Healthier Communities of Phytoplankton and Bacteria Achieved via the Application of Modified Clay in Shrimp Aquaculture Ponds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Preparation of the MC
2.3. Sample Collection
2.4. Water Quality Analysis
2.5. DNA Extraction, PCR Amplification, and Illumina Sequencing
2.6. Data Processing and Analysis
3. Results
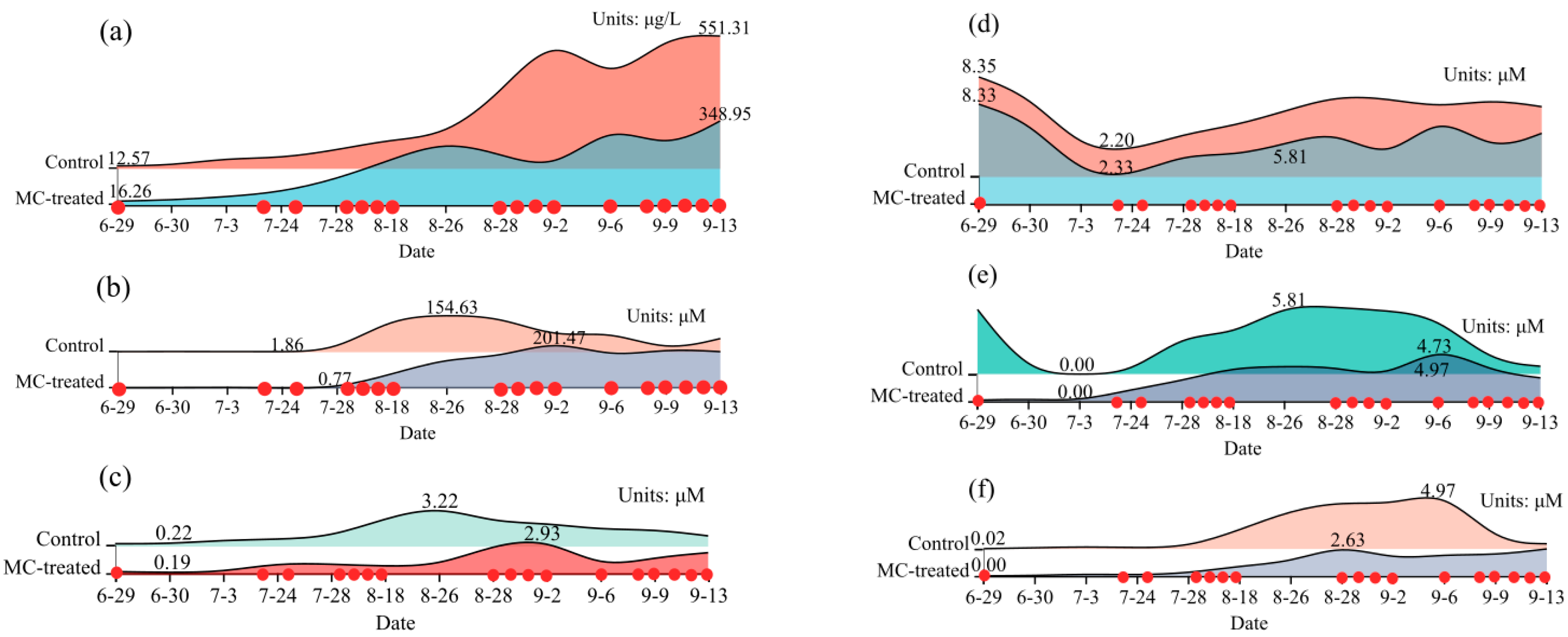
3.1. Water Quality Analysis
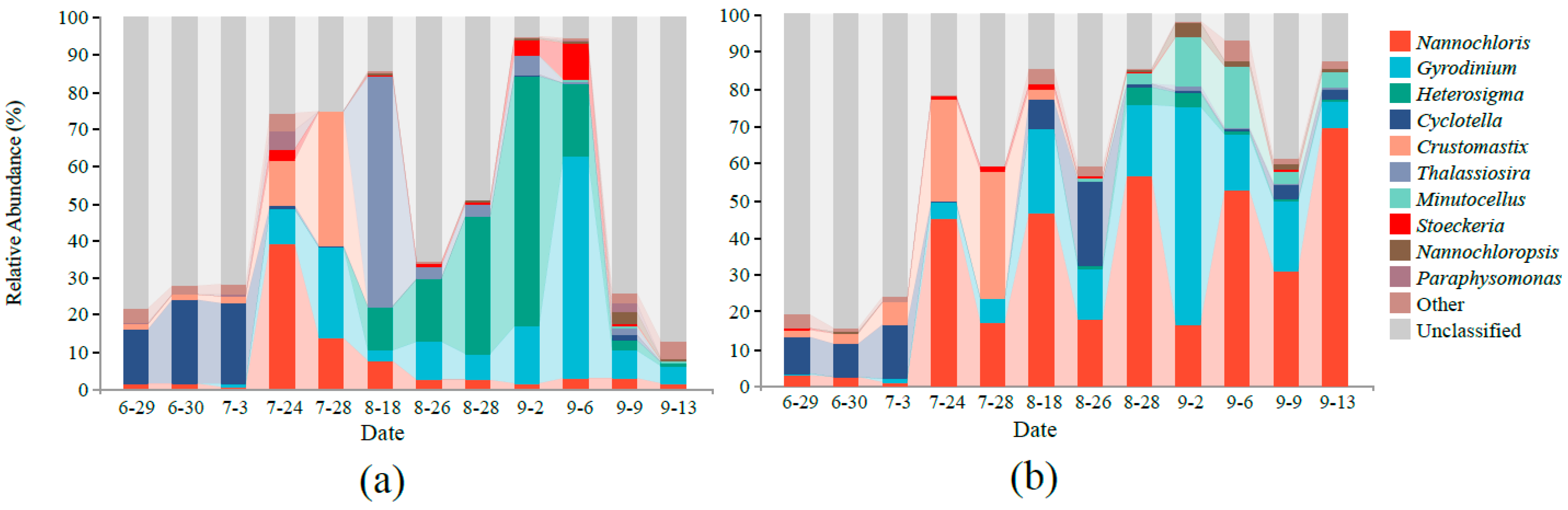
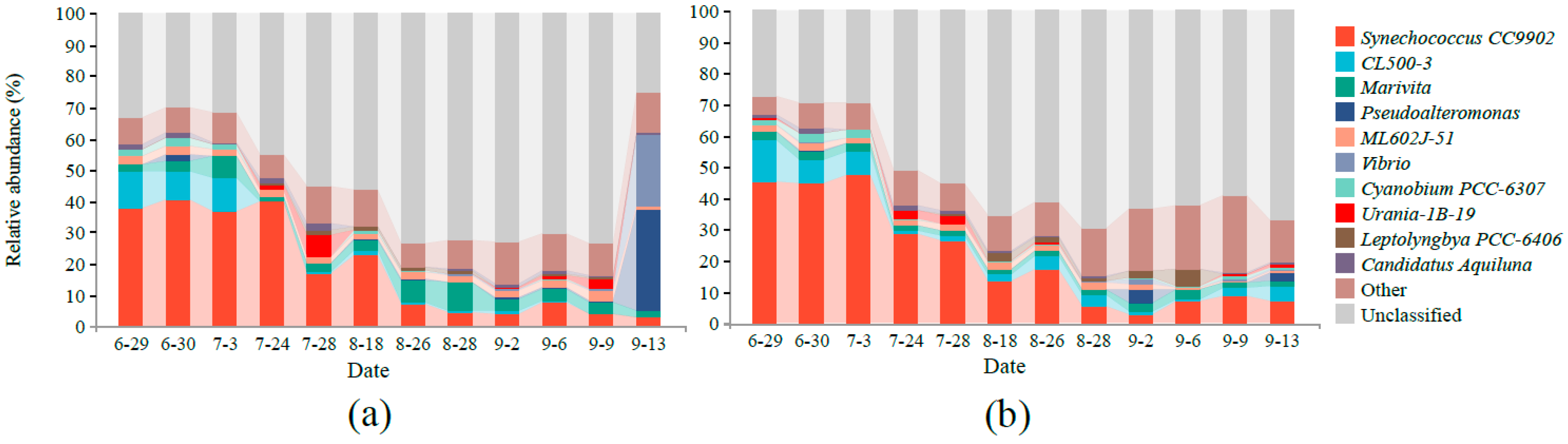
3.2. Microbial Community Composition
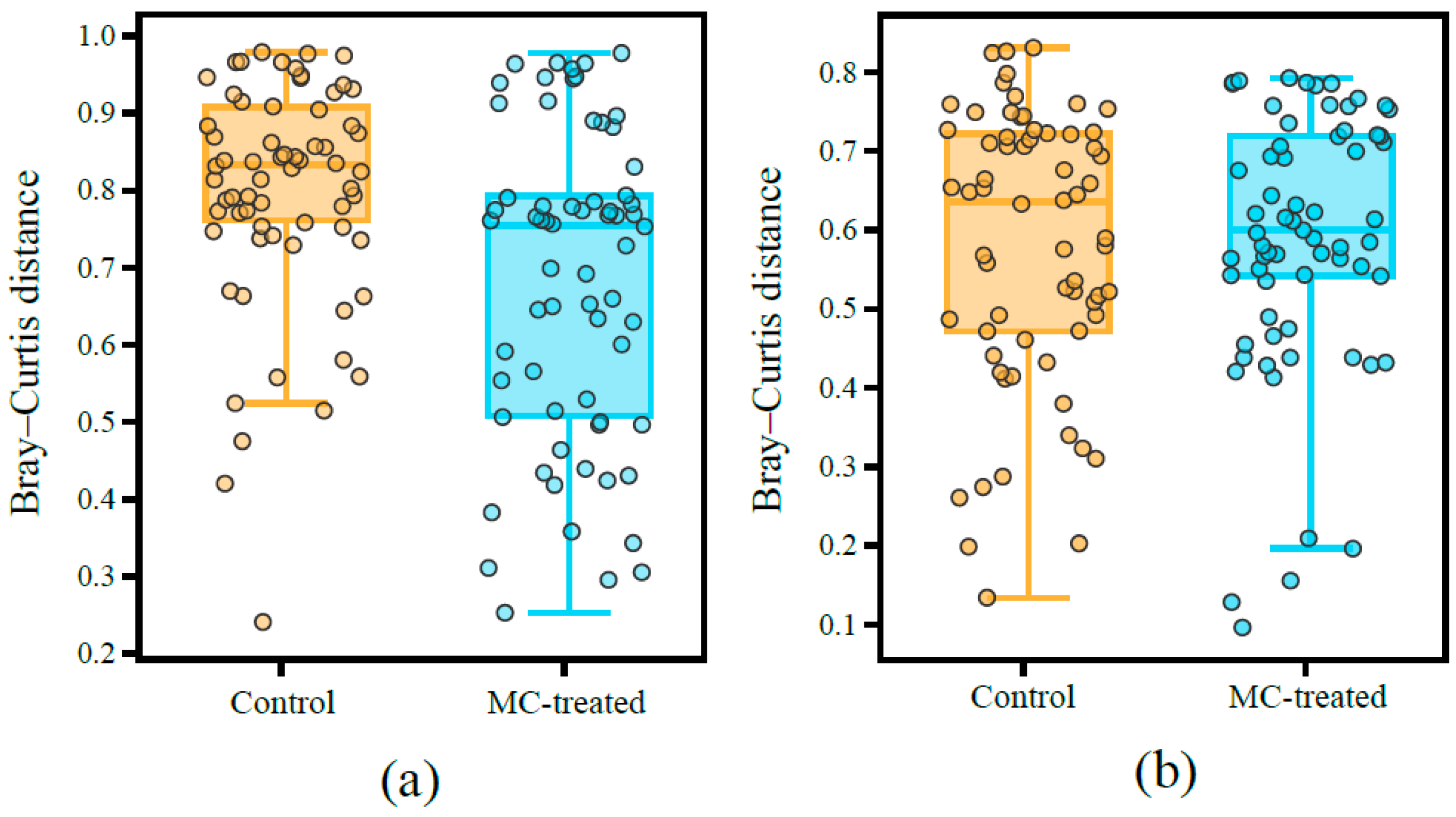
3.3. Diversity of Microbial Communities
3.4. Network Structure of Microbial Communities
3.5. Key Microbial Populations of Molecular Ecological Network
4. Discussion
4.1. Effect of MC on Phytoplankton Community Structure in Aquaculture Water
4.2. Effect of MC on Bacteria Community Structure in Aquaculture Water
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guppy, J.L.; Jones, D.B.; Kjeldsen, S.R.; Le Port, A.; Khatkar, M.S.; Wade, N.M.; Sellars, M.J.; Steinig, E.J.; Raadsma, H.W.; Jerry, D.R.; et al. Development and validation of a RAD-Seq target-capture based genotyping assay for routine application in advanced black tiger shrimp (Penaeus monodon) breeding programs. BMC Genom. 2020, 21, 541. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.C.; Bonetti, C.; Seiffert, W.Q. Hydrological and Water Quality Indices as management tools in marine shrimp culture. Aquaculture 2011, 318, 425–433. [Google Scholar] [CrossRef]
- Xiong, J.; Zhu, J.; Zhang, D. The application of bacterial indicator phylotypes to predict shrimp health status. Appl. Microbiol. Biotechnol. 2014, 98, 8291. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Rodríguez, R.; Páez-Osuna, F. Nutrients, phytoplankton and harmful algal blooms in shrimp ponds: A review with special reference to the situation in the Gulf of California. Aquaculture 2003, 219, 317–336. [Google Scholar] [CrossRef]
- Azanza, R.V.; Fukuyo, Y.; Yap, L.G.; Takayama, H. Prorocentrum minimum bloom and its possible link to a massive fish kill in Bolinao, Pangasinan, Northern Philippines. Harmful Algae 2005, 4, 519–524. [Google Scholar] [CrossRef]
