Pan-Genome Analysis of Oral Bacterial Pathogens to Predict a Potential Novel Multi-Epitopes Vaccine Candidate
Abstract
1. Introduction
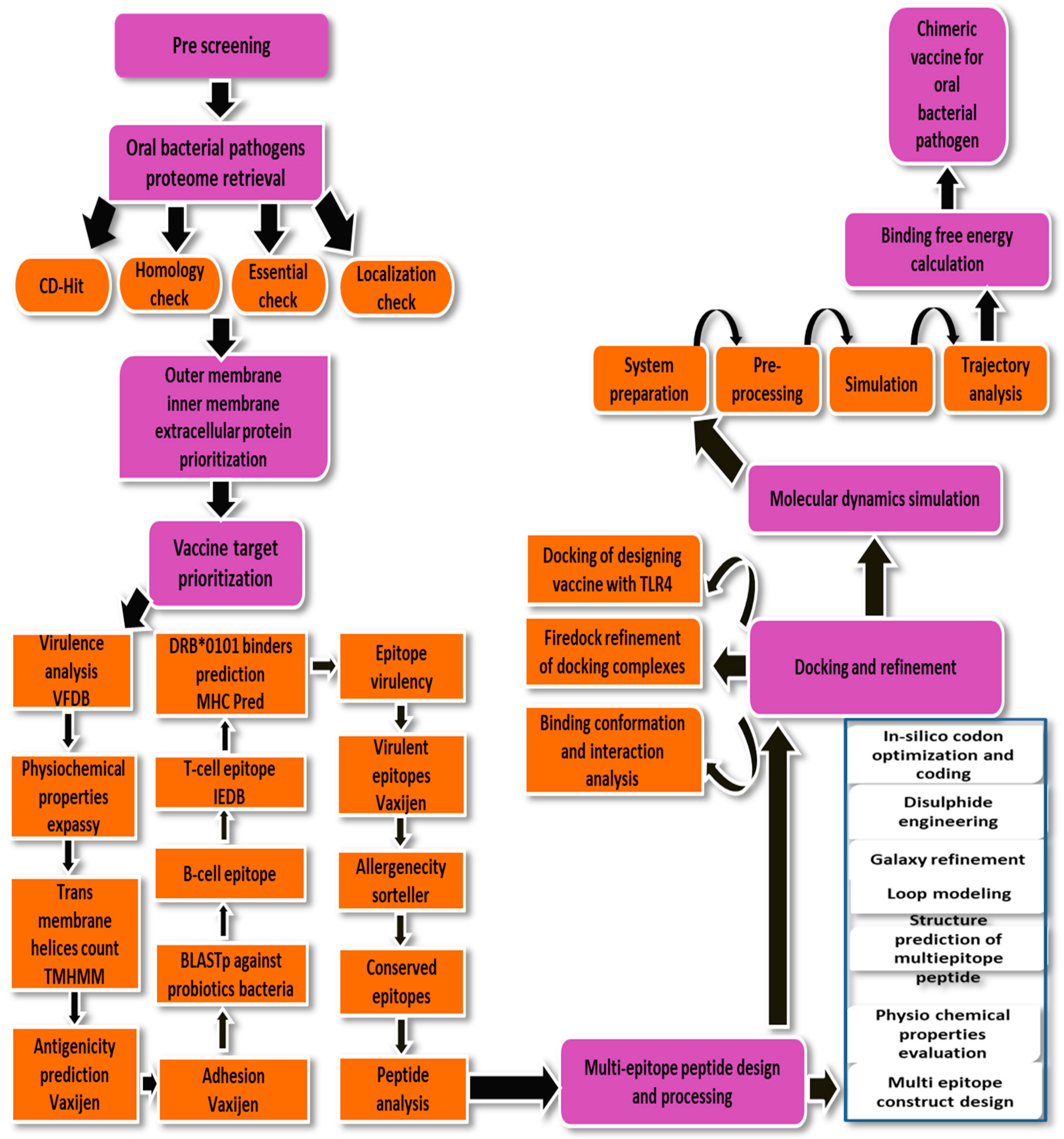
2. Research Methodology
2.1. Subtractive Proteome and Reverse Vaccinology Phase
2.2. Pre-Screening Phase
2.3. Cluster Database at High Identity with Tolerance (CD-HIT) Analysis
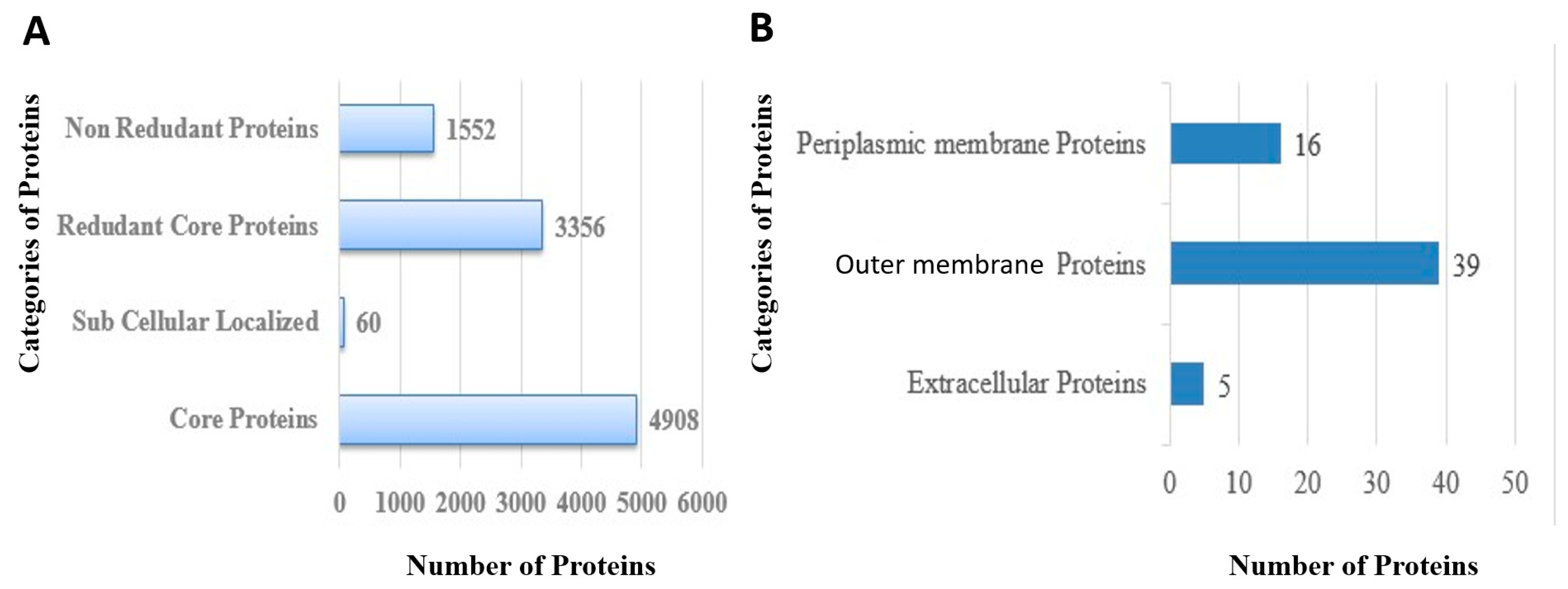
2.4. Sub-Cellular Localization Phase
2.5. Vaccine Candidate’s Prioritization Phase
2.6. Physiochemical Properties Analysis
2.7. Analysis of Transmembrane Helices
2.8. Antigenicity, Allergenicity, and Adhesion Probability Prediction
2.9. Prediction of Immune Cell Epitopes
2.10. MHCPred Analysis
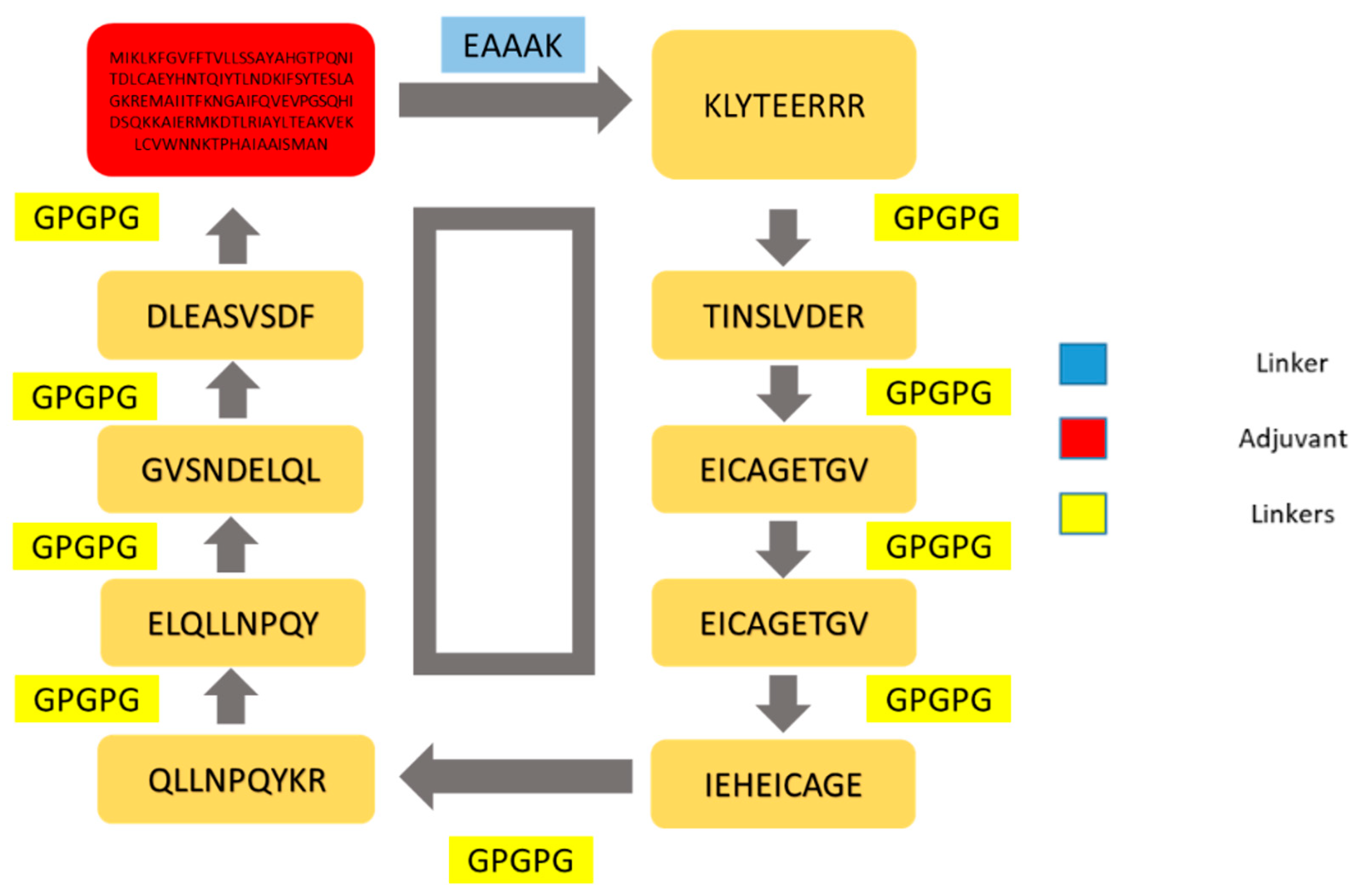
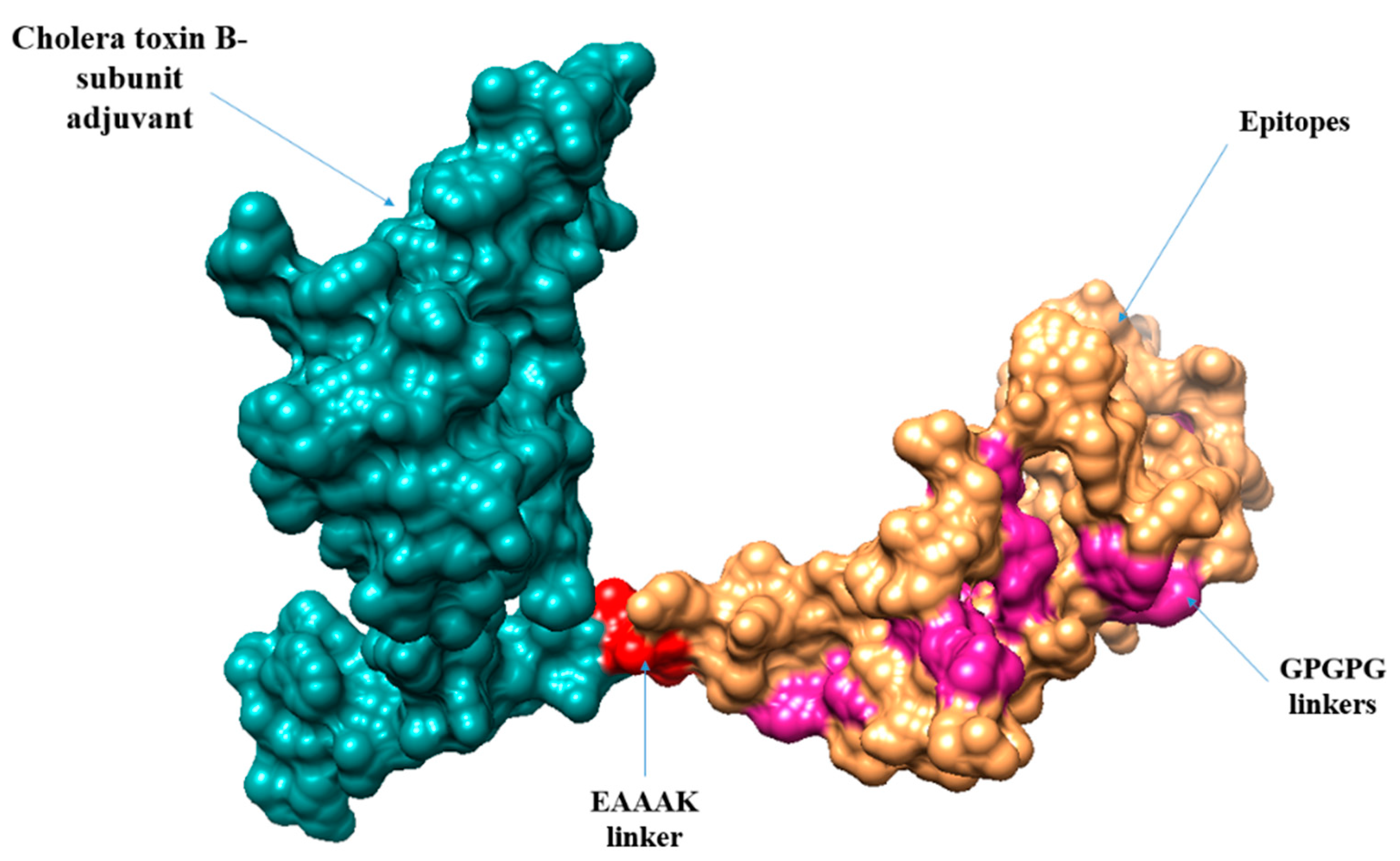
2.11. Multi-Epitopes Vaccine Design
2.12. Loop Modeling and Vaccine Refinement
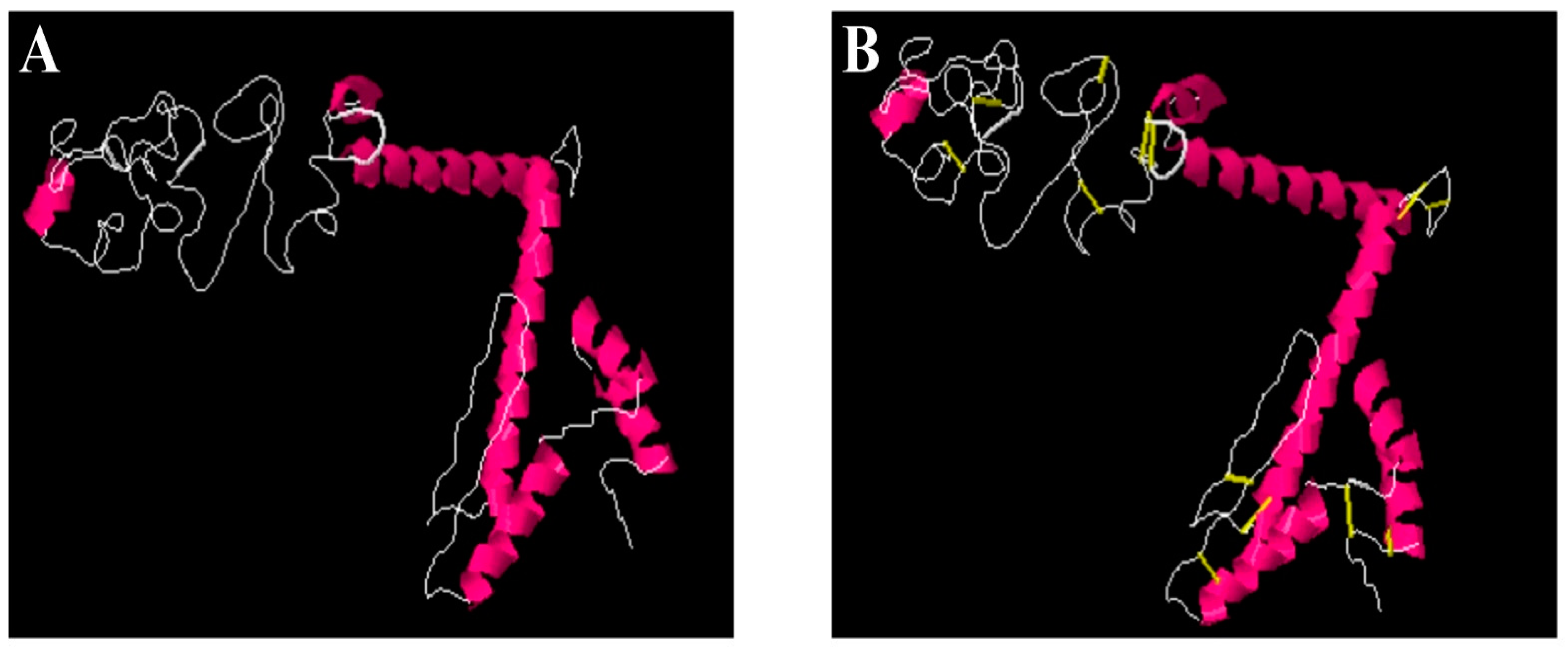
2.13. Disulfide Engineering and Codon Optimization
2.14. Docking and Refinement
2.15. Molecular Dynamics Simulation
2.16. MM-GBSA Binding Free Energies
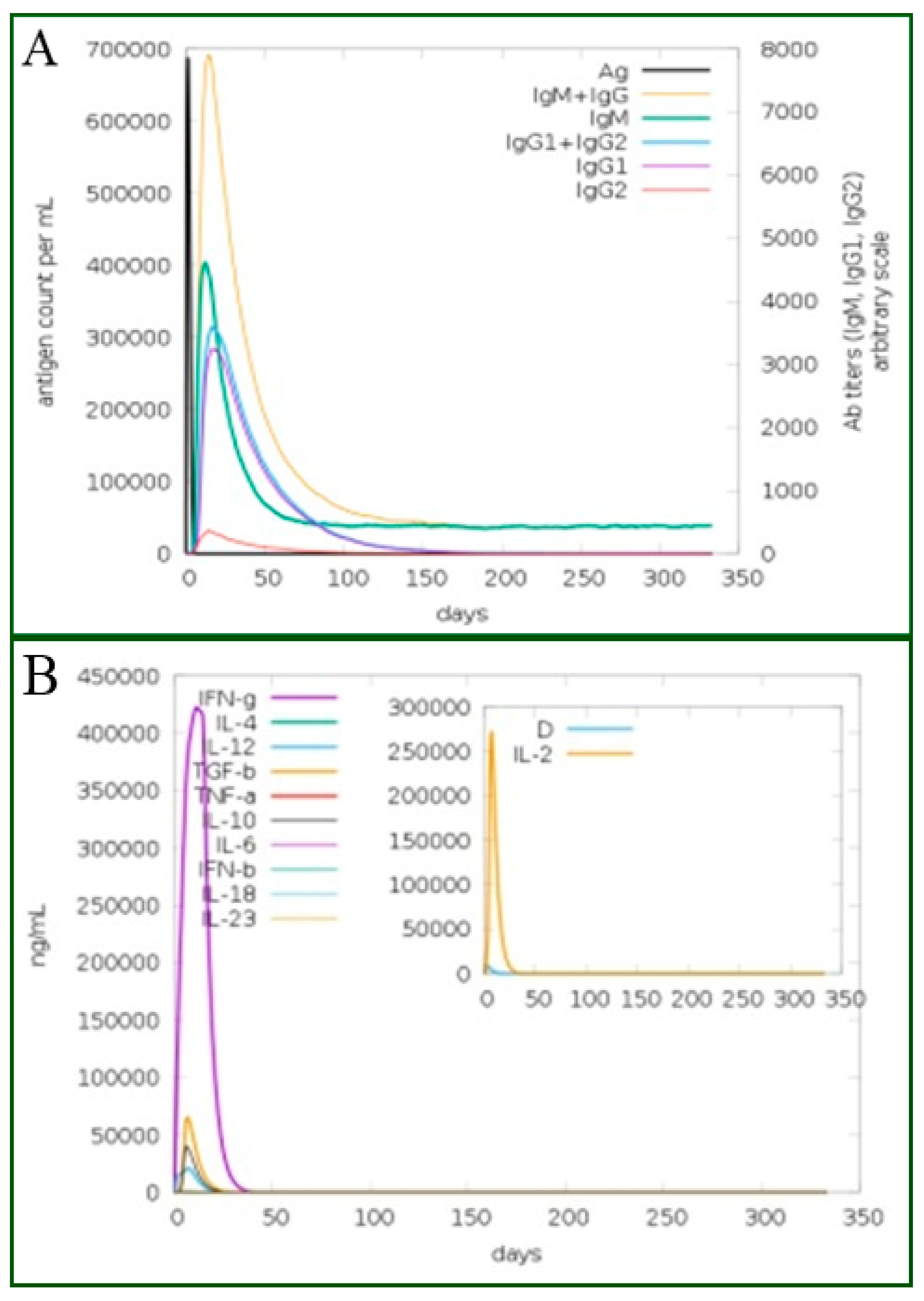
2.17. Immune Simulation
3. Results
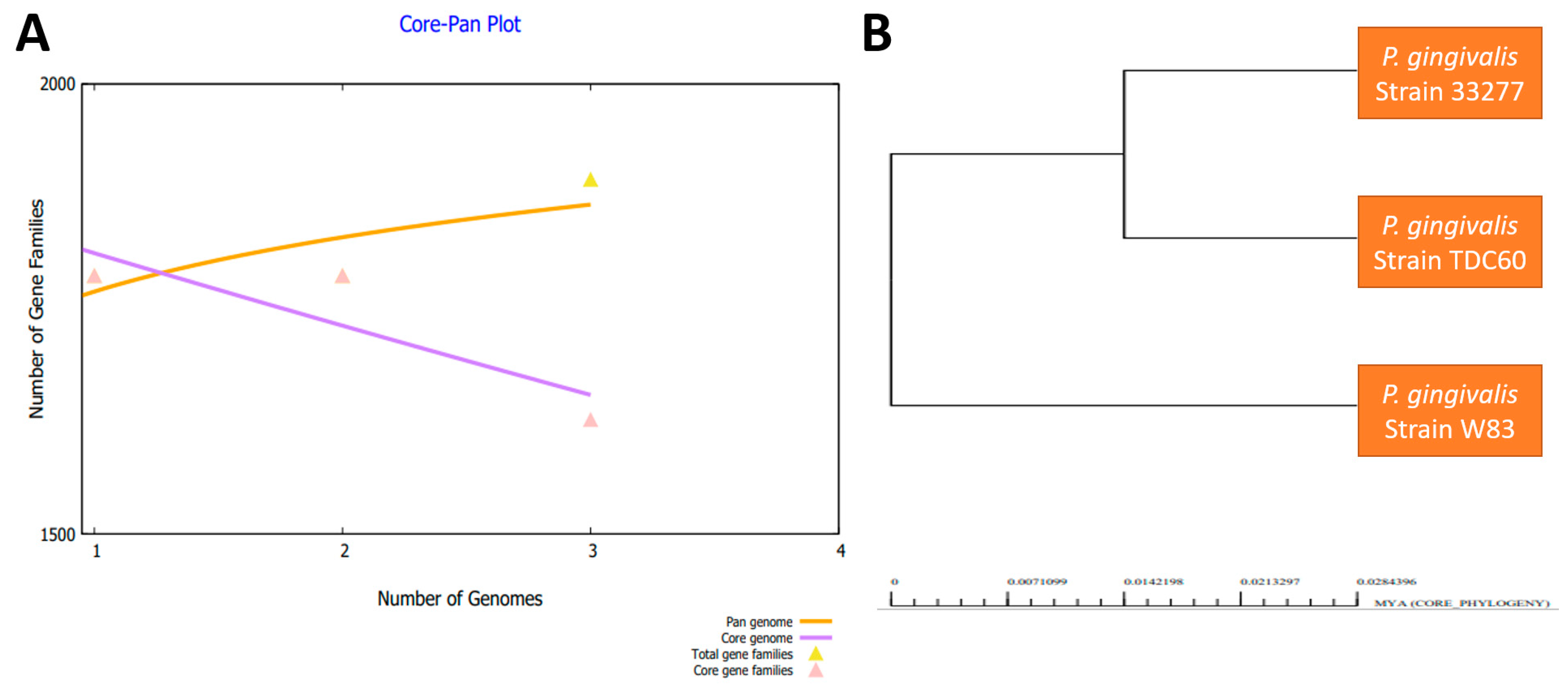
3.1. Genomes Retrieval of P. gingivalis
3.2. Bacterial Pan-Genome Analysis
3.3. CD-HIT Analysis
3.4. Proteins Subcellular Localization
3.5. VFDB Analysis
3.6. Transmembrane Helices and Physiochemical Analysis
