Novel Chimeric Vaccine Candidate Development against Leptotrichia buccalis
Abstract
1. Introduction
2. Research Methodology
2.1. Complete Genome Retrieval
2.2. Subcellular Localization and Virulent Protein Analysis
2.3. Epitope Prediction
2.4. Epitopes Prioritization Phase
2.5. Vaccine Construct Design
2.6. Structure Modelling of the Vaccine Construct
2.7. Disulfide Engineering
2.8. Molecular Docking
2.9. Molecular Dynamic Simulation
3. Results and Discussion
3.1. Complete Sequenced Genome Retrieval
3.2. CD-HIT Analysis
3.3. Subcellular Localization
3.4. Virulent Factor Database Analysis
3.5. B- and T-Cell Epitope Prediction
3.6. Epitope Properties Analyses
3.7. Vaccine Construct Design
3.8. D Structure, Loop Modelling and Refinement
3.9. Disulfide Engineering
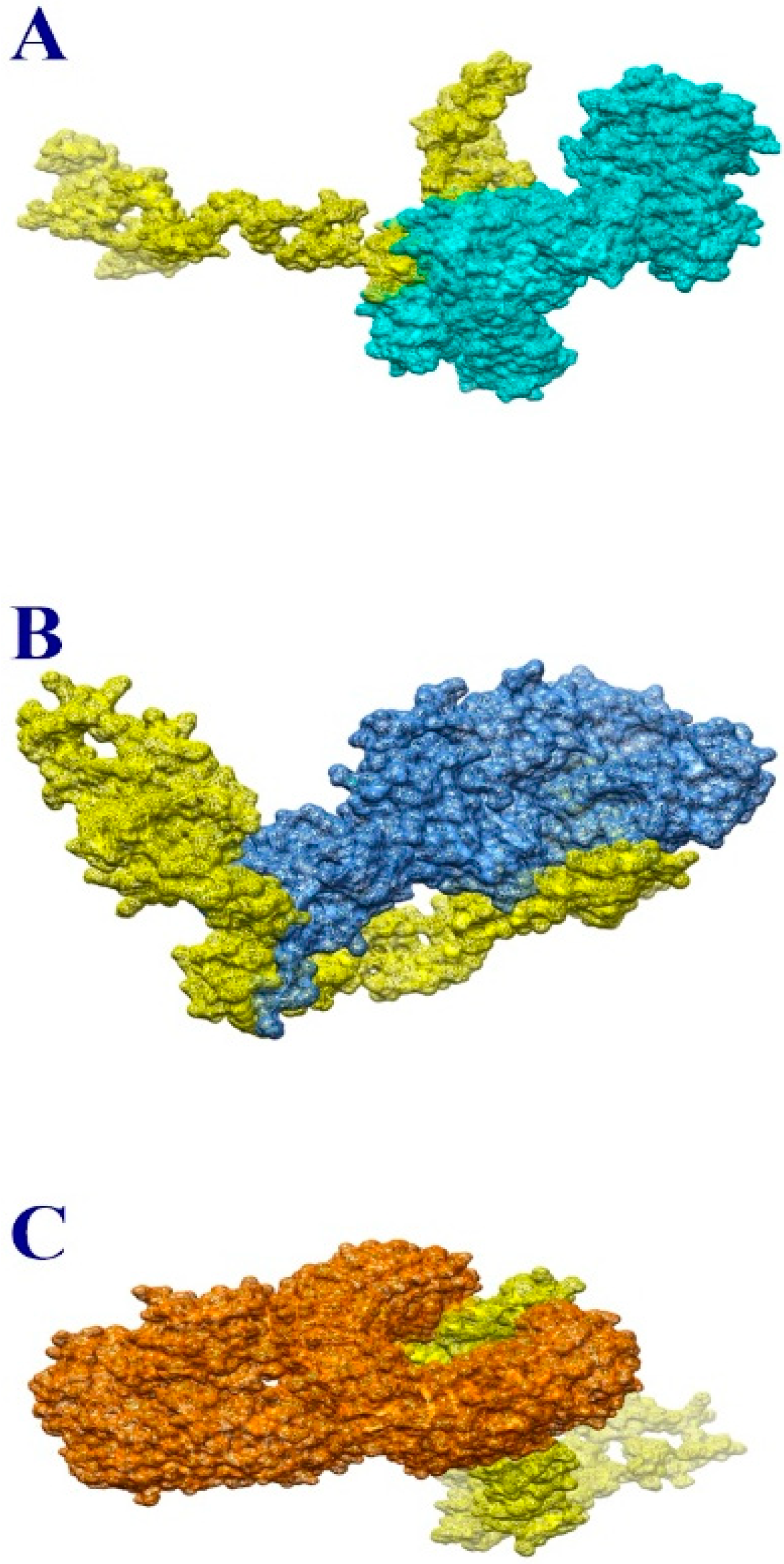
3.10. Molecular Docking
3.11. Molecular Dynamic Simulation
3.12. Estimation of Binding Free Energy
4. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Z.; Li, W. Deciphering the Role of Human Gastrointestinal Microbiota in the Pathogenesis of Vaginal Infection and Cervical Cancer. J. Environ. Pathol. Toxicol. Oncol. 2020, 39, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Fonti, V.; Di Cesare, A.; Šangulin, J.; Del Negro, P.; Celussi, M. Antibiotic Resistance Genes and Potentially Pathogenic Bacteria in the Central Adriatic Sea: Are They Connected to Urban Wastewater Inputs? Water 2021, 13, 3335. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Ullah, A.; Ahmad, S.; Ismail, S.; Afsheen, Z.; Khurram, M.; Tahir ul Qamar, M.; AlSuhaymi, N.; Alsugoor, M.H.; Allemailem, K.S. Towards A Novel Multi-Epitopes Chimeric Vaccine for Simulating Strong Immune Responses and Protection against Morganella Morganii. Int. J. Environ. Res. Public Health 2021, 18, 10961. [Google Scholar] [CrossRef]
- Yero, D.; Conchillo-Solé, O.; Daura, X. Antigen Discovery in Bacterial Panproteomes. In Vaccine Delivery Technology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 43–62. [Google Scholar]
- Reche, P.A.; Fernandez-Caldas, E.; Flower, D.R.; Fridkis-Hareli, M.; Hoshino, Y. Peptide-Based Immunotherapeutics and Vaccines. J. Immunol. Res. 2014, 2014, 256784. [Google Scholar] [CrossRef]
- Smith, K.A. Edward Jenner and the Small Pox Vaccine. Front. Immunol. 2011, 2, 21. [Google Scholar] [CrossRef]
- Rappuoli, R. Reverse Vaccinology, a Genome-Based Approach to Vaccine Development. Vaccine 2001, 19, 2688–2691. [Google Scholar] [CrossRef]