- Pitois, S.; Jackson, M.H.; Wood, B.J. Sources of the eutrophication problems associated with toxic algae: An overview. J. Environ. Health 2001, 64, 25–32. [Google Scholar]
- Zhou, J.; Fang, W.; Yang, X.; Zhou, S.; Hu, L.; Li, X.; Qi, X.; Su, H.; Xie, L. A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS ONE 2012, 7, e29961. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, C.; Zheng, Z.; Wei, Y.; Lu, K.; Zhu, J. Nutrient enrichment during shrimp cultivation alters bacterioplankton assemblies and destroys community stability. Ecotoxicol. Environ. Saf. 2018, 156, 366–374. [Google Scholar] [CrossRef]
- Austin, B.; Zhang, X. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 2010, 43, 119–124. [Google Scholar] [CrossRef]
- Bauer, J.; Teitge, F.; Neffe, L.; Adamek, M.; Jung, A.; Peppler, C.; Steinhagen, D.; Jung-Schroers, V. Recommendations for identifying pathogenic Vibrio spp. as part of disease surveillance programmes in recirculating aquaculture systems for Pacific white shrimps (Litopannaeus vannamei). J. Fish. Dis. 2018, 41, 1877–1897. [Google Scholar] [CrossRef] [PubMed]
- Roux, F.L.; Wegner, K.M.; Baker-Austin, C.; Vezzulli, L.; Osorio, C.R.; Amaro, C.; Ritchie, J.M.; Defoirdt, T.; Destoumieux-Garzón, D.; Blokesch, M. The emergence of Vibrio pathogens in Europe: Ecology, evolution, and pathogenesis (Paris, 11–12th March 2015). Front. Microbiol. 2015, 6, 830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, X.; Xiong, J.; Zhu, J.; Wang, Y.; Zhao, Q.; Chen, H.; Guo, A.; Wu, J.; Dai, H. Bacterioplankton assemblages as biological indicators of shrimp health status. Ecol. Indic. 2014, 38, 218–224. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Yu, Z. An eco-environmental assessment of harmful algal bloom mitigation using modified clay. Harmful Algae 2021, 107, 102067. [Google Scholar] [CrossRef]
- Yu, Z.; Song, X.; Cao, X.; Liu, Y. Mitigation of harmful algal blooms using modified clays: Theory, mechanisms, and applications. Harmful Algae 2017, 69, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhiming, Y.U.; Song, X.; Cao, X. Impact of Modified Clays on the Infant Oyster (Crassostrea gigas). Mar. Sci. Bull. 2007, 26, 53–60. (In Chinese) [Google Scholar]