3.7. Similarity with Human Genome and Prediction of Antigenicity and Allergenicity
3.8. Homology Check of Normal Flora
3.9. B-Cell Epitopes Prediction
3.10. MHC-I and MHC-II Epitopes Prediction
3.11. Epitope Prioritization Phase
3.12. MHCPred, Allergenicity, Antigenicity, Solubility and Toxicity Analysis
3.13. Multi-Epitopes Vaccine Designing
3.14. Vaccine Structure Prediction, Loops Modeling and Refinement
3.15. Disulfide Engineering and Codon Optimization
3.16. Analysis of Molecular Docking
3.17. Docked Complexes Refinement
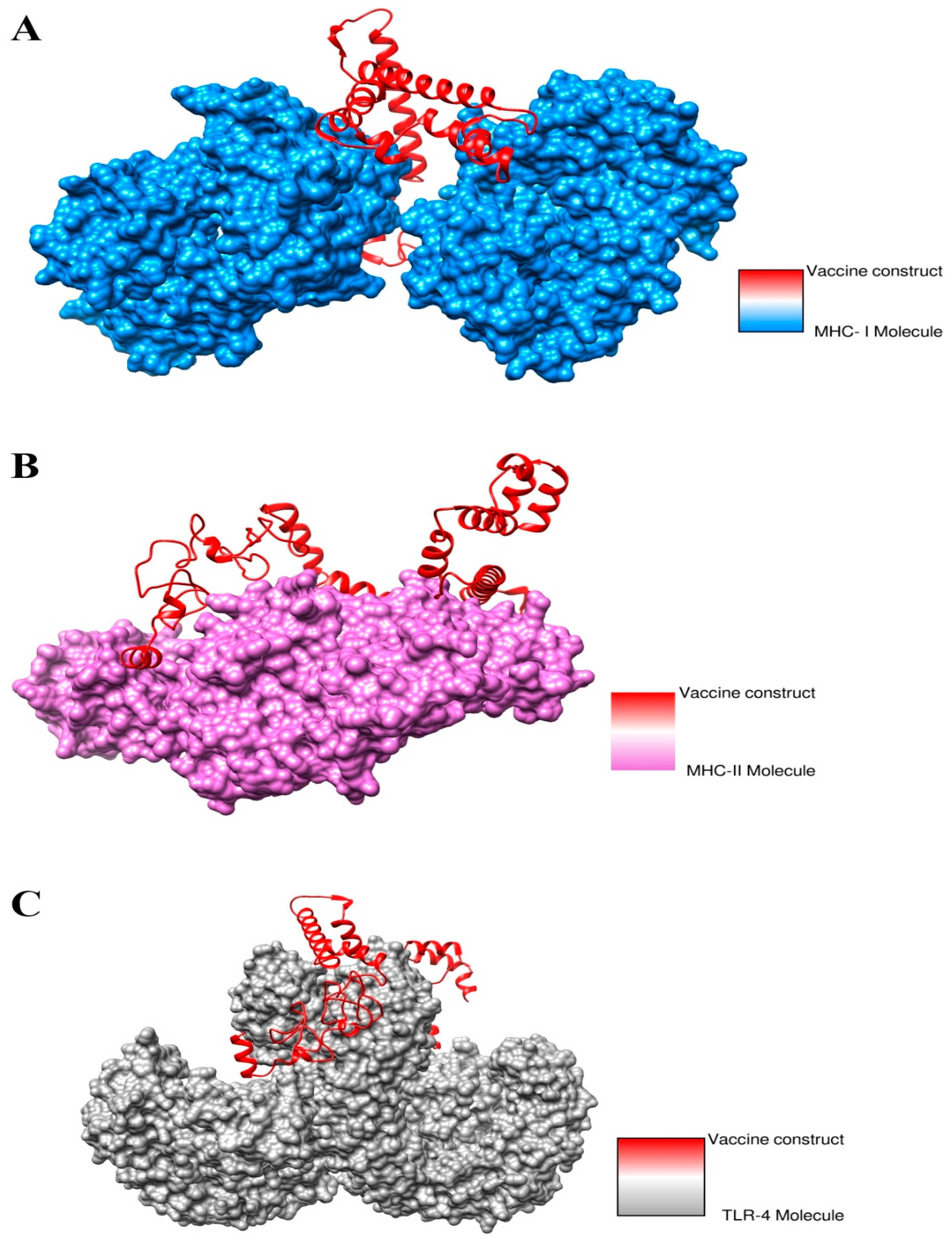
3.18. Docked Conformation of Vaccine with Immune Receptors
3.19. Interactions of Vaccine to Immune Receptors
3.20. Molecular Dynamics Simulation
3.21. Calculation of Binding Free Energies
3.22. Immune Stimulations
4. Discussion
5. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2018, 84, 41–44. [Google Scholar] [CrossRef]
- MacLean, R.C.; San Millan, A. The evolution of antibiotic resistance. Science 2019, 365, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015, a population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Bank, W. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank: Washington, DC, USA, 2017. [Google Scholar]
- O’Neill, J. Review on antimicrobial resistance: Tackling drug-resistant infections globally: Final report and recommendations. In Review on Antimicrobial Resistance: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; CABI: Wallingford, UK, 2016. [Google Scholar]