- Ali, A.; Naz, A.; Soares, S.C.; Bakhtiar, M.; Tiwari, S.; Hassan, S.S.; Hanan, F.; Ramos, R.; Pereira, U.; Barh, D.; et al. Pan-Genome Analysis of Human Gastric Pathogen, H. Pylori: Comparative Genomics and Pathogenomics Approaches to Identify Regions Associated with Pathogenicity and Prediction of Potential Core Therapeutic Targets. Biomed Res. Int. 2015, 2015, 139580. [Google Scholar] [CrossRef]
- Bagchi, A.; Saha, P.; Biswas, A.; Islam, S.M. Application of Microbes in Vaccine Production. In Application of Microbes in Environmental and Microbial Biotechnology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 573–585. [Google Scholar]
- Naz, K.; Naz, A.; Ashraf, S.T.; Rizwan, M.; Ahmad, J.; Baumbach, J.; Ali, A. PanRV: Pangenome-Reverse Vaccinology Approach for Identifications of Potential Vaccine Candidates in Microbial Pangenome. BMC Bioinform. 2019, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Adu-Bobie, J.; Capecchi, B.; Serruto, D.; Rappuoli, R.; Pizza, M. Two Years into Reverse Vaccinology. Vaccine 2003, 21, 605–610. [Google Scholar] [CrossRef]
- Ehsan, N.; Ahmad, S.; Azam, S.S.; Rungrotmongkol, T.; Uddin, R. Proteome-Wide Identification of Epitope-Based Vaccine Candidates against Multi-Drug Resistant Proteus Mirabilis. Biologicals 2018, 55, 27–37. [Google Scholar] [CrossRef]
- Gul, S.; Ahmad, S.; Ullah, A.; Ismail, S.; Khurram, M.; Tahir ul Qamar, M.; Hakami, A.R.; Alkhathami, A.G.; Alrumaihi, F.; Allemailem, K.S. Designing a Recombinant Vaccine against Providencia Rettgeri Using Immunoinformatics Approach. Vaccines 2022, 10, 189. [Google Scholar] [CrossRef]
- Asad, Y.; Ahmad, S.; Rungrotmongkol, T.; Ranaghan, K.E.; Azam, S.S. Immuno-Informatics Driven Proteome-Wide Investigation Revealed Novel Peptide-Based Vaccine Targets against Emerging Multiple Drug Resistant Providencia Stuartii. J. Mol. Graph. Model. 2018, 80, 238–250. [Google Scholar] [CrossRef]
- Barh, D.; Barve, N.; Gupta, K.; Chandra, S.; Jain, N.; Tiwari, S.; Leon-Sicairos, N.; Canizalez-Roman, A.; dos Santos, A.R.; Hassan, S.S.; et al. Exoproteome and Secretome Derived Broad Spectrum Novel Drug and Vaccine Candidates in Vibrio Cholerae Targeted by Piper Betel Derived Compounds. PLoS ONE 2013, 8, e52773. [Google Scholar] [CrossRef]