- Lu, G.; Song, X.; Yu, Z.; Cao, X. Application of PAC-modified kaolin to mitigate Prorocentrum donghaiense: Effects on cell removal and phosphorus cycling in a laboratory setting. J. Appl. Phycol. 2017, 29, 1–12. [Google Scholar] [CrossRef]
- Lu, G.; Song, X.; Yu, Z.; Cao, X.; Yuan, Y. Environmental effects of modified clay flocculation on Alexandrium tamarense and paralytic shellfish poisoning toxins (PSTs). Chemosphere 2015, 127, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Song, X.; Cao, X.; Yu, Z. Variation of bacteria in flocs after removal of Karenia mikimotoi by different types of modified clay. Mar. Sci. 2021, 45, 46–56. (In Chinese) [Google Scholar]
- Attramadal, K.J.K.; Tøndel, B.; Salvesen, I.; Øie, G.; Vadstein, O.; Olsen, Y. Ceramic clay reduces the load of organic matter and bacteria in marine fish larval culture tanks. Aquac. Eng. 2012, 49, 23–34. [Google Scholar] [CrossRef]
- Yu, Z.; Zou, J.; Ma, X. A new method to improve the capability of clays for removing red tide organisms. Oceanol. Et Limnol. Sin. 1994, 25, 226–232. (In Chinese) [Google Scholar]
- Liu, Y.; Cao, X.; Yu, Z.; Song, X.; Qiu, L. Controlling harmful algae blooms using aluminum-modified clay. Mar. Pollut. Bull. 2016, 103, 211–219. [Google Scholar] [CrossRef]
- Paterson, J.S.; Nayar, S.; Mitchell, J.G.; Seuront, L. Population-specific shifts in viral and microbial abundance within a cryptic upwelling. J. Mar. Syst. 2013, 113–114, 52–61. [Google Scholar] [CrossRef]
- Guo, M.; Wu, F.; Hao, G.; Qi, Q.; Rong, L.; Li, N.; Wei, L.; Chai, T. Bacillus subtilis Improves Immunity and Disease Resistance in Rabbits. Front. Immunol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.K.; Au, C.H.; Chu, K.H.; Kwan, H.S.; Wong, C.K. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 2010, 4, 1053. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Jia, G. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The smallest of all worlds: Pollination networks. J. Theor. Biol. 2006, 240, 270–276. [Google Scholar] [CrossRef]
- Guimerà, R.; Sales-Pardo, M.; Amaral, L. Classes of complex networks defined by role-to-role connectivity profiles. Nat. Phys. 2006, 3, 63. [Google Scholar] [CrossRef] [PubMed]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed]
