Electrophoretic Determination of Trimethylamine (TMA) in Biological Samples as a Novel Potential Biomarker of Cardiovascular Diseases Methodological Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Standards
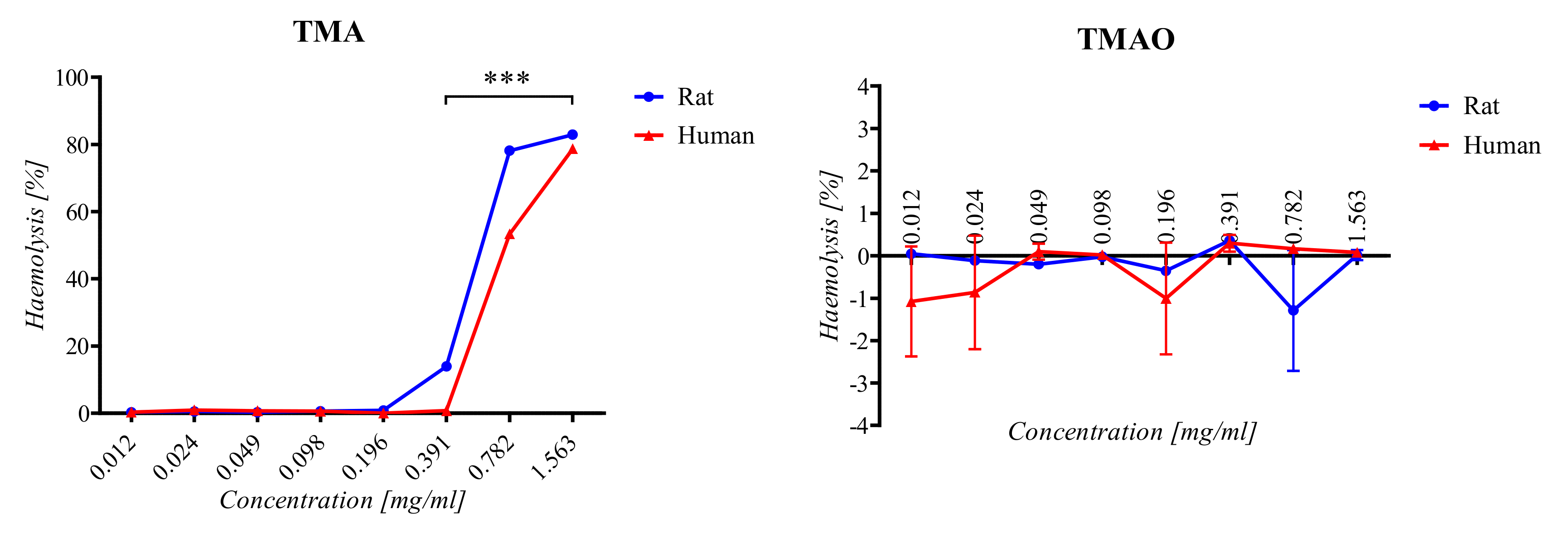
2.2. Haemolysis Assay
2.3. Cell Culture
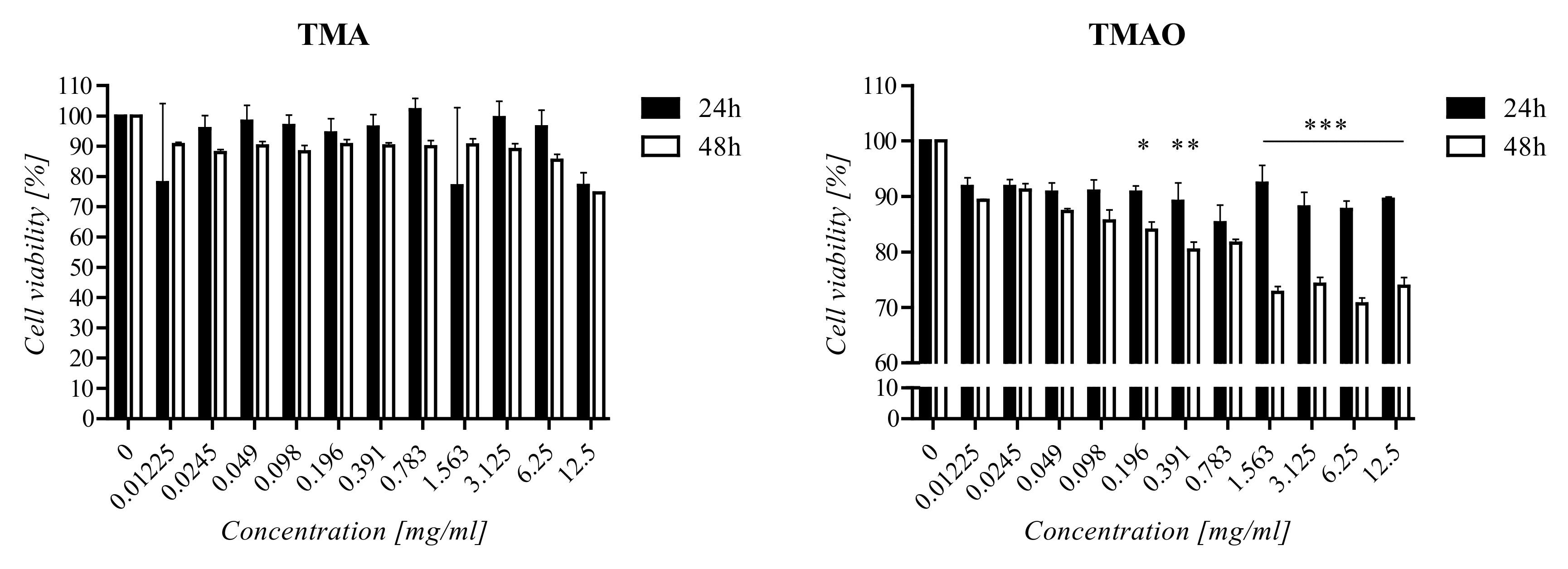
2.4. Cell Viability Assay
2.5. Samples
2.6. Sample Preparation
2.7. Instrumentation: Capillary Zone Electrophoresis
3. Results
3.1. Haemolysis Assay
3.2. Cell Viability Assay
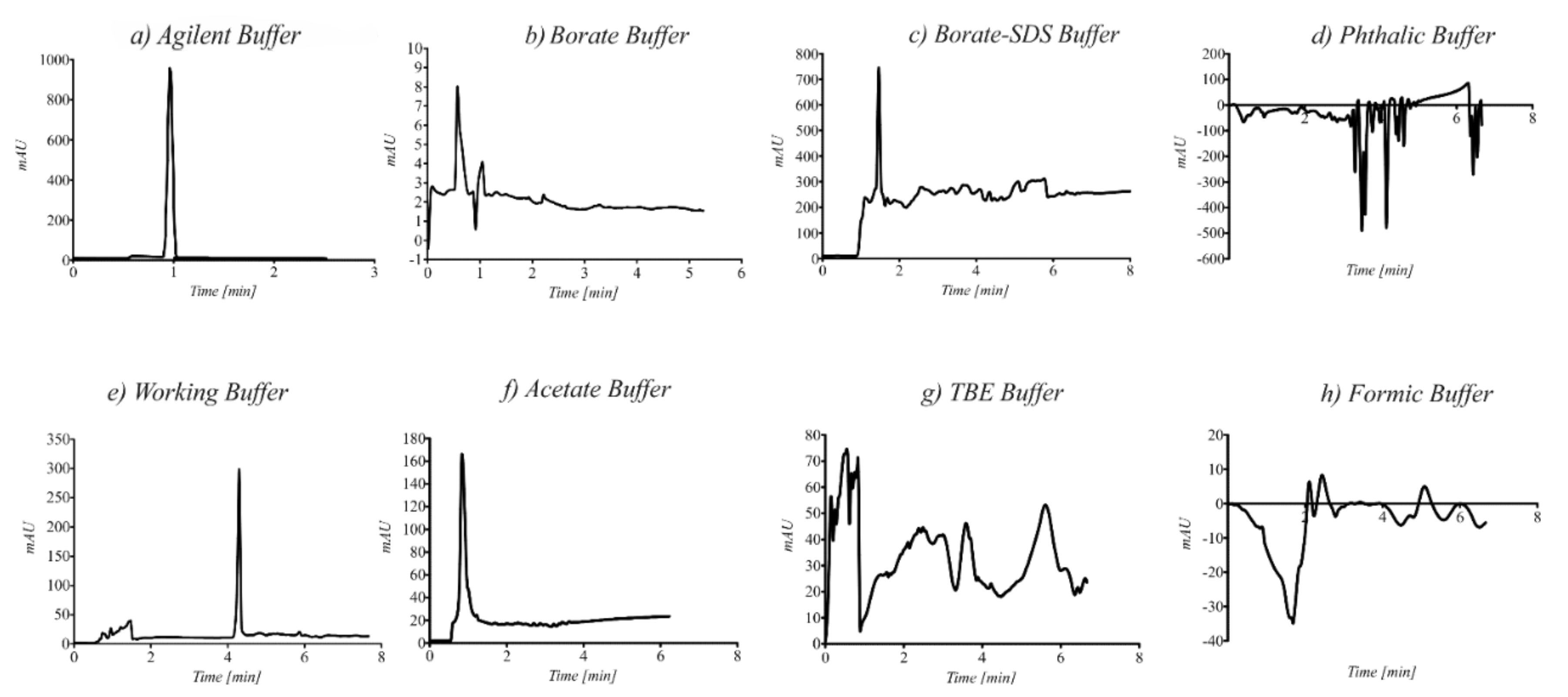
3.3. Optimization of CE Conditions
3.4. Method Validation
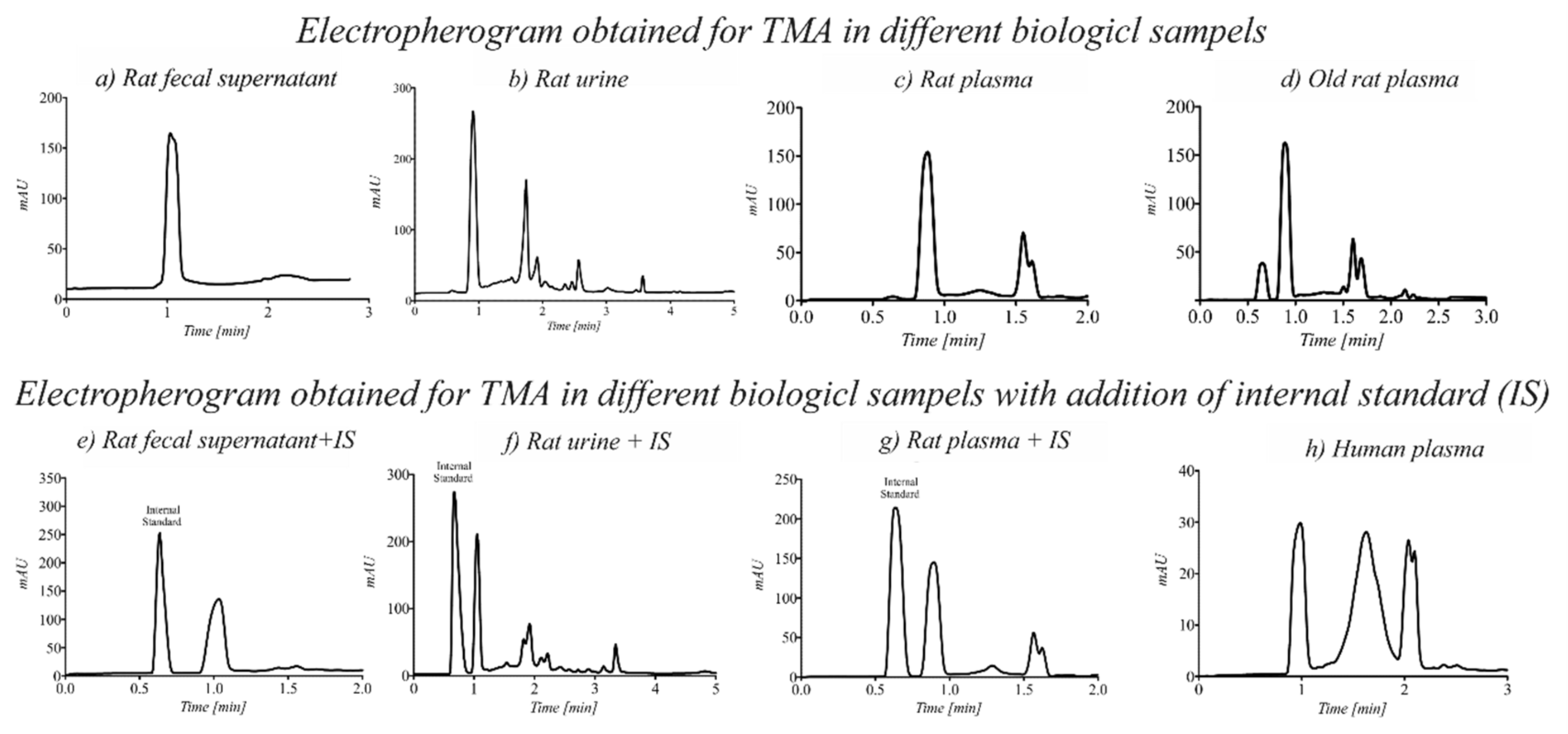
3.5. Applicability of the Method
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaworska, K.; Hering, D.; Mosieniak, G.; Bielak-Zmijewska, A.; Pilz, M.; Konwerski, M.; Gasecka, A.; Kaplon-Cieślicka, A.; Filipiak, K.; Sikora, E.; et al. TMA, a forgotten uremic toxin, but not TMAO, is involved in cardiovascular pathology. Toxins 2019, 11, 490. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Gaur, A.; Singhal, V.; Parakh, N.; Bhargava, B.; Sharma, A. The complex metabolism of trimethylamine in humans: Endogenous and exogenous sources. Expert Rev. Mol. Med. 2016, 18, e8. [Google Scholar] [CrossRef]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. 2014, 20, 16062. [Google Scholar]
- Falony, G.; Vieira-Silva, S.; Raes, J. Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine. Annu. Rev. Microbiol. 2015, 69, 305–321. [Google Scholar] [CrossRef]
- Institute of Medicine. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline; National Academies Press: Washington, DC, USA, 1998. [Google Scholar]
- Chester, D.N.; Goldman, J.D.; Ahuja, J.K.; Moshfegh, A.J. Dietary Intakes of Choline: What We Eat in America, NHANES 2007–2008; Food Surveys Research Group Dietary Data Brief No 9; US Department of Agriculture: Washington, DC, USA, 2011.