- Yu, C.-S.; Cheng, C.-W.; Su, W.-C.; Chang, K.-C.; Huang, S.-W.; Hwang, J.-K.; Lu, C.-H. CELLO2GO: A Web Server for Protein SubCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Saadi, M.; Karkhah, A.; Nouri, H.R. Development of a Multi-Epitope Peptide Vaccine Inducing Robust T Cell Responses against Brucellosis Using Immunoinformatics Based Approaches. Infect. Genet. Evol. 2017, 51, 227–234. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A Server for in Silico Prediction of Allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S.; Consortium, O.S.D.D. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Li, W.; Joshi, M.; Singhania, S.; Ramsey, K.; Murthy, A. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef]
- Baldauf, K.J.; Royal, J.M.; Hamorsky, K.T.; Matoba, N. Cholera Toxin B: One Subunit with Many Pharmaceutical Applications. Toxins 2015, 7, 974–996. [Google Scholar] [CrossRef]
- Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Combating Tigecycline Resistant Acinetobacter Baumannii: A Leap Forward towards Multi-Epitope Based Vaccine Discovery. Eur. J. Pharm. Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: A Protein Structure and Structural Feature Prediction Server. Nucleic Acids Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB Server for Protein Structure Prediction and Refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Craig, D.B.; Dombkowski, A.A. Disulfide by Design 2.0: A Web-Based Tool for Disulfide Engineering in Proteins. BMC Bioinform. 2013, 14, 346. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A Web Server for Fast Interaction Refinement in Molecular Docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Karplus, M. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.; Brozell, S.R.; Cerutti, D.; Cheatham, T.; Cruzeiro, V.W.D.; Darden, T.; Duke, R.E.; Giambasu, G.; et al. Amber 2020; University of California: Los Angeles, CA, USA, 2020. [Google Scholar]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Antechamber: An Accessory Software Package for Molecular Mechanical Calculations. J. Am. Chem. Soc 2001, 222, U403. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Ahmad, S.; Raza, S.; Abro, A.; Liedl, K.R.; Azam, S.S. Toward Novel Inhibitors against KdsB: A Highly Specific and Selective Broad-Spectrum Bacterial Enzyme. J. Biomol. Struct. Dyn. 2019, 37, 1326–1345. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Significance of Root-Mean-Square Deviation in Comparing Three-Dimensional Structures of Globular Proteins. J. Mol. Biol. 1994, 235, 625–634. [Google Scholar] [CrossRef]