- Godhe, A.; Asplund, M.E.; Harnstrom, K.; Saravanan, V.; Tyagi, A.; Karunasagar, I. Quantification of Diatom and Dinoflagellate Biomasses in Coastal Marine Seawater Samples by Real-Time PCR. Appl. Environ. Microbiol. 2008, 74, 7174–7182. [Google Scholar] [CrossRef]
- Chen, S.-N.; Shang, P.-L.; Kang, P.-L.; Du, M.-M. Metabolic Functional Community Diversity of Associated Bacteria during the Degradation of Phytoplankton from a Drinking Water Reservoir. Int J. Env. Res. Public Health 2020, 17, 1687. [Google Scholar] [CrossRef]
- Armbrecht, L.; Hallegraeff, G.; Bolch, C.J.S.; Woodward, C.; Cooper, A. Hybridisation capture allows DNA damage analysis of ancient marine eukaryotes. Sci. Rep. 2021, 11, 3220. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Z.; Lu, T.; Peijnenburg, W.J.G.M.; Gillings, M.; Yang, X.; Chen, J.; Penuelas, J.; Zhu, Y.-G.; Zhou, N.-Y.; et al. Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems. Commun. Biol. 2020, 3, 737. [Google Scholar] [CrossRef]
- Liang, Z. Effects of Alexandrium tamarense and Heterosigma akashiide on the Early Development of Several Fishery Organisms. Master’s Thesis, Ocean University of China, Qingdao, China, 2004. (In Chinese). [Google Scholar]
- Vasseur, C.; Bougaran, G.; Garnier, M.; Hamelin, J.; Leboulanger, C.; Chevanton, M.L.; Mostajir, B.; Sialve, B.; Steyer, J.P.; Fouilland, E. Carbon conversion efficiency and population dynamics of a marine algae-bacteria consortium growing on simplified synthetic digestate: First step in a bioprocess coupling algal production and anaerobic digestion. Bioresour. Technol. 2012, 119, 79–87. [Google Scholar] [CrossRef]
- Witt, U.; Koske, P.H.; Kuhlmann, D.; Lenz, J.; Nellen, W. Production of Nannochloris spec. (Chlorophyceae) in large-scale outdoor tanks and its use as a food organism in marine aquaculture. Aquaculture 1981, 23, 171–181. [Google Scholar] [CrossRef]
- Kim, E.J.; Park, S.; Hong, H.J.; Choi, Y.E.; Yang, J.W. Biosorption of chromium (Cr(III)/Cr(VI)) on the residual microalga Nannochloris oculata after lipid extraction for biodiesel production. Bioresour. Technol. 2011, 102, 11155–11160. [Google Scholar] [CrossRef]
- Black, C.K.; Mihai, D.M.; Washington, I. The Photosynthetic Eukaryote Nannochloris eukaryotum as an Intracellular Machine To Control and Expand Functionality of Human Cells. Nano Lett. 2014, 14, 2720–2725. [Google Scholar] [CrossRef]