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. 2010, 7, 30. [Google Scholar]
- Rebouche, C.J.; Chenard, C.A. Metabolic fate of dietary carnitine in human adults: Identification and quantification of urinary and fecal metabolites. J. Nutr. 1991, 121, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Craciun, S.; Balskus, E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 21307–21312. [Google Scholar] [CrossRef] [PubMed]
- Unemoto, T.; Hayashi, M.; Miyaki, K.; Hayashi, M. Formation of trimethylamine from dl-carnitine by Serratia marcescens. Biochim. Biophys. Acta BBA Gen. Subj. 1966, 121, 220–222. [Google Scholar] [CrossRef]
- Seim, H.; Löster, H.; Claus, R.; Kleber, H.P.; Strack, E. Splitting of the C-N bond in carnitine by an enzyme (trimethylamine forming) from membranes of Acinetobacter calcoaceticus. FEMS Microbiol. Lett. 1982, 15, 165–167. [Google Scholar] [CrossRef][Green Version]
- Koeth, R.A.; Levison, B.S.; Culley, M.K.; Buffa, J.A.; Wang, Z.; Gregory, J.C.; Org, E.; Wu, Y.; Li, L.; Smith, J.D.; et al. γ-butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014, 20, 799–812. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, Z.; Zhou, J.; Gong, S.; Wang, W.; Chen, Y. Novel fiber coated with amide bridged-calix[4]arene used for solid-phase microextraction of aliphatic amines. J. Chromatogr. A 2004, 1041, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mills, G.A.; Walker, V.; Mughal, H. Quantitative determination of trimethylamine in urine by solid-phase microextraction and gas chromatography-mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 1999, 723, 281–285. [Google Scholar] [CrossRef]
- Namieśnik, J.; Jastrzebska, A.; Zygmunt, B. Determination of volatile aliphatic amines in air by solid-phase microextraction coupled with gas chromatography with flame ionization detection. J. Chromatogr. A 2003, 1016, 1–9. [Google Scholar] [CrossRef]