- Klemm, E.J.; Wong, V.K.; Dougan, G. Emergence of dominant multidrug-resistant bacterial clades: Lessons from history and whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, 12872–12877. [Google Scholar] [CrossRef] [PubMed]
- The White House. National Action Plan for Combating Antibiotic-Resistant Bacteria; The White House: Washington, DC, USA, 2015; Volume 62.
- National Institutes of Health. NIAID’s Antibacterial Resistance Program: Current Status and Future Directions; National Institutes of Health: Washington, DC, USA, 2014.
- Gagneux-Brunon, A.; Lucht, F.; Launay, O.; Berthelot, P.; Botelho-Nevers, E. Vaccines for healthcare-associated infections: Present, future, and expectations. Expert Rev. Vaccines 2018, 17, 421–433. [Google Scholar] [CrossRef]
- Brooks, B.D.; Brooks, A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 2, management strategies and new agents. Pharm. Ther. 2015, 40, 344. [Google Scholar]
- The White House. National Strategy for Combating Antibiotic Resistant Bacteria; The White House: Washington, DC, USA, 2014.
- Reddick, L.E.; Alto, N.M. Bacteria Fighting Back: How Pathogens Target and Subvert the Host Innate Immune System. Mol. Cell 2014, 54, 321–328. [Google Scholar] [CrossRef]
