Predicting Physical Exercise Adherence in Fitness Apps Using a Deep Learning Approach
Abstract
:1. Introduction
2. Materials and Methods
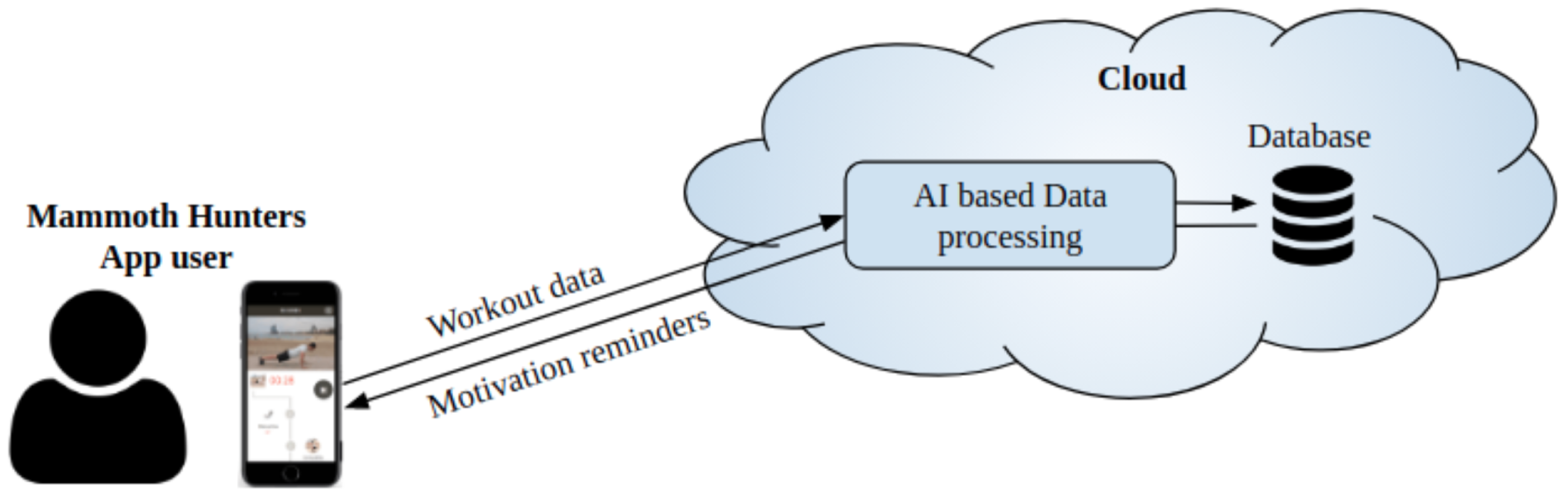
2.1. Data Acquisition
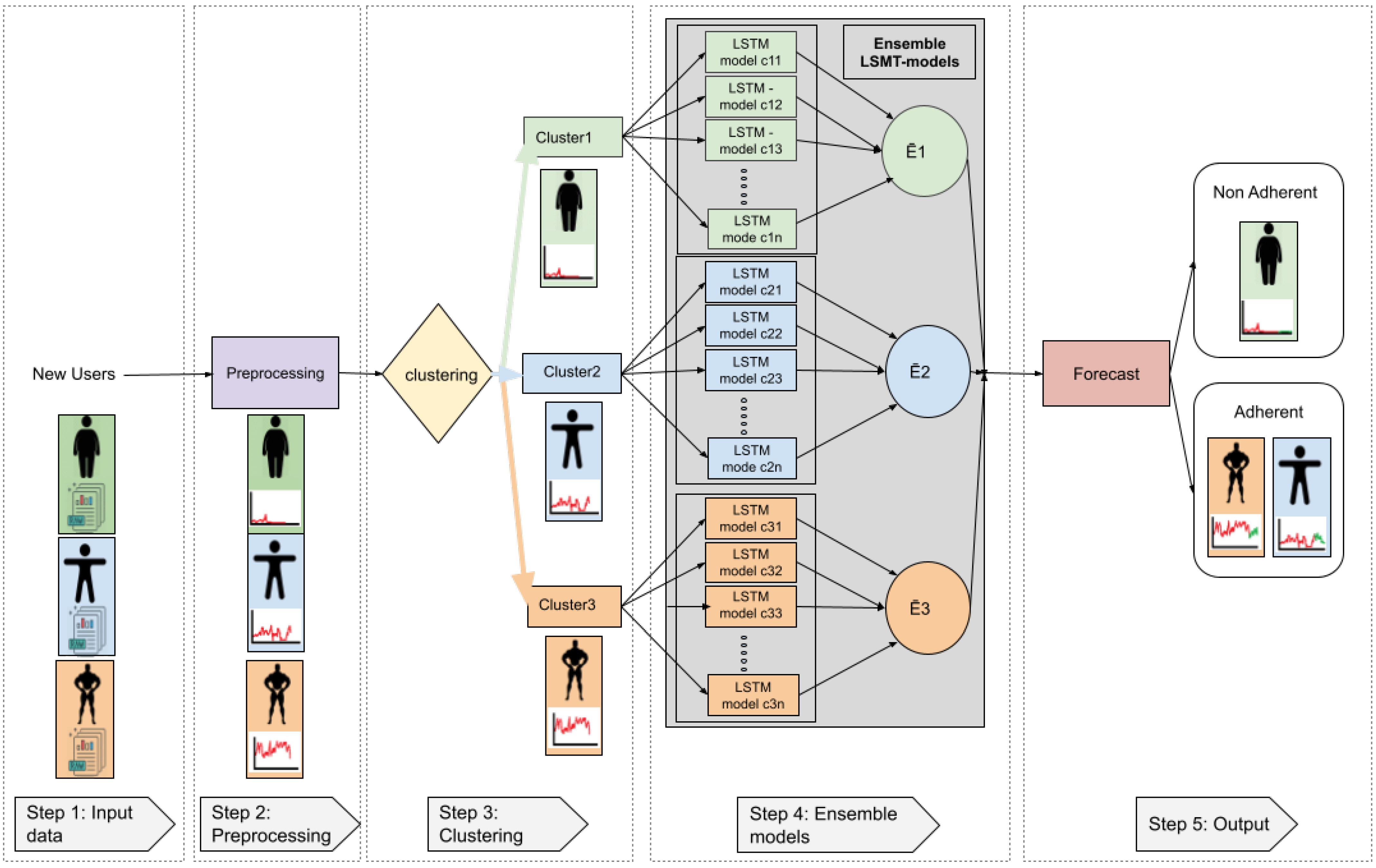
2.2. Proposed Framework
2.2.1. Input Data
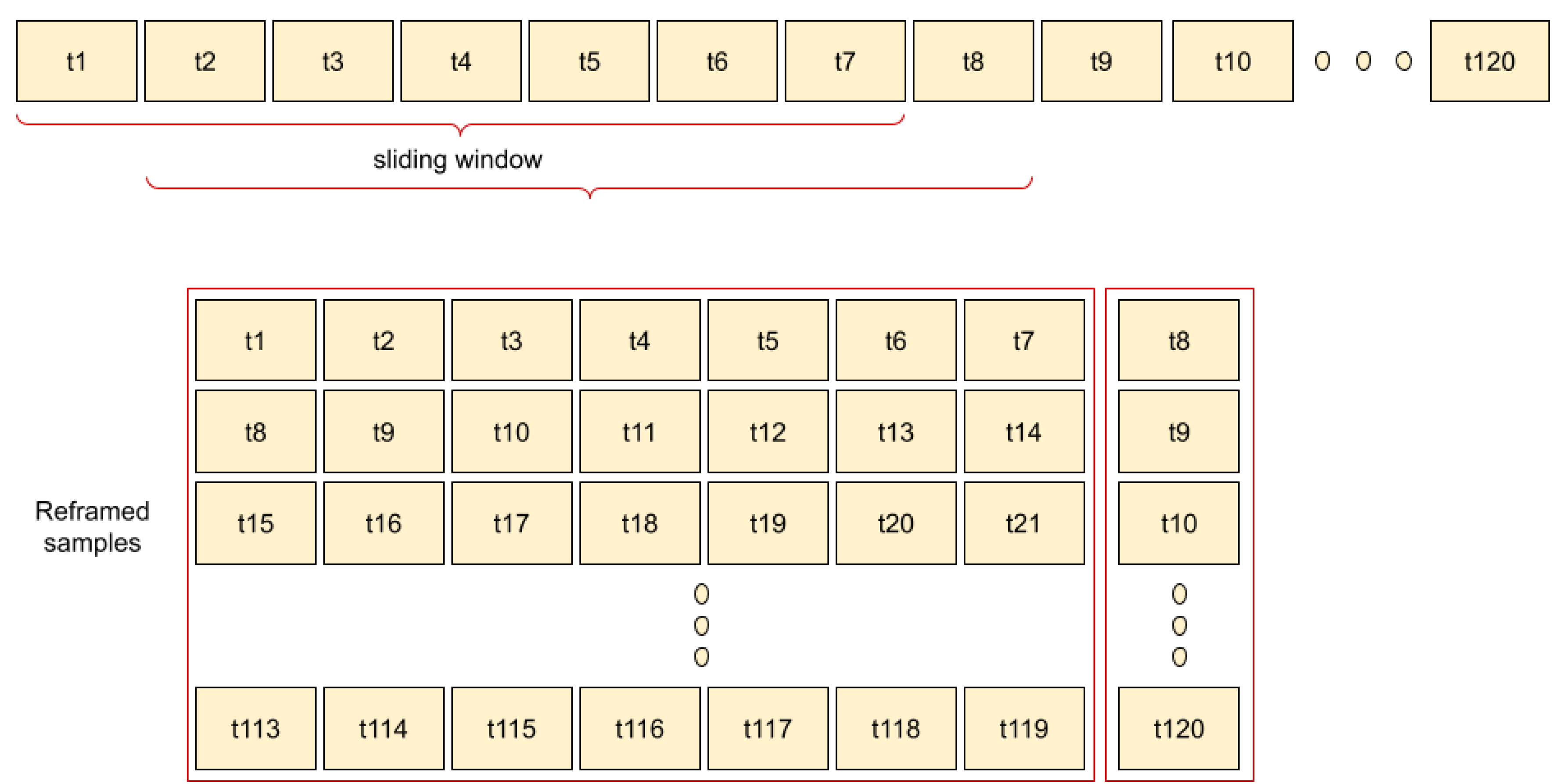
2.2.2. Pre-Processing
2.2.3. Clustering
- K-means: K-means is one of the preferred methods in unsupervised ML strategies. It is widely used in manufacturing, education and business [34], and is based on minimization of the sum of distances between each object and the centroid of its group or cluster. Once the number of clusters (K) is chosen, the first initialisation step is to establish K centroids among the data. Then, samples are assigned to their closest centroids. Next, the positions of the centroids are updated, so that the distances between the elements of each cluster may be minimized. The assignment and update process is then repeated until no points change clusters, or equivalently, until the centroids stay the same. In order to assign a point to the closest centroid, a proximity measure that quantifies the notion of closest is required. Commonly, it is the Euclidean distance that is used for this, and so the goal is to find the objective function which minimizes the squared distance of each point to its closest centroid [35]. The sum of squared errors (SSE) calculates the error of each point (i.e., Euclidean distance from each point to the closest centroid) [35,36]. The SSE is defined by the Equation (1):where is the standard Euclidean distance, x is an object, is the cluster and is the centroid (mean) for cluster what minimizes the SSE of the cluster is the mean, defined by the Equation (2):where n corresponds to the number of objects in the cluster. The Basic K-means functionality is described in Table 8.