- Ahmad, S.; Raza, S.; Uddin, R.; Azam, S.S. Binding Mode Analysis, Dynamic Simulation and Binding Free Energy Calculations of the MurF Ligase from Acinetobacter Baumannii. J. Mol. Graph. Model. 2017, 77, 72–85. [Google Scholar] [CrossRef]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen Bonds in Proteins: Role and Strength. eLS 2001. [Google Scholar] [CrossRef]
- Turner, P.J. XMGRACE, Version 5.1.19 ed; Center for Coastal and Land-Margin Research Oregon Graduate Institute of Science and Technology Beaverton: Beaverton, OR, USA, 2005. [Google Scholar]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Modeling 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Brandies, P.; Peel, E.; Hogg, C.J.; Belov, K. The Value of Reference Genomes in the Conservation of Threatened Species. Genes 2019, 10, 846. [Google Scholar] [CrossRef]
- Sanober, G.; Ahmad, S.; Azam, S.S. Identification of Plausible Drug Targets by Investigating the Druggable Genome of MDR Staphylococcus Epidermidis. Gene Rep. 2017, 7, 147–153. [Google Scholar] [CrossRef]
- Allemailem, K.S. A Comprehensive Computer Aided Vaccine Design Approach to Propose a Multi-Epitopes Subunit Vaccine against Genus Klebsiella Using Pan-Genomics, Reverse Vaccinology, and Biophysical Techniques. Vaccines 2021, 9, 1087. [Google Scholar] [CrossRef]
- Naz, A.; Awan, F.M.; Obaid, A.; Muhammad, S.A.; Paracha, R.Z.; Ahmad, J.; Ali, A. Identification of Putative Vaccine Candidates against Helicobacter Pylori Exploiting Exoproteome and Secretome: A Reverse Vaccinology Based Approach. Infect. Genet. Evol. 2015, 32, 280–291. [Google Scholar] [CrossRef]
- Dorosti, H.; Eslami, M.; Negahdaripour, M.; Ghoshoon, M.B.; Gholami, A.; Heidari, R.; Dehshahri, A.; Erfani, N.; Nezafat, N.; Ghasemi, Y. Vaccinomics Approach for Developing Multi-Epitope Peptide Pneumococcal Vaccine. J. Biomol. Struct. Dyn. 2019, 37, 3524–3535. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, Y.-K.; Goo, Y.-K. Humoral and Cellular Immune Response to Plasmodium Vivax VIR Recombinant and Synthetic Antigens in Individuals Naturally Exposed to P. Vivax in the Republic of Korea. Malar. J. 2021, 20, 288. [Google Scholar] [CrossRef] [PubMed]
- Krummel, M.F.; Bartumeus, F.; Gérard, A. T Cell Migration, Search Strategies and Mechanisms. Nat. Rev. Immunol. 2016, 16, 193–201. [Google Scholar] [CrossRef]