- Adhoum, N.; Monser, L.; Sadok, S.; El-Abed, A.; Greenway, G.M.; Uglow, R.F. Flow injection potentiometric detection of trimethylamine in seafood using tungsten oxide electrode. Anal. Chim. Acta 2003, 478, 53–58. [Google Scholar] [CrossRef]
- Gupta, S.; Misra, T.N. Manganese phthalocyanine for the detection of fish freshness by its trimethylamine emission. Sens. Actuators B Chem. 1997, 41, 199–202. [Google Scholar] [CrossRef]
- Loechel, C.; Basran, A.; Basran, J.; Scrutton, N.S.; Hall, E.A.H. Using trimethylamine dehydrogenase in an enzyme linked amperometric electrode. Part 1. Wild-type enzyme redox mediation. Analyst 2003, 128, 166–172. [Google Scholar] [CrossRef]
- Siddiqi, Z.M.; Pathania, D. Rapid, selective and direct spectrophotometeric determination of aliphatic amines with m-dinitrobenzene. Talanta 2003, 60, 1197–1203. [Google Scholar] [CrossRef]
- García-Garrido, J.A.; de Castro, M.D.L. Determination of Trimethylamine in Fish by Pervaporation and Photometric Detection. Analyst 1997, 122, 663–666. [Google Scholar] [CrossRef]
- Lee, M.B.; Storer, M.K.; Blunt, J.W.; Lever, M. Validation of (1)H NMR spectroscopy as an analytical tool for methylamine metabolites in urine. Clin. Chim. Acta 2006, 365, 264–269. [Google Scholar] [CrossRef]
- Herráez-Hernández, R.; Cháfer-Pericás, C.; Verdú-Andrés, J.; Campíns-Falcó, P. An evaluation of solid phase microextraction for aliphatic amines using derivatization with 9-fluorenylmethyl chloroformate and liquid chromatography. J. Chromatogr. A 2006, 1104, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Cháfer-Pericás, C.; Herráez-Hernández, R.; Campíns-Falcó, P. Liquid chromatographic determination of trimethylamine in water. J. Chromatogr. A 2004, 1023, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chang, H. Indirect Fluorescence of Amines in Capillary Electrophoresis, Using Cresyl Violet. J. Liq. Chromatogr. Relat. Technol. 2003, 26, 3387–3400. [Google Scholar] [CrossRef]