- Qamar, M.T.U.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A.; et al. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.E.; Black, S.; Salisbury, D.; Rappuoli, R. Antimicrobial resistance and the role of vaccines. Proc. Natl. Acad. Sci. USA 2018, 115, 12868–12871. [Google Scholar] [CrossRef]
- Manonmanipavithra, R.; Ari, G.; Rajendran, S.; Mahendra, J. An Overview On Porphyromonas Gingivalis—An Important Periodontopathic Pathogen. Ann. Rom. Soc. Cell Biol. 2020, 24, 416–421. [Google Scholar]
- Gibson, F.C.; Genco, C.A. Prevention of Porphyromonas gingivalis-Induced Oral Bone Loss following Immunization with Gingipain R1. Infect. Immun. 2001, 69, 7959–7963. [Google Scholar] [CrossRef][Green Version]
- Khalaf, H.; Palm, E.; Bengtsson, T. Cellular Response Mechanisms in Porphyromonas gingivalis Infection; Books on Demand: Norderstedt, Germany, 2017; pp. 45–68. [Google Scholar] [CrossRef]
- Carrouel, F.; Viennot, S.; Santamaria, J.; Veber, P.; Bourgeois, D. Quantitative molecular detection of 19 major pathogens in the interdental biofilm of periodontally healthy young adults. Front. Microbiol. 2016, 7, 840. [Google Scholar] [CrossRef] [PubMed]
- Vaernewyck, V.; Arzi, B.; Sanders, N.N.; Cox, E.; Devriendt, B. Mucosal Vaccination Against Periodontal Disease: Current Status and Opportunities. Front. Immunol. 2021, 12, 768397. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, NCBI Resource. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017, 45, D12–D17. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.; Awan, F.M.; Obaid, A.; Muhammad, S.A.; Paracha, R.Z.; Ahmad, J.; Ali, A. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect. Genet. Evol. 2015, 32, 280–291. [Google Scholar] [CrossRef]
- Hassan, A.; Naz, A.; Obaid, A.; Paracha, R.Z.; Naz, K.; Awan, F.M.; Muhmmad, S.A.; Janjua, H.A.; Ahmad, J.; Ali, A. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genom. 2016, 17, 732. [Google Scholar] [CrossRef]
- Baseer, S.; Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Towards a peptide-based vaccine against Shigella sonnei: A subtractive reverse vaccinology based approach. Biologicals 2017, 50, 87–99. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Tiwari, S.; Jamal, S.B.; Barh, D.; Azevedo, V.; Soares, S.C. An In Silico Identification of Common Putative Vaccine Candidates against Treponema pallidum: A Reverse Vaccinology and Subtractive Genomics Based Approach. Int. J. Mol. Sci. 2017, 18, 402. [Google Scholar] [CrossRef]
- Naz, K.; Naz, A.; Ashraf, S.T.; Rizwan, M.; Ahmad, J.; Baumbach, J.; Ali, A. PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinform. 2019, 20, 123. [Google Scholar] [CrossRef]
- Blast, N. Basic Local Alignment Search Tool; National Library of Medicine, National Center for Biotechnology Information: Bethesda, ML, USA, 2015.