- BIRCH: This is a non-supervised algorithm. Due to its ability to find good clustering with only a single data scan, it is especially suitable for larger datasets or streaming data [37]. This characteristic was especially relevant to our research, since we expect to obtain a larger dataset in the near future, and we were in need of a process that would facilitate the upscaling of our application.In order to understand how the BIRCH algorithm works, the concept of cluster feature (CF) needs to be introduced. CF is a set of three summary statistics which represent a single cluster, from a set of data points. The first statistic, count, quantifies how many data values are present in the cluster. The second, linear sum, is a measurement which represents cluster location. Finally, squared sum refers to the sum of the squared coordinates that represents the spread of the clusters. The last two statistics are equivalents to mean and variance of the data point [37]. BIRCH is frequently explained in two steps: (1) building the CF tree, and (2) global clustering.Phase 1–Building the CF Tree: Firstly, the data is loaded into the memory by building a CF Tree, for which purpose a sequential clustering approach is used. Thus, the algorithm simultaneously scans and records the data, and then determines whether a point should be added to an existing cluster, or a new cluster should be created.Phase 2–Global clustering: Secondly, an existing clustering algorithm is applied to the sub-clusters (the CCF lead nodes), so as to assemble these sub-clusters into clusters. This could, for instance, be achieved using the agglomerative hierarchical method.The basic BIRCH algorithm is described in Table 9.