- Li, M.; Lee, S.H. Determination of trimethylamine in fish by capillary electrophoresis with electrogenerated tris(2,2’-bipyridyl)ruthenium(II) chemiluminescence detection. Luminescence 2007, 22, 588–593. [Google Scholar] [CrossRef]
- Sirkisoon, L.R.; Makamba, H.C.; Saito, S.; Colyer, C.L. Carbon Dot-Mediated Capillary Electrophoresis Separations of Metallated and Demetallated Forms of Transferrin Protein. Molecules 2019, 24, 1916. [Google Scholar] [CrossRef] [PubMed]
- Klepárník, K.; Boček, P. Electrophoresis today and tomorrow: Helping biologists’ dreams come true. Bioessays 2010, 32, 218–226. [Google Scholar] [CrossRef]
- Stolz, A.; Jooß, K.; Höcker, O.; Römer, J.; Schlecht, J.; Neusüß, C. Recent advances in capillary electrophoresis-mass spectrometry: Instrumentation, methodology and applications. Electrophoresis 2019, 40, 79–112. [Google Scholar] [CrossRef]
- Sieradzka, E.; Witt, K.; Milnerowicz, H. The application of capillary electrophoresis techniques in toxicological analysis. Biomed. Chromatogr. 2014, 28, 1507–1513. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.-Y.; Ding, G.-S.; Liu, F.-J.; Tang, A.-N. Water-compatible surface molecularly imprinted silica nanoparticles as pseudostationary phase in electrokinetic chromatography for the enantioseparation of tryptophan. J. Chromatogr. A 2013, 1311, 176–182. [Google Scholar] [CrossRef]
- Turiel, E.; Martin-Esteban, A. Molecular imprinting technology in capillary electrochromatography. J. Sep. Sci. 2005, 28, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Guggenheim, E.J.; Briffa, S.M.; Thorn, J.A.; Lynch, I.; Valsami-Jones, E. Current Application of Capillary Electrophoresis in Nanomaterial Characterisation and Its Potential to Characterise the Protein and Small Molecule Corona. Nanomaterials 2018, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- García-Cañas, V.; Aznar, E.; Simó, C. Screening gut microbial trimethylamine production by fast and cost-effective capillary electrophoresis. Anal. Bioanal. Chem. 2019, 411, 2697–2705. [Google Scholar] [CrossRef] [PubMed]
