTTCTGGACTTGGAAC | GACGGTATGCAACAGGACAT | CAATATTAAGACACTGTAACTCAGGAATGGATAATAGCTC |
| Mitophagy | |||
| PINK1 | GTTGCTTGGGACCTCTCTTG | TGAACACAATGAGCCAGGAG | TGTAAGTGACTGCTCCATACTCCCCA |
| PARK2 | GCTTGGTGGTTTTCTTGATGG | TTGAAGCCTCAGGAACAACT | CCTGCTCGGCGGCTCTTTCA |
| BNIP3 | CCACTAACGAACCAAGTCAGAC | CATCTCTGCTGCTCTCTCAT | AAAGGTGCTGGTGGAGGTTGTCA |
| BNIP3 L | CAAACATGATCTGCCCATCTTC | TCCTCATCCTCCATCCACAA | TCTCACTGTGACAGCCCTTCGC |
| RT | COLD | |
|---|---|---|
| Blood Velocity (m‧s–1) | 0.64 ± 0.13 | 0.62 ± 0.17 |
| Arterial Diameter (cm) | 0.62 ± 0.05 | 0.60 ± 0.05 * |
| Blood Flow (mL‧min–1) | 275.0 ± 54.0 | 259.1 ± 69.0 |
| Shear Rate (s–1) | 412 ± 27 | 412 ± 34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robins, L.; Kwon, M.; McGlynn, M.L.; Rosales, A.M.; Pekas, E.J.; Collins, C.; Park, S.-Y.; Slivka, D.R. Influence of Local Muscle Cooling on Mitochondrial-Related Gene Expression at Rest. Int. J. Environ. Res. Public Health 2022, 19, 12028. https://doi.org/10.3390/ijerph191912028
Robins L, Kwon M, McGlynn ML, Rosales AM, Pekas EJ, Collins C, Park S-Y, Slivka DR. Influence of Local Muscle Cooling on Mitochondrial-Related Gene Expression at Rest. International Journal of Environmental Research and Public Health. 2022; 19(19):12028. https://doi.org/10.3390/ijerph191912028
Chicago/Turabian StyleRobins, Larry, Monica Kwon, Mark L. McGlynn, Alejandro M. Rosales, Elizabeth J. Pekas, Christopher Collins, Song-Young Park, and Dustin R. Slivka. 2022. "Influence of Local Muscle Cooling on Mitochondrial-Related Gene Expression at Rest" International Journal of Environmental Research and Public Health 19, no. 19: 12028. https://doi.org/10.3390/ijerph191912028
APA StyleRobins, L., Kwon, M., McGlynn, M. L., Rosales, A. M., Pekas, E. J., Collins, C., Park, S.-Y., & Slivka, D. R. (2022). Influence of Local Muscle Cooling on Mitochondrial-Related Gene Expression at Rest. International Journal of Environmental Research and Public Health, 19(19), 12028. https://doi.org/10.3390/ijerph191912028