- Wang, X. Research on the Relationship between Phytoplankton and Chemical Measurement of Water Factors in the Ponds of Litopenaeus vannamei. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2018. [Google Scholar]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic Molecular Ecological Network of Soil Microbial Communities in Response to Elevated CO2. mBio 2011, 2, e00122-11. [Google Scholar] [CrossRef]
- Jeanbille, M.; Gury, J.; Duran, R.; Tronczynski, J.; Agogué, H.; Ben Saïd, O.; Ghiglione, J.; Auguet, J. Response of Core Microbial Consortia to Chronic Hydrocarbon Contaminations in Coastal Sediment Habitats. Front. Microbiol 2016, 7, 1637. [Google Scholar] [CrossRef]
- Xiong, J.; Chen, H.; Hu, C.; Ye, X.; Kong, D.; Zhang, D. Evidence of bacterioplankton community adaptation in response to long-term mariculture disturbance. Sci. Rep. 2015, 5, 15274. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.S.; Wei, J.B.; Song, X.Y.; Liu, S.Y. Effects of Different Land Use Typess on the Molecular Ecological Network of Soil Bacteria. Environ. Sci. 2020, 41, 1456–1465. (In Chinese) [Google Scholar]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling Interactions in the Microbiome: A Network Perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Yang, Y.; Hang, B. Responses of Phytoplankton Communities to N/P Ratio Changes in Phosphorus-limited Subtropical Reservoir in Southern China. J. Trop. Subtrop. Bot. 2021, 29, 20–30. (In Chinese) [Google Scholar]
- Wang, R.; Guo, Z.; Tang, Y.; Kuang, J.; Duan, Y.; Lin, H.; Jiang, S.; Shu, H.; Huang, J. Effects on development and microbial community of shrimp Litopenaeus vannamei larvae with probiotics treatment. AMB Express 2020, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, C.; Chen, Y.; Zhang, D.; Zhao, M.; Li, H.; Guo, P. Microbiome Analysis Reveals Microecological Balance in the Emerging Rice-Crayfish Integrated Breeding Mode. Front. Microbiol. 2021, 12, 669570. [Google Scholar] [CrossRef]
- Xiong, J.; Wang, K.; Wu, J.; Qiuqian, L.; Yang, K.; Qian, Y.; Zhang, D. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotechnol. 2015, 99, 6911–6919. [Google Scholar] [CrossRef]