- Mazzarino, L.; Loch-Neckel, G.; dos Santos Bubniak, L.; Ourique, F.; Otsuka, I.; Halila, S.; Pedrosa, R.C.; Santos-Silva, M.C.; Lemos-Senna, E.; Muniz, E.C.; et al. Nanoparticles made from xyloglucan-block-polycaprolactone copolymers: Safety assessment for drug delivery. Toxicol. Sci. 2015, 147, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.A.; Shea, J.W. Dietary choline and betaine and risk of cvd: A systematic review and meta-analysis of prospective studies. Nutrients 2017, 9, 711. [Google Scholar] [CrossRef]
- Bertoia, M.L.; Pai, J.K.; Cooke, J.P.; Joosten, M.M.; Mittleman, M.A.; Rimm, E.B.; Mukamal, K.J. Plasma homocysteine, dietary B vitamins, betaine, and choline andrisk of peripheral artery disease. Atherosclerosis 2014, 269, 29–34. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Rimm, E.B.; Hu, F.B.; Albert, C.M.; Rexrode, K.M.; Manson, J.A.E.; Qi, L. Dietary phosphatidylcholine and risk of all-cause and cardiovascular-specific mortality among US women and men. Am. J. Clin. Nutr. 2016, 104, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Millard, H.R.; Musani, S.K.; Dibaba, D.T.; Talegawkar, S.A.; Taylor, H.A.; Tucker, K.L.; Bidulescu, A. Dietary choline and betaine; associations with subclinical markers of cardiovascular disease risk and incidence of CVD, coronary heart disease and stroke: The Jackson Heart Study. Eur. J. Nutr. 2018, 57, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Bidulescu, A.; Chambless, L.E.; Siega-Riz, A.M.; Zeisel, S.H.; Heiss, G. Usual choline and betaine dietary intake and incident coronary heart disease: The Atherosclerosis Risk in Communities (ARIC) Study. BMC Cardiovasc. Disord. 2007, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dalmeijer, G.W.; Olthof, M.R.; Verhoef, P.; Bots, M.L.; van der Schouw, Y.T. Prospective study on dietary intakes of folate, betaine, and choline and cardiovascular disease risk in women. Eur. J. Clin. Nutr. 2008, 62, 386–394. [Google Scholar] [CrossRef]