- Zhang, R.; Ou, H.; Zhang, C. DEG: A database of essential genes. Nucleic Acids Res. 2004, 32, D271–D272. [Google Scholar] [CrossRef] [PubMed]
- Sanober, G.; Ahmad, S.; Azam, S.S. Identification of plausible drug targets by investigating the druggable genome of MDR Staphylococcus epidermidis. Gene Rep. 2017, 7, 147–153. [Google Scholar] [CrossRef]
- Ali, A.; Naz, A.; Soares, S.C.; Bakhtiar, M.; Tiwari, S.; Hassan, S.S.; Hanan, F.; Ramos, R.; Pereira, U.; Barh, D.; et al. Pan-genome analysis of human gastric pathogen H. pylori: Comparative genomics and pathogenomics approaches to identify regions associated with pathogenicity and prediction of potential core therapeutic targets. BioMed Res. Int. 2015, 2015, 139580. [Google Scholar] [CrossRef]
- Johri, S.; Solanki, J.; Cantu, V.A.; Fellows, S.R.; Edwards, R.A.; Moreno, I.; Vyas, A.; Dinsdale, E.A. ‘Genome skimming’ with the MinION hand-held sequencer identifies CITES-listed shark species in India’s exports market. Sci. Rep. 2019, 9, 4476. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Naz, A.; Ahmad, J.; Naz, K.; Obaid, A.; Parveen, T.; Ahsan, M.; Ali, A. VacSol: A high throughput in silico pipeline to predict potential therapeutic targets in prokaryotic pathogens using subtractive reverse vaccinology. BMC Bioinform. 2017, 18, 106. [Google Scholar] [CrossRef]
- Azam, S.S.; Shamim, A. An insight into the exploration of druggable genome of Streptococcus gordonii for the identification of novel therapeutic candidates. Genomics 2014, 104, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Sikic, K.; Carugo, O. Protein sequence redundancy reduction: Comparison of various method. Bioinformation 2010, 5, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idrees, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2012, 12, 53–62. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Combating tigecycline resistant Acinetobacter baumannii: A leap forward towards multi-epitope based vaccine discovery. Eur. J. Pharm. Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0, improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Barve, N.; Gupta, K.; Chandra, S.; Jain, N.; Tiwari, S.; Leon-Sicairos, N.; Canizalez-Roman, A.; Santos, A.; Hassan, S.S.; et al. Exoproteome and Secretome Derived Broad Spectrum Novel Drug and Vaccine Candidates in Vibrio cholerae Targeted by Piper betel Derived Compounds. PLoS ONE 2013, 8, e52773. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.I.; Naz, A.; Ali, A.; Andleeb, S. Prediction of vaccine candidates against Pseudomonas aeruginosa: An integrated genomics and proteomics approach. Genomics 2017, 109, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Nain, Z.; Abdullah, F.; Rahman, M.M.; Karim, M.M.; Khan, S.A.; Bin Sayed, S.; Mahmud, S.; Rahman, S.M.R.; Sheam, M.; Haque, Z.; et al. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J. Biomol. Struct. Dyn. 2020, 38, 4850–4867. [Google Scholar] [CrossRef]
- ProtParam E. ExPASy-ProtParam Tool. 2017. Available online: https://web.expasy.org/protparam/ (accessed on 3 March 2022).
- Sajjad, R.; Ahmad, S.; Azam, S.S. In silico screening of antigenic B-cell derived T-cell epitopes and designing of a multi-epitope peptide vaccine for Acinetobacter nosocomialis. J. Mol. Graph. Model. 2019, 94, 107477. [Google Scholar] [CrossRef]
- Saidijam, M.; Azizpour, S.; Patching, S.G. Comprehensive analysis of the numbers, lengths and amino acid compositions of transmembrane helices in prokaryotic, eukaryotic and viral integral membrane proteins of high-resolution structure. J. Biomol. Struct. Dyn. 2017, 36, 443–464. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, P.; Luo, J.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Wadood, A.; Jamal, A.; Riaz, M.; Khan, A.; Uddin, R.; Jelani, M.; Azam, S.S. Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb. Pathog. 2018, 115, 194–198. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiang, Z.; Mobley, H.L.T. Vaxign: The First Web-Based Vaccine Design Program for Reverse Vaccinology and Applications for Vaccine Development. J. Biomed. Biotechnol. 2010, 2010, 297505. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2018, 47, D339–D343. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0, improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Vashi, Y.; Jagrit, V.; Kumar, S. Understanding the B and T cell epitopes of spike protein of severe acute respiratory syndrome coronavirus-2, A computational way to predict the immunogens. Infect. Genet. Evol. 2020, 84, 104382. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.; Ahmad, S.; Azam, S.S. Vaccinomics to design a novel single chimeric subunit vaccine for broad-spectrum immunological applications targeting nosocomial Enterobacteriaceae pathogens. Eur. J. Pharm. Sci. 2020, 146, 105258. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef]
- Nezafat, N.; Karimi, Z.; Eslami, M.; Mohkam, M.; Zandian, S.; Ghasemi, Y. Designing an efficient multi-epitope peptide vaccine against Vibrio cholerae via combined immunoinformatics and protein interaction based approaches. Comput. Biol. Chem. 2016, 62, 82–95. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.Z.; Sweredoski, M.J.; Baldi, P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005, 33, W72–W76. [Google Scholar] [CrossRef]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [PubMed]
- Dombkowski, A.A.; Sultana, K.Z.; Craig, D.B. Protein disulfide engineering. FEBS Lett. 2014, 588, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. In Molecular Modeling of Proteins; Springer: Berlin/Heidelberg, Germany, 2008; pp. 365–382. [Google Scholar]
- Solanki, V.; Tiwari, M.; Tiwari, V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 5240. [Google Scholar] [CrossRef]
- Ohto, U.; Yamakawa, N.; Akashi-Takamura, S.; Miyake, K.; Shimizu, T. Structural Analyses of Human Toll-like Receptor 4 Polymorphisms D299G and T399I. J. Biol. Chem. 2012, 287, 40611–40617. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Takehara, M.; Kobayashi, K.; Nagahama, M. Toll-Like Receptor 4 Protects Against Clostridium perfringens Infection in Mice. Front. Cell. Infect. Microbiol. 2021, 11, 633440. [Google Scholar] [CrossRef]