- Dar, H.A.; Ismail, S.; Waheed, Y.; Ahmad, S.; Jamil, Z.; Aziz, H.; Hetta, H.F.; Muhammad, K. Designing a Multi-Epitope Vaccine against Mycobacteroides Abscessus by Pangenome-Reverse Vaccinology. Sci. Rep. 2021, 11, 11197. [Google Scholar] [CrossRef]
- Dombkowski, A.A.; Sultana, K.Z.; Craig, D.B. Protein Disulfide Engineering. FEBS Lett. 2014, 588, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Garg, N.; Shukla, G.; Capalash, N.; Sharma, P. Immunoprotective Efficacy of Acinetobacter Baumannii Outer Membrane Protein, FilF, Predicted in Silico as a Potential Vaccine Candidate. Front. Microbiol. 2016, 7, 158. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.; Seixas, A.M.M.; Mandal, M.; Rodríguez-Ortega, M.J.; Leitão, J.H. Characterization of the Burkholderia Cenocepacia J2315 Surface-Exposed Immunoproteome. Vaccines 2020, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Chintoan-Uta, C.; Cassady-Cain, R.L.; Al-Haideri, H.; Watson, E.; Kelly, D.J.; Smith, D.G.E.; Sparks, N.H.C.; Kaiser, P.; Stevens, M.P. Superoxide Dismutase SodB Is a Protective Antigen against Campylobacter Jejuni Colonisation in Chickens. Vaccine 2015, 33, 6206–6211. [Google Scholar] [CrossRef][Green Version]
- Solanki, V.; Tiwari, M.; Tiwari, V. Prioritization of Potential Vaccine Targets Using Comparative Proteomics and Designing of the Chimeric Multi-Epitope Vaccine against Pseudomonas Aeruginosa. Sci. Rep. 2019, 9, 5240. [Google Scholar] [CrossRef]
- Seib, K.L.; Zhao, X.; Rappuoli, R. Developing Vaccines in the Era of Genomics: A Decade of Reverse Vaccinology. Clin. Microbiol. Infect. 2012, 18, 109–116. [Google Scholar] [CrossRef] [PubMed]


| Proteins | B-Cell Epitopes |
|---|---|
| WP_012806254.1 (trypsin-like peptidase domain-containing protein) | NFMKKGNKKFALF |
| KNDTKSNSTENMANVEQTKSISQEELQKYTKNAVQTQDA | |
| KTVTVNTYNPLEEMLFGRSGGQEKRESGS | |
| RSSLGIEQI | |
| TPNALQQQQIIQQRQQQQQQE | |
| WP_015769552.1 (sel1 repeat family protein) | DVKSKATIEQKENNRIK |
| GIDTKIDYKKAMEW | |
| GFGVKKDYKQ | |
| EKGLGVEKSFDS | |
| EMAGDYAKA | |
| GKGVKKNLKEASE | |
| WP_015769221.1 (TrbI/VirB10 family protein) | NDYFETSEDDFTEQKEEEISLEDEGNGTIKNSK |
| KKNMKSNEQEKVDSISTGTELDINDAVNTQANKNPQVSETIAQSGTENINTASDKTGTPNLSQYDSQLGSDYNNDNFDSSYGASTSPPSFSNVSENENTNTSATSVAPSEKYKEWRKSSIGFDKGVSTQTPQVPEQYQEQQPASQNTQQQNENDTDQNKQKSKSLFLKQKQDSFYSTNLKNPAIGKYELK | |
| QGVDLLGNAGLKGKTNNH | |
| EGLNVNIETGSRSRVNIGTG |
| MHC-I | Percentile SCORE | MHC-II | Percentile Score |
|---|---|---|---|
| FMKKGNKKF | 0.06 | FMKKGNKKFAL | 2.2 |