- Nagata, C.; Wada, K.; Tamura, T.; Konishi, K.; Kawachi, T.; Tsuji, M.; Nakamura, K. Choline and betaine intakes are not associated with cardiovascular disease mortality risk in Japanese men and women. J. Nutr. 2015, 145, 1787–1792. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, C.; Moré, M.; Bellamine, A. Trimethylamine n-oxide in relation to cardiometabolic health—Cause or effect? Nutrients 2020, 12, 1330. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, Q.; Jiang, H. Gut microbiota in atherosclerosis: Focus on trimethylamine N-oxide. APMIS 2020, 128, 353–366. [Google Scholar] [CrossRef] [PubMed]
| Characterization Parameter | TMA [µg/mL] |
|---|---|
| Calibration range | 6.1–3125.0 |
| Calibration curve equation | y = 0.0118x + 9.5357 |
| Linearity (R2) | 0.9950 |
| LOD | 1.2 |
| LOQ | 3.6 |
| Real Urine Samples | Real Plasma Samples | Real Faces Samples | |||
|---|---|---|---|---|---|
| Sample Name | Average TMA Concentration ± U [µg/mL] * | Sample Name | Average TMA Concentration ± U [µg/mL] * | Sample Name | Average TMA Concentration ± U [µg/mL] * |
| U1 | 1.0 × 102 ± 1.7 | P1 | 3.4 × 10 ± 3.6 | F1 | 266.6 ± 3.2 |
| U2 | 6.8 × 10 ± 1.0 | P2 | nd ** | F2 | 429.9 ± 3.2 |
| U3 | 9.0 × 10 ± 8.3 | P3 | nd ** | F3 | nd ** |
| U4 | 2.9 × 10 ± 3.5 | P4 | 1.25 × 102 ± 0.19 | F4 | 562.1 ± 6.7 |
| U5 | 1.1 × 103 ± 3.6 × 10 | P5 | nd ** | F5 | 61.7 ± 1.8 |
| U6 | 1.3 × 102 ± 1.0 × 10 | P6 | 2.1 × 10 ± 3.2 | F6 | 28.6 ± 1.8 |
| U7 | 3.1 × 102 ± 4.1 × 10 | P7 | 7.8 × 10 ± 5.3 | F7 | nd ** |
| U8 | 1.2 × 102 ± 1.0 × 10 | P8 | nd ** | F8 | 578.6 ± 1.1 |
| U9 | 7.9 × 10 ± 1.2 × 10 | P9 | nd ** | F9 | 31.1 ± 1.8 |
| U10 | 3.5 × 102 ± 2.8 × 10 | P10 | nd ** | F10 | 71.4 ± 2.1 |
| U11 | 3.5 × 102 ± 3.1 × 10 | P11 | nd ** | F11 | 244.0 ± 9.6 |
| U12 | 4.0 × 102 ± 4.4 × 10 | P12 | nd ** | F12 | 277.6 ± 3.2 |