- Mukherjee, S.; Karmakar, S.; Babu, S.P.S. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins Struct. Funct. Bioinform. 2007, 69, 139–159. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Andleeb, S.; Imtiaz-Ud-Din, I.-U.; Rauf, M.K.; Azam, S.S.; Badshah, A.; Sadaf, H.; Raheel, A.; Tahir, M.N.; Raza, S. A one-pot multicomponent facile synthesis of dihydropyrimidin-2(1H)-thione derivatives using triphenylgermane as a catalyst and its binding pattern validation. RSC Adv. 2016, 6, 79651–79661. [Google Scholar] [CrossRef]
- Case, D.A.; Cerutti, D.S.; Cheateham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER16 Package; University of California San Francisco: San Francisco, CA, USA, 2016. [Google Scholar]
- Brice, A.R.; Dominy, B.N. Examining Electrostatic Influences on Base-Flipping: A Comparison of TIP3P and GB Solvent Models. Commun. Comput. Phys. 2013, 13, 223–237. [Google Scholar] [CrossRef]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.J. XMGRACE, Version 5.1. 19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- Shey, R.A.; Ghogomu, S.M.; Esoh, K.K.; Nebangwa, N.D.; Shintouo, C.M.; Nongley, N.F.; Asa, B.F.; Ngale, F.N.; Vanhamme, L.; Souopgui, J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep. 2019, 9, 4409. [Google Scholar] [CrossRef]
- Yadav, S.; Kapley, A. Antibiotic resistance: Global health crisis and metagenomics. Biotechnol. Rep. 2021, 29, e00604. [Google Scholar] [CrossRef]
- Uddin, R.; Jamil, F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput. Biol. Chem. 2018, 74, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Raoufi, E.; Hemmati, M.; Eftekhari, S.; Khaksaran, K.; Mahmodi, Z.; Farajollahi, M.M.; Mohsenzadegan, M. Epitope Prediction by Novel Immunoinformatics Approach: A State-of-the-art Review. Int. J. Pept. Res. Ther. 2019, 26, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Albagi, S.; Ahmed, O.H.; Gumaa, M.A.; Abd_elrahman, K.A.; Abu-Haraz, A.H. Immunoinformatics-Peptide Driven Vaccine and In silico Modeling for Duvenhage Rabies Virus Glycoprotein G. J. Clin. Cell. Immunol. 2017, 8, 2. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein–Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Abraham Peele, K.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Venkateswarulu, T.C. Design of multi-epitope vaccine candidate against SARS-CoV-2, A in-silico study. J. Biomol. Struct. Dyn. 2021, 39, 3793–3801. [Google Scholar] [CrossRef]
- Javadi, M.; Oloomi, M.; Bouzari, S. In Silico Design of a Poly-epitope Vaccine for Urinary Tract Infection Based on Conserved Antigens by Modern Vaccinology. Int. J. Pept. Res. Ther. 2021, 27, 909–921. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Kataoka, K.; Kawabata, S.; Koyanagi, K.; Hashimoto, Y.; Miyake, T.; Fujihashi, K. Respiratory FimA-Specific Secretory IgA Antibodies Upregulated by DC-Targeting Nasal Double DNA Adjuvant Are Essential for Elimination of Porphyromonas gingivalis. Front. Immunol. 2021, 12, 190. [Google Scholar] [CrossRef]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Griffin, C.; et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Kulik, E.M.; Thurnheer, T.; Karygianni, L.; Walter, C.; Sculean, A.; Eick, S. Antibiotic Susceptibility Patterns of Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis Strains from Different Decades. Antibiotics 2019, 8, 253. [Google Scholar] [CrossRef]
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef] [PubMed]
- Lombard, M.; Pastoret, P.-P.; Moulin, A. A brief history of vaccines and vaccination. OIE Rev. Sci. Tech. 2007, 26, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Enayatkhani, M.; Hasaniazad, M.; Faezi, S.; Gouklani, H.; Davoodian, P.; Ahmadi, N.; Einakian, M.A.; Karmostaji, A.; Ahmadi, K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19, an in silico study. J. Biomol. Struct. Dyn. 2021, 39, 2857–2872. [Google Scholar] [CrossRef] [PubMed]
- Tahir ul Qamar, M.; Shokat, Z.; Muneer, I.; Ashfaq, U.A.; Javed, H.; Anwar, F.; Bari, A.; Zahid, B.; Saari, N. Multiepitope-Based Subunit Vaccine Design and Evaluation against Respiratory Syncytial Virus Using Reverse Vaccinology Approach. Vaccines 2020, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Bruno, L.; Cortese, M.; Rappuoli, R.; Merola, M. Lessons from Reverse Vaccinology for viral vaccine design. Curr. Opin. Virol. 2015, 11, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Yi, N.-N.; Kim, U.-S.; Choi, J.-S.; Kim, S.-J.; Choi, J.-I. Porphyromonas gingivalis heat shock protein vaccine reduces the alveolar bone loss induced by multiple periodontopathogenic bacteria. J. Periodontal Res. 2006, 41, 10–14. [Google Scholar] [CrossRef]
- Ross, B.C.; Czajkowski, L.; Hocking, D.; Margetts, M.; Webb, E.; Rothel, L.; Patterson, M.; Agius, C.; Camuglia, S.; Reynolds, E.; et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine 2001, 19, 4135–4142. [Google Scholar] [CrossRef]

| Organism Name | Strain | Size (Mb) | GC% |
|---|---|---|---|
| P. gingivalis | ATCC 33277 | 2.35 | 48.4 |
| P. gingivalis | TDC60 | 2.34 | 48.3 |
| P. gingivalis | W83 | 2.34 | 48.3 |
| Protein ID | Bit Score | Sequence Identity |
|---|---|---|
| Outer membrane | ||
| >core/451/1/Org1_Gene734 | 117 | 35% |
| >core/207/1/Org1_Gene488 | 1303 | 99% |
| Extracellular membrane | ||
| >core/12/1/Org1_Gene1814 | 208 | 39% |
| Periplasmic membrane | ||
| >core/1384/1/Org1_Gene274 | 213 | 53% |
| >core/351/1/Org1_Gene1345 | 281 | 39% |
| >core/1029/1/Org1_Gene254 | 112 | 36% |
| Vaccine Target | T.M.H | Physiochemical Properties | Human Blast | Antigenicity | Allergenicity | |||
|---|---|---|---|---|---|---|---|---|
| Extracellular membrane | HMMTOP | TMHMM | M.W | T.PI | (I.I) | |||
| >core/12/1/Org1_Gene1814 | 1 | 0 | 149.58 | 5.13 | 25.75 | Non-Similar | 0.42 | Non-allergen |
| Outer membrane | ||||||||
| >core/207/1/Org1_Gene488 | 5 | 0 | 73.12 | 8.9 | 8.9 | Non-similar | 0.54 | Non-allergen |
| >core/451/1/Org1_Gene734 | 0 | 0 | 50.89 | 9.72 | 9.72 | Non-similar | 0.60 | |
| Periplasmic membrane | ||||||||
| >core/351/1/Org1_Gene1345 | 2 | 1 | 52.79 | 6.43 | 19.98 | −0.092 | 0.59 | Non-allergen |
| >core/1029/1/Org1_Gene254 | 1 | 0 | 30.28 | 6.65 | 23.64 | Non-similar | 0.47 | Non-allergen |