| MKKGNKKFAL | 0.73 | VEQTKSISQEE | 2 |
| QTKSISQEE | 3.3 | KNDTKSNSTENMANV | 26 |
| VEQTKSISQ | 2.5 | ELQKYTKNAVQTQDA | 6.1 |
| NSTENMANV | 0.13 | ||
| KNDTKSNST | 18 | VEQTKSISQEEL | 2.5 |
| LQKYTKNAV | 1.1 | KNDTKSNSTENMANV | 26 |
| KNAVQTQDA | 16 | ELQKYTKNAVQTQDA | 6.1 |
| QTKSISQEEL | 0.91 | ||
| VEQTKSISQ | 2.5 | LQQQQIIQQRQQQQ | 0.13 |
| NSTENMANV | 0.13 | TPNALQQQQIIQQRQ | 0.38 |
| KNDTKSNST | 18 | ||
| LQKYTKNAV | 1.1 | TIEQKENNRIK | 3.6 |
| KNAVQTQDA | 16 | DVKSKATIEQK | 4 |
| QQQQIIQQR | 0.11 | ||
| IIQQRQQQQ | 9.1 | IDTKIDYKKAM | 5.4 |
| NALQQQQII | 0.08 | IDTKIDYKKAMEW | 11 |
| TPNALQQQQI | 0.94 | ||
| QQQQIIQQRQ | 5 | KGLGVEKSFDS | 13 |
| IEQKENNRI | 0.39 | ||
| KSKATIEQK | 0.01 | KGVKKNLKEAS | 6.4 |
| DVKSKATIE | 4.3 | ||
| DTKIDYKKAM | 0.52 | DYFETSEDDFT | 0.62 |
| KIDYKKAMEW | 0.07 | EQKEEEISLEDEG | 0.89 |
| IDTKIDYKK | 6 | EDEGNGTIKNSK | 18 |
| KGLGVEKSF | 0.48 | ||
| LGVEKSFDS | 45 | ASQNTQQQNENDTDQ | 100 |
| GVKKNLKEA | 0.78 | ASTSPPSFSNVSENE | 100 |
| KGVKKNLKEA | 1.4 | ENENTNTSATSVAPS | 97 |
| DYFETSEDDF | 0.61 | KKNMKSNEQEKVDSI | 93 |
| EQKEEEISL | 0.25 | STNLKNPAIGKYELK | 92 |
| EEISLEDEG | 4.8 | KSKSLFLKQKQDSFY | 95 |
| EGNGTIKNSK | 1.5 | NENDTDQNKQKSKSL | 95 |
| EDEGNGTIK | 11 | NENDTDQNKQKSKSL | 100 |
| ASQNTQQQN | 5.7 | ||
| QQNENDTDQ | 15 | GVDLLGNAGLKGKT | 5 |
| STSPPSFSNV | 0.04 | DLLGNAGLKGKTNNH | 14 |
| SFSNVSENE | 6.5 | ||
| NTNTSATSV | 0.1 | GLNVNIETGSRSRV | 22 |
| ENENTNTSAT | 7.1 | NIETGSRSRVNIGTG | 66 |
| TSATSVAPS | 5.4 | ||
| NEQEKVDSI | 0.29 | ||
| KKNMKSNEQ | 13 | ||
| NPAIGKYEL | 0.08 | ||
| STNLKNPAI | 0.73 | ||
| FLKQKQDSF | 0.01 | ||
| KSKSLFLKQK | 0.04 | ||
| QNKQKSKSL | 0.02 | ||
| NENDTDQNK | 1.3 | ||
| LLGNAGLKGK | 0.7 | ||
| GVDLLGNAGL | 4.6 | ||
| LLGNAGLKGK | 0.7 | ||
| GLKGKTNNH | 1.7 | ||
| NVNIETGSR | 0.34 | ||
| GLNVNIETG | 12 | ||
| NIETGSRSR | 0.88 | ||
| RSRVNIGTG | 2 |
| Pairs of Amino Acid Residues | Chi3 Value | Energy in kcal/mol |
|---|---|---|
| Ile2-Lys5 | −91.05 | 2.75 |
| Phe9-Thr22 | 102.16 | 0.56 |
| Val22-Gly21 | −64.27 | 3.7 |
| Ser16-Ala19 | −64.37 | 6.37 |
| His20-Pro23 | −111.46 | 5.49 |
| Thr36-Lue41 | −109.49 | 7.37 |
| Gln37-Val108 | −86.81 | 2.82 |
| Phe46-Arg56 | −112.11 | 5.11 |
| Tyr97-Ala101 | −86.93 | 4.38 |
| Asn111-Pro114 | 83.18 | 1.33 |
| Gln136-Gly141 | 95.28 | 4.64 |
| Gln149-Leu152 | −111.28 | 3.38 |
| Gly157-Thr163 | −61.86 | 5.43 |
| Gln162-Asp165 | −101.08 | 4.65 |
| Asn178-Pro184 | −67.1 | 5.64 |