- Holben, W.E.; Williams, P.; Saarinen, M.; Srkilahti, L.K.; Apajalahti, J. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 2002, 44, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Skjermo, J.; Salvesen, I.; Øie, G.; Olsen, Y.; Vadstein, O. Microbially matured water: A technique for selection of a non-opportunistic bacterial flora in water that may improve performance of marine larvae. Aquac. Int. 1997, 5, 13–28. [Google Scholar] [CrossRef]
- Attramadal, K.; Truong, T.; Bakke, I.; Skjermo, J.; Olsen, Y.; Vadstein, O. RAS and microbial maturation as tools for K-selection of microbial communities improve survival in cod larvae. Aquaculture 2014, 432, 483–490. [Google Scholar] [CrossRef]
- Dai, W.; Yu, W.; Zhang, J.; Zhu, J.; Tao, Z.; Xiong, J. The gut eukaryotic microbiota influences the growth performance among cohabitating shrimp. Appl. Microbiol. Biotechnol. 2017, 101, 6447–6457. [Google Scholar] [CrossRef]
- Xiong, J.; Zhu, J.; Dai, W.; Dong, C.; Qiu, Q.; Li, C. Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ. Microbiol. 2017, 19, 1490–1501. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 8. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Leveque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High Microbial Diversity Promotes Soil Ecosystem Functioning. Appl. Environ. Microbiol. 2018, 84, 13. [Google Scholar] [CrossRef]
- Trivedi, C.; Delgado-Baquerizo, M.; Hamonts, K.; Lai, K.T.; Reich, P.B.; Singh, B.K. Losses in microbial functional diversity reduce the rate of key soil processes. Soil Biol. Biochem. 2019, 135, 267–274. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA. 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [PubMed]
- Isbell, F.; Craven, D.; Connolly, J.; Loreau, M.; Schmid, B.; Beierkuhnlein, C.; Bezemer, T.M.; Bonin, C.; Bruelheide, H.; de Luca, E.; et al. Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 2015, 526, 574–577. [Google Scholar] [CrossRef]
- Naeem, S.; Li, S.B. Biodiversity enhances ecosystem reliability. Nature 1997, 390, 507–509. [Google Scholar] [CrossRef]
- Tilman, D.; Reich, P.B.; Knops, J.M.H. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 2006, 441, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, N.; Tamames, J.; Linz, A.M.; Pedros-Alio, C.; Puente-Sanchez, F. Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J. 2019, 13, 2969–2983. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. Fems. Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef]