- Affinity propagation: this clustering method was proposed by Fred and Dueck in 2007, and works with the similarity matrix. Points that appear close to each other have high similarity while those that are furthest have low similarity [38]. Unlike others, the AP method is not required as a parameter, although it is commonly used in experiments where many clusters are needed. AP works with three matrices: similarity matrix, responsibility matrix and availability matrix.Similarity matrix: this is the first matrix obtained, and is calculated by negating the sum of the squares of the difference between participants [39]. Thus, the elements in the main diagonal of the similarity matrix equal 0 (zero) and a value needs to be selected in order to fill these. Consequently, the algorithm will converge around a few clusters if the selected value is low, and vice-versa, insofar as the algorithm will converge with many clusters, in the case of high selected values.Responsibility matrix: once the similarity matrix has been calculated, the next step is to calculate the responsibility matrix, given by the Equation (3).where i corresponds to the number of rows and k to the number of columns in the associated matrix.Availability matrix: the availability matrix is then calculated. All elements are set to zero, and Equations (4) and (5) are then used to calculate elements off the diagonal.In essence, the Equation (4) corresponds to the sum of all values in the columns that are above 0, except for values which are identical for both rows and the given column.Criterion matrix: Finally, the algorithm calculates the criterion matrix. This equals the sum of the availability matrix and the responsibility matrix at that location, and is given by (6).The highest criterion value of each row is designated as the exemplar. The pseudocode of AP can be seen in Table 10.