| U13 | 1.6 × 102 ± 2.4 × 10 | P13 | nd ** | F13 | nd ** |
| U14 | 1.5 × 102 ± 2.1 × 10 | P14 | nd ** | F14 | 522.5 ± 27.9 |
| U15 | 2.6 × 102 ± 1.1 × 10 | P15 | 1.1 × 103 ± 1.1 × 102 | F15 | 102.3 ± 1.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konop, M.; Rybka, M.; Waraksa, E.; Laskowska, A.K.; Nowiński, A.; Grzywacz, T.; Karwowski, W.J.; Drapała, A.; Kłodzińska, E.M. Electrophoretic Determination of Trimethylamine (TMA) in Biological Samples as a Novel Potential Biomarker of Cardiovascular Diseases Methodological Approach. Int. J. Environ. Res. Public Health 2021, 18, 12318. https://doi.org/10.3390/ijerph182312318
Konop M, Rybka M, Waraksa E, Laskowska AK, Nowiński A, Grzywacz T, Karwowski WJ, Drapała A, Kłodzińska EM. Electrophoretic Determination of Trimethylamine (TMA) in Biological Samples as a Novel Potential Biomarker of Cardiovascular Diseases Methodological Approach. International Journal of Environmental Research and Public Health. 2021; 18(23):12318. https://doi.org/10.3390/ijerph182312318
Chicago/Turabian StyleKonop, Marek, Mateusz Rybka, Emilia Waraksa, Anna K. Laskowska, Artur Nowiński, Tomasz Grzywacz, Wojciech J. Karwowski, Adrian Drapała, and Ewa Maria Kłodzińska. 2021. "Electrophoretic Determination of Trimethylamine (TMA) in Biological Samples as a Novel Potential Biomarker of Cardiovascular Diseases Methodological Approach" International Journal of Environmental Research and Public Health 18, no. 23: 12318. https://doi.org/10.3390/ijerph182312318
APA StyleKonop, M., Rybka, M., Waraksa, E., Laskowska, A. K., Nowiński, A., Grzywacz, T., Karwowski, W. J., Drapała, A., & Kłodzińska, E. M. (2021). Electrophoretic Determination of Trimethylamine (TMA) in Biological Samples as a Novel Potential Biomarker of Cardiovascular Diseases Methodological Approach. International Journal of Environmental Research and Public Health, 18(23), 12318. https://doi.org/10.3390/ijerph182312318