- Wang, F.; Tang, Y. Determination of key species in the food web and their impact on the robustness. Biodivers. Sci. 2019, 27, 1132–1137. (In Chinese) [Google Scholar]
- Simon, M.; Scheuner, C.; Meier-Kolthoff, J.P.; Brinkhoff, T.; Wagner-Döbler, I.; Ulbrich, M.; Klenk, H.-P.; Schomburg, D.; Petersen, J.; Göker, M. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017, 11, 1483–1499. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, V.; Zucker, F.; Moraru, C. Marine Bacteriophages. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 322–341. [Google Scholar]
- Mu, D.S.; Wang, S.; Liang, Q.Y.; Du, Z.Z.; Du, Z. Bradymonabacteria, a novel bacterial predator with versatile survival strategies in saline environments. Microbiome 2020, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. The Family Cryomorphaceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014. [Google Scholar]
- Ivanova, E.P.; Ng, H.J.; Webb, H.K. The Family Pseudoalteromonadaceae. In The Prokaryotes: Gammaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014. [Google Scholar]
- Chen, H.; Athar, R.; Zheng, G.; Williams, H.N. Prey bacteria shape the community structure of their predators. ISME J. 2011, 5, 1314–1322. [Google Scholar] [CrossRef]
- Ye, X.; Zhao, H.; Chen, M.; Li, H.; He, X. Isolation, identification and prey range of Halobacteriovorax sp. BALOs10. Acta Microbiol. Sin. 2019, 59, 1331–1341. (In Chinese) [Google Scholar]
- Topolska, J.; Latowski, D.; Kaschabek, S.; Manecki, M.; Merkel, B.J.; Rakovan, J. Pb remobilization by bacterially mediated dissolution of pyromorphite Pb5(PO4)3Cl in presence of phosphate-solubilizing Pseudomonas putida. Environ. Sci. Pollut. Res. 2014, 21, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Guangyuan, L.U.; Song, X.; Zhiming, Y.U.; Cao, X.; Yuan, Y. Effects of modified clay flocculation on major nutrients and diatom aggregation during Skeletonema costatum blooms in the laboratory. Chin. J. Oceanol. Limnol. 2015, 33, 1007–1019. [Google Scholar]
- Spietz, R.L.; Williams, C.M.; Rocap, G.; Horner-Devine, M.C. A Dissolved Oxygen Threshold for Shifts in Bacterial Community Structure in a Seasonally Hypoxic Estuary. PLoS ONE 2015, 10, e0135731. [Google Scholar] [CrossRef]