| Ser203-Glu208 | 81.74 | 0.99 |
| Lys204-Ile207 | 78.02 | 5.86 |
| Thr218-Gly225 | 79.29 | 4.37 |
| Pro224-Phe229 | 126.17 | 2.01 |
| Gly225-Tyr228 | 118.82 | 3.29 |
| Ser232-Gly239 | 125.75 | 3.07 |
| Gly267-Asn278 | 121.18 | 5.67 |
| Asp289-Pro294 | 101.78 | 2.12 |
| Asp331-Gly339 | −81.35 | 2.33 |
| Gly358-Pro364 | 99.08 | 1.47 |
| His376-Gly386 | −82.67 | 5.08 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 6 | −12.17 | −8.14 | 1.36 | −0.66 | −1.44 |
| 2 | 7 | 3.70 | −31.78 | 18.19 | 10.92 | −5.88 |
| 3 | 4 | 15.71 | −9.34 | 2.03 | 2.73 | −1.13 |
| 4 | 1 | 20.24 | −17.21 | 3.42 | 9.96 | −0.90 |
| 5 | 3 | 21.66 | −27.06 | 33.12 | 12.45 | −2.59 |
| 6 | 9 | 77.86 | −26.71 | 81.40 | 13.56 | −5.32 |
| 7 | 2 | 164.85 | −58.38 | 246.65 | 24.17 | −8.25 |
| 8 | 8 | 257.49 | −18.25 | 306.00 | 6.56 | −4.93 |
| 9 | 5 | 1003.52 | −46.18 | 1289.89 | 17.46 | −6.73 |
| 10 | 10 | 2759.81 | −34.85 | 3473.08 | 3.77 | −5.51 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 9 | −1.17 | −7.24 | 1.98 | 1.78 | −0.71 |
| 2 | 5 | 23.89 | −3.24 | 0.00 | 0.80 | −0.77 |
| 3 | 7 | 31.61 | −0.77 | 0.00 | 1.39 | 0.00 |
| 4 | 3 | 241.30 | −42.08 | 354.56 | 8.23 | −4.07 |
| 5 | 2 | 460.31 | −32.37 | 594.32 | 8.59 | −4.83 |
| 6 | 6 | 473.83 | −13.18 | 598.99 | 4.82 | 0.00 |
| 7 | 8 | 1395.95 | −38.28 | 1774.07 | 16.30 | −3.72 |
| 8 | 10 | 2536.69 | −60.68 | 3213.80 | 16.12 | −12.38 |
| 9 | 1 | 3964.79 | −27.95 | 5027.40 | −4.47 | −5.94 |
| 10 | 4 | 3973.45 | −52.13 | 5057.12 | 6.76 | −7.93 |
| Rank | Solution Number | Global Energy | Attractive VdW | Repulsive VdW | ACE | HB |
|---|---|---|---|---|---|---|
| 1 | 8 | −4.52 | −2.14 | 1.11 | 1.84 | −1.11 |
| 2 | 5 | 3.89 | −2.10 | 1.00 | 1.90 | −0.77 |
| 3 | 7 | 1.61 | −2.87 | 2.00 | 2.45 | 0.00 |
| 4 | 3 | 214.00 | −41.00 | 381.00 | 9.16 | −4.07 |
| 5 | 2 | 160.00 | −35.08 | 594.32 | 8.59 | −4.83 |
| 6 | 6 | 273.00 | −17.10 | 598.99 | 4.82 | 0.00 |
| 7 | 9 | 335.00 | −39.22 | 1774.07 | 16.30 | −3.72 |
| 8 | 10 | 236.78 | −61.69 | 3213.80 | 16.12 | −12.38 |
| 9 | 1 | 961.10 | -28.10 | 5027.40 | −4.47 | −5.94 |
| 10 | 4 | 978.36 | -50.00 | 6059.10 | 8.18 | −6.10 |
| MHC-I-Vaccine Complex | MHC-II-Vaccine Complex | TLR-4-Vaccine Complex |
|---|---|---|
| ARG17 | ARG4 | PRO23 |
| GLY16 | THR3 | GLU24 |
| GLU19 | PRO5 | SER25 |
| PRO15 | ARG100 | TYR26 |
| ARG14 | LYS126 | ASP50 |
| GLU89 | GLU158 | PRO49 |
| ARG75 | GLY125 | SER76 |
| VAL76 | ARG189 | ILE48 |