| >core/1384/1/Org1_Gene274 | 0 | 0 | 21.50 | 5.97 | 19.27 | Non-Similar | 0.50 | Non-allergen |
| S.NO | Shortlisted Proteins | L. casei | L. johnsonii | L. rhamnosus | Bacteroides |
|---|---|---|---|---|---|
| 1 | >core/451/1/Org1_Gene734 (Lytic transglycosylase domain-containing protein) | No Similarity Found | |||
| 2 | >core/1029/1/Org1_Gene254 (FKBP-type peptidyl-propyl cis-trans isomerase) | ||||
| 3 | >core/1384/1/Org1_Gene274 (Superoxide dismutase protein) | ||||
| Selected Epitopes | DRB*01 01 IC50 Score | Antigenicity | Allergenicity | Solubility | Toxin-Pred |
|---|---|---|---|---|---|
| KLYTEERRR | 12.11 | 0.1368 | Non-allergen | Good water soluble | Non-toxin |
| TINSLVDER | 9.95 | 0.6485 | |||
| EICAGETGV | 93.76 | 0.8637 | |||
| EICAGETGV | 93.76 | 0.8637 | |||
| IEHEICAGE | 52.48 | 0.6314 | |||
| QLLNPQYKR | 10.26 | 0.574 | |||
| ELQLLNPQY | 19.54 | 1.2269 | |||
| GVSNDELQL | 27.35 | 1.3654 | |||
| DLEASVSDF | 17.14 | 0.687 |
| S. N | A. A | Sequence Number | A. A | Chi3 | Energy | Sum B-Factors |
|---|---|---|---|---|---|---|
| 6 | Phe | 34 | His | 103.11 | 4.15 | 0 |
| 57 | Glu | 69 | Phe | 111.41 | 7.45 | 0 |
| 74 | Pro | 77 | Gln | 117.49 | 3.97 | 0 |
| 104 | Glu | 110 | Asn | 94.29 | 2.09 | 0 |
| 104 | Glu | 113 | Thr | 98.57 | 4.81 | 0 |
| 104 | Glu | 114 | Pro | 89.56 | 4.62 | 0 |
| 150 | Asp | 154 | Pro | 84.4 | 4.89 | 0 |
| Rank | Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|---|
| 1 | 5 | −13.83 | −7.21 | 2.90 | −5.95 | −0.81 |
| 2 | 9 | −5.42 | −32.42 | 25.47 | 10.50 | −1.38 |
| 3 | 1 | 1.38 | −0.17 | 0.00 | 0.37 | 0.00 |
| 4 | 7 | 15.35 | −0.04 | 0.00 | 0.45 | 0.00 |
| 5 | 2 | 28.20 | −7.73 | 6.54 | 6.57 | −0.93 |
| 6 | 3 | 39.32 | −11.69 | 46.34 | 4.65 | −2.39 |
| 7 | 6 | 62.01 | −21.28 | 93.15 | 6.42 | −5.06 |
| 8 | 4 | 86.39 | −25.68 | 125.90 | 7.34 | −3.11 |
| 9 | 10 | 236.35 | −25.31 | 300.30 | 5.74 | −2.77 |
| 10 | 8 | 1229.01 | −57.59 | 1598.19 | 3.99 | −5.68 |
| Rank | Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|---|
| 1 | 2 | 11.10 | −1.42 | 0.26 | −0.41 | 0.00 |
| 2 | 3 | 77.74 | −20.90 | 84.35 | 16.05 | −1.80 |
| 3 | 8 | 275.98 | −34.08 | 352.40 | 8.64 | −1.46 |
| 4 | 9 | 283.33 | −25.69 | 372.45 | 18.43 | −3.55 |
| 5 | 7 | 515.10 | −9.49 | 643.79 | 3.88 | −0.66 |
| 6 | 5 | 2625.03 | −43.38 | 3325.36 | 13.08 | −3.76 |
| 7 | 4 | 3591.01 | −54.69 | 4575.96 | 3.90 | −2.12 |
| 8 | 10 | 4292.57 | −74.36 | 5482.30 | 5.14 | −10.62 |
| 9 | 6 | 5698.37 | −73.32 | 7235.58 | 12.17 | −7.72 |
| 10 | 1 | 6855.32 | −82.75 | 8713.45 | 8.65 | −11.19 |
| Rank | Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|---|
| 1 | 9 | −13.10 | −25.20 | 9.57 | 11.76 | −3.90 |
| 2 | 10 | 5.35 | −0.18 | 0.00 | 0.06 | 0.00 |
| 3 | 1 | 79.08 | −35.14 | 152.47 | 2.38 | −3.03 |
| 4 | 3 | 998.05 | −36.87 | 1285.61 | 20.01 | −3.89 |
| 5 | 5 | 1020.25 | −34.41 | 1304.89 | 21.14 | −9.75 |
| 6 | 2 | 3071.74 | −70.38 | 3963.53 | 11.31 | −10.56 |
| 7 | 7 | 3176.30 | −74.94 | 4102.83 | 14.27 | −10.73 |
| 8 | 8 | 4986.99 | −83.86 | 6372.21 | 10.54 | −11.68 |
| 9 | 6 | 5724.53 | −49.93 | 7242.80 | 0.76 | −12.72 |
| 10 | 4 | 11,424.45 | −116.65 | 14,451.06 | 29.22 | −14.28 |
| Vaccine Complex | Interactive Residues |
|---|---|
| MHC-I | Ala128, Ala135, Arg157, Arg181, Arg51, Asn155, Asp76, Asp129, Asp 83, Asp238, Glu128, Glu148, Glu154, Gln120, Gly79, His151, Leu23, Lys64, Lys 104, Lys144, Lys197, Phe22, Phe152, Pro50, Ser207, Ser132, Thr240, Thr240, Val9, Val152, |
| MHC-II | Arg44, Asn19, Asp66, Arg189, Cys174, Glu4, Glu10, Glu22, Glu187, Glu214, Gln10, Gln92, Gln174, Gly20, Leu8, Leu11, Leu45, Leu215, Lys197, Lys93, Pro183, Pro124, Pro142, Ser182, Ser126, Thr185,Thr100, Lys98, Thr21, Thr172, Tyr83,Thr181, Val99, Val91,Val86 |
| TLR-4 | Asp428, Asn464, Asn464, Asn6, Asn86, Arg606, Cys542, Leu485, Gln588,Gln588, Gln484, Gln510, Glu136, Glu509, Glu605, Glu485, Gly183, His529, His555, His557, Len462, Lys 4, Lys244, Lys560, Leu87, Phe228, Phe 463, Phe487, Phe538, Pro88, Pro489, Ser569, Tyr79, Thr548, Thr584, Val461 |
| Energy Parameter | TLR-4-Vaccine Complex | MHC-I-Vaccine Complex | MHC-II-Vaccine Complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −90.14 | −80.96 | −70.46 |
| EEL | −85.88 | −59.00 | −42.26 |
| Delta G gas | −176.02 | −139.96 | −112.72 |
| Delta G solv | 40.29 | 38.64 | 36.55 |
| Delta Total | −135.73 | −101.32 | −76.17 |
| MM-PBSA | |||
| VDWAALS | −90.14 | −80.96 | −70.46 |
| EEL | −85.88 | −59.00 | −42.26 |
| Delta G gas | −176.02 | −139.96 | −112.72 |
| Delta G solv | 43.16 | 39.87 | 42.59 |
| Delta Total | −132.86 | −100.09 | −70.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rida, T.; Ahmad, S.; Ullah, A.; Ismail, S.; Tahir ul Qamar, M.; Afsheen, Z.; Khurram, M.; Saqib Ishaq, M.; Alkhathami, A.G.; Alatawi, E.A.; et al. Pan-Genome Analysis of Oral Bacterial Pathogens to Predict a Potential Novel Multi-Epitopes Vaccine Candidate. Int. J. Environ. Res. Public Health 2022, 19, 8408. https://doi.org/10.3390/ijerph19148408
Rida T, Ahmad S, Ullah A, Ismail S, Tahir ul Qamar M, Afsheen Z, Khurram M, Saqib Ishaq M, Alkhathami AG, Alatawi EA, et al. Pan-Genome Analysis of Oral Bacterial Pathogens to Predict a Potential Novel Multi-Epitopes Vaccine Candidate. International Journal of Environmental Research and Public Health. 2022; 19(14):8408. https://doi.org/10.3390/ijerph19148408
Chicago/Turabian StyleRida, Tehniyat, Sajjad Ahmad, Asad Ullah, Saba Ismail, Muhammad Tahir ul Qamar, Zobia Afsheen, Muhammad Khurram, Muhammad Saqib Ishaq, Ali G. Alkhathami, Eid A. Alatawi, and et al. 2022. "Pan-Genome Analysis of Oral Bacterial Pathogens to Predict a Potential Novel Multi-Epitopes Vaccine Candidate" International Journal of Environmental Research and Public Health 19, no. 14: 8408. https://doi.org/10.3390/ijerph19148408
APA StyleRida, T., Ahmad, S., Ullah, A., Ismail, S., Tahir ul Qamar, M., Afsheen, Z., Khurram, M., Saqib Ishaq, M., Alkhathami, A. G., Alatawi, E. A., Alrumaihi, F., & Allemailem, K. S. (2022). Pan-Genome Analysis of Oral Bacterial Pathogens to Predict a Potential Novel Multi-Epitopes Vaccine Candidate. International Journal of Environmental Research and Public Health, 19(14), 8408. https://doi.org/10.3390/ijerph19148408