- Zhou, J.; Lyu, Y.; Richlen, M.; Anderson, D.M.; Cai, Z. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. CRC Crit. Rev. Plant. Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef]


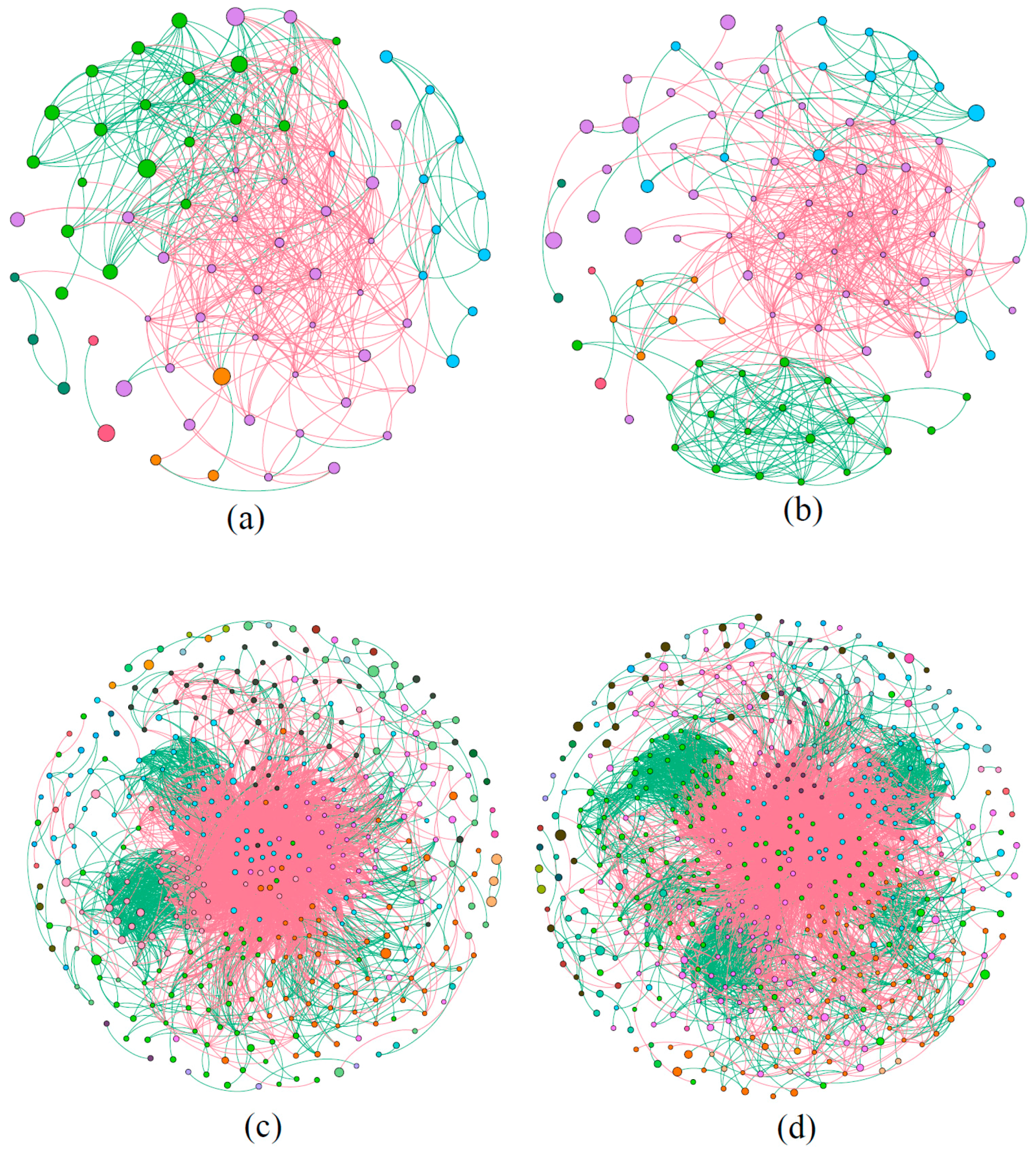
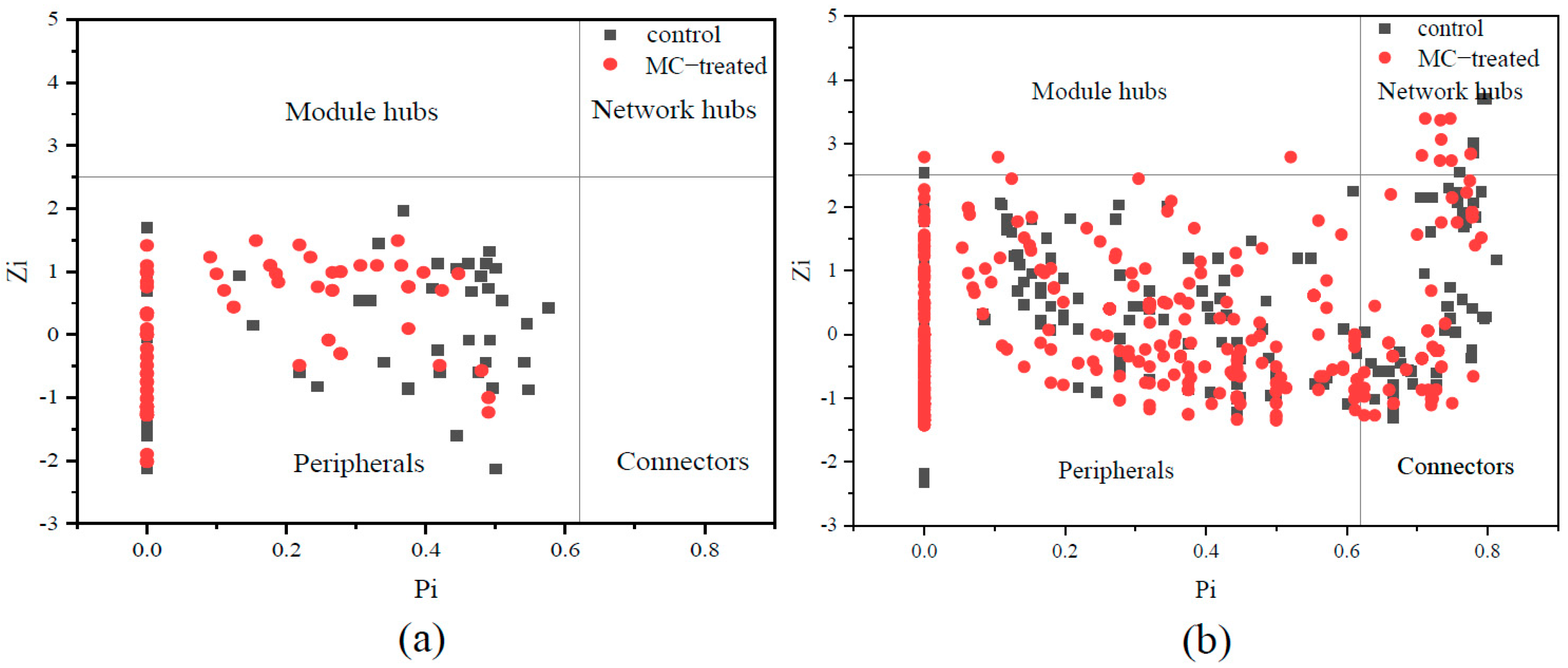
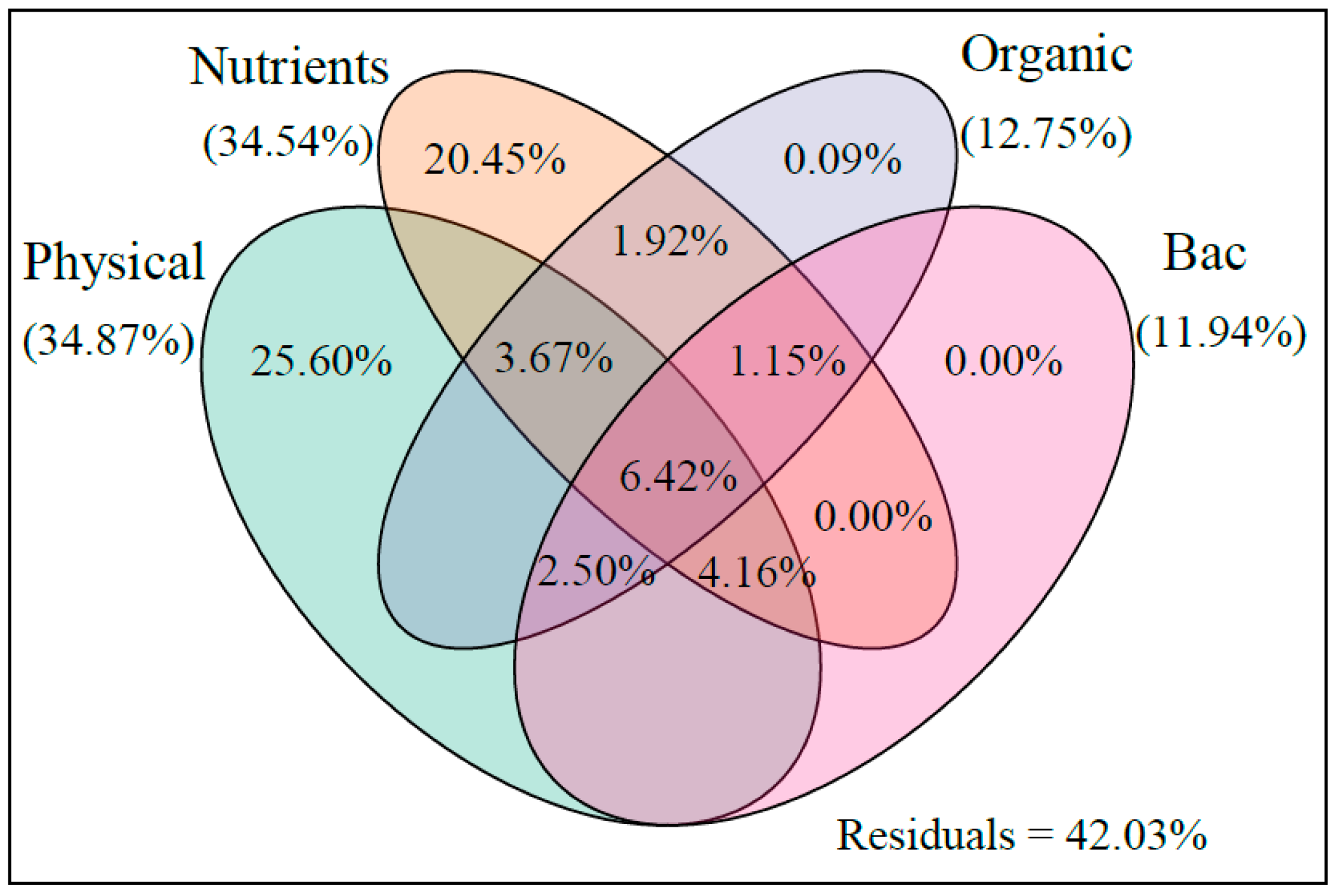

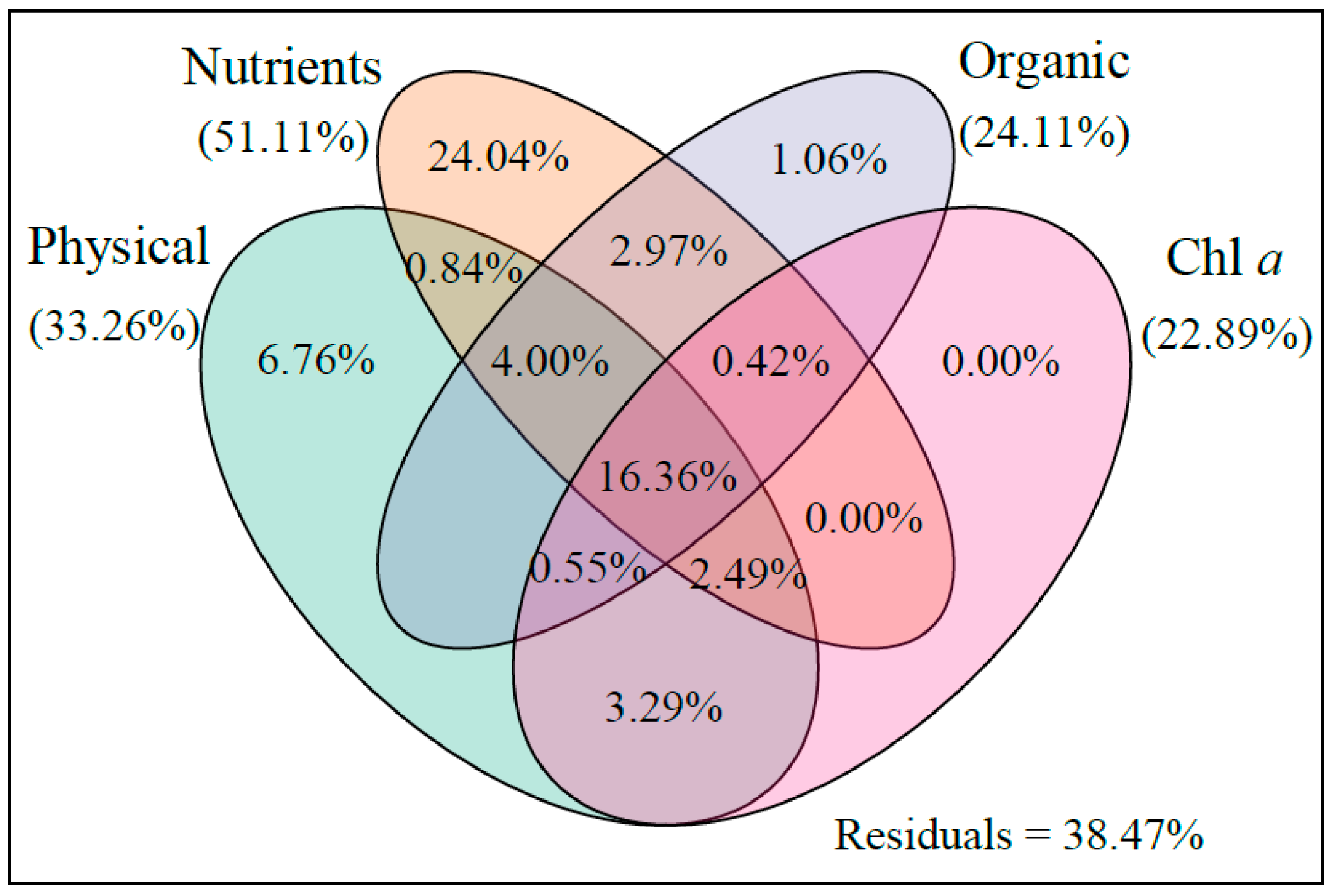
| Network Indexes | Phytoplankton | Bacteria | Explanation | ||
|---|---|---|---|---|---|
| Control | MC-Treated | Control | MC-Treated | ||
| Total nodes | 74 | 94 | 397 | 500 | Network scale |
| Total links | 444 | 501 | 3791 | 4590 | Species interaction complexity |
| Average clustering coefficient | 0.385 | 0.375 | 0.253 | 0.224 | Node connection degree |
| Average path distance | 2.464 | 2.833 | 3.150 | 3.315 | Response speed |
| Positive link percentage (%) | 36.26 | 38.52 | 23.48 | 30.98 | Cooperation and mutualisms |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Song, X.; Cao, X.; He, L.; Liu, S.; Yu, Z. Healthier Communities of Phytoplankton and Bacteria Achieved via the Application of Modified Clay in Shrimp Aquaculture Ponds. Int. J. Environ. Res. Public Health 2021, 18, 11569. https://doi.org/10.3390/ijerph182111569
Ding Y, Song X, Cao X, He L, Liu S, Yu Z. Healthier Communities of Phytoplankton and Bacteria Achieved via the Application of Modified Clay in Shrimp Aquaculture Ponds. International Journal of Environmental Research and Public Health. 2021; 18(21):11569. https://doi.org/10.3390/ijerph182111569
Chicago/Turabian StyleDing, Yu, Xiuxian Song, Xihua Cao, Liyan He, Shanshan Liu, and Zhiming Yu. 2021. "Healthier Communities of Phytoplankton and Bacteria Achieved via the Application of Modified Clay in Shrimp Aquaculture Ponds" International Journal of Environmental Research and Public Health 18, no. 21: 11569. https://doi.org/10.3390/ijerph182111569
APA StyleDing, Y., Song, X., Cao, X., He, L., Liu, S., & Yu, Z. (2021). Healthier Communities of Phytoplankton and Bacteria Achieved via the Application of Modified Clay in Shrimp Aquaculture Ponds. International Journal of Environmental Research and Public Health, 18(21), 11569. https://doi.org/10.3390/ijerph182111569