| SER88 | THR120 | PHE74 |
| THR86 | PRO127 | SER73 |
| ALA139 | ARG125 | TYR72 |
| THR142 | ASP159 | LEU69 |
| MET138 | ASP78 | ALA97 |
| THR73 | TYR79 | GLY96 |
| GLU47 | PRO81 | ASP95 |
| ARG45 | LYS120 | ILE93 |
| LYS48 | LYS121 | GLY120 |
| ASP38 | GLY28 | LEU117 |
| ASP29 | GLN115 | |
| GLU30 | ILE114 | |
| ARG93 | PRO113 | |
| ARG94 | HIS148 | |
| VAL160 | ILE149 | |
| ASN124 | LYS150 | |
| PHE7 | ALA133 | |
| THR87 | LYS130 | |
| HIS86 | TYR65 | |
| PHE87 | PHE64 | |
| ALA97 | ALA107 | |
| ASN98 | ARG106 |
| Energy Parameter | TLR-4-Vaccine Complex | MHC-I-Vaccine Complex | MHC-II-Vaccine Complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −351.10 | −310.51 | −284.36 |
| Electrostatic | −190.99 | −139.02 | −111.01 |
| Delta G solv | 25.30 | 49.74 | 36.00 |
| Delta Total | −516.79 | −399.79 | −359.37 |
| MM-PBSA | |||
| VDWAALS | −351.10 | −310.51 | −284.36 |
| EEL | −190.99 | −139.02 | −111.01 |
| Delta G solv | 26.32 | 47.10 | 39.17 |
| Delta Total | −515.77 | −402.43 | −356.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammari, A.; Alasmari, A.F.; Alharbi, M.; Ali, N.; Muhseen, Z.T.; Ashfaq, U.A.; Ud-din, M.; Ullah, A.; Arshad, M.; Ahmad, S. Novel Chimeric Vaccine Candidate Development against Leptotrichia buccalis. Int. J. Environ. Res. Public Health 2022, 19, 10742. https://doi.org/10.3390/ijerph191710742
Alshammari A, Alasmari AF, Alharbi M, Ali N, Muhseen ZT, Ashfaq UA, Ud-din M, Ullah A, Arshad M, Ahmad S. Novel Chimeric Vaccine Candidate Development against Leptotrichia buccalis. International Journal of Environmental Research and Public Health. 2022; 19(17):10742. https://doi.org/10.3390/ijerph191710742
Chicago/Turabian StyleAlshammari, Abdulrahman, Abdullah F. Alasmari, Metab Alharbi, Nemat Ali, Ziyad Tariq Muhseen, Usman Ali Ashfaq, Miraj Ud-din, Asad Ullah, Muhammad Arshad, and Sajjad Ahmad. 2022. "Novel Chimeric Vaccine Candidate Development against Leptotrichia buccalis" International Journal of Environmental Research and Public Health 19, no. 17: 10742. https://doi.org/10.3390/ijerph191710742
APA StyleAlshammari, A., Alasmari, A. F., Alharbi, M., Ali, N., Muhseen, Z. T., Ashfaq, U. A., Ud-din, M., Ullah, A., Arshad, M., & Ahmad, S. (2022). Novel Chimeric Vaccine Candidate Development against Leptotrichia buccalis. International Journal of Environmental Research and Public Health, 19(17), 10742. https://doi.org/10.3390/ijerph191710742