2.2.4. Regression Models
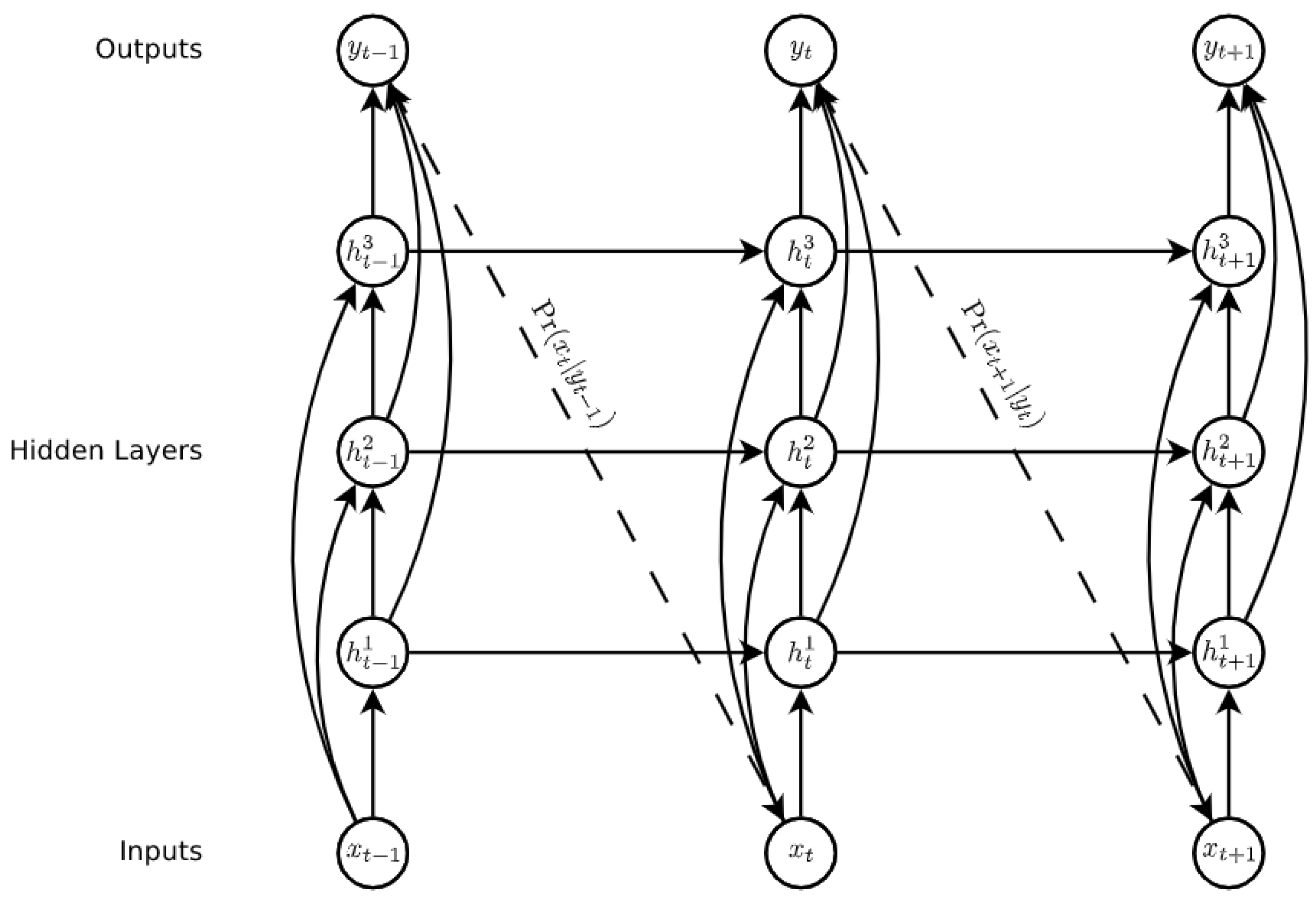
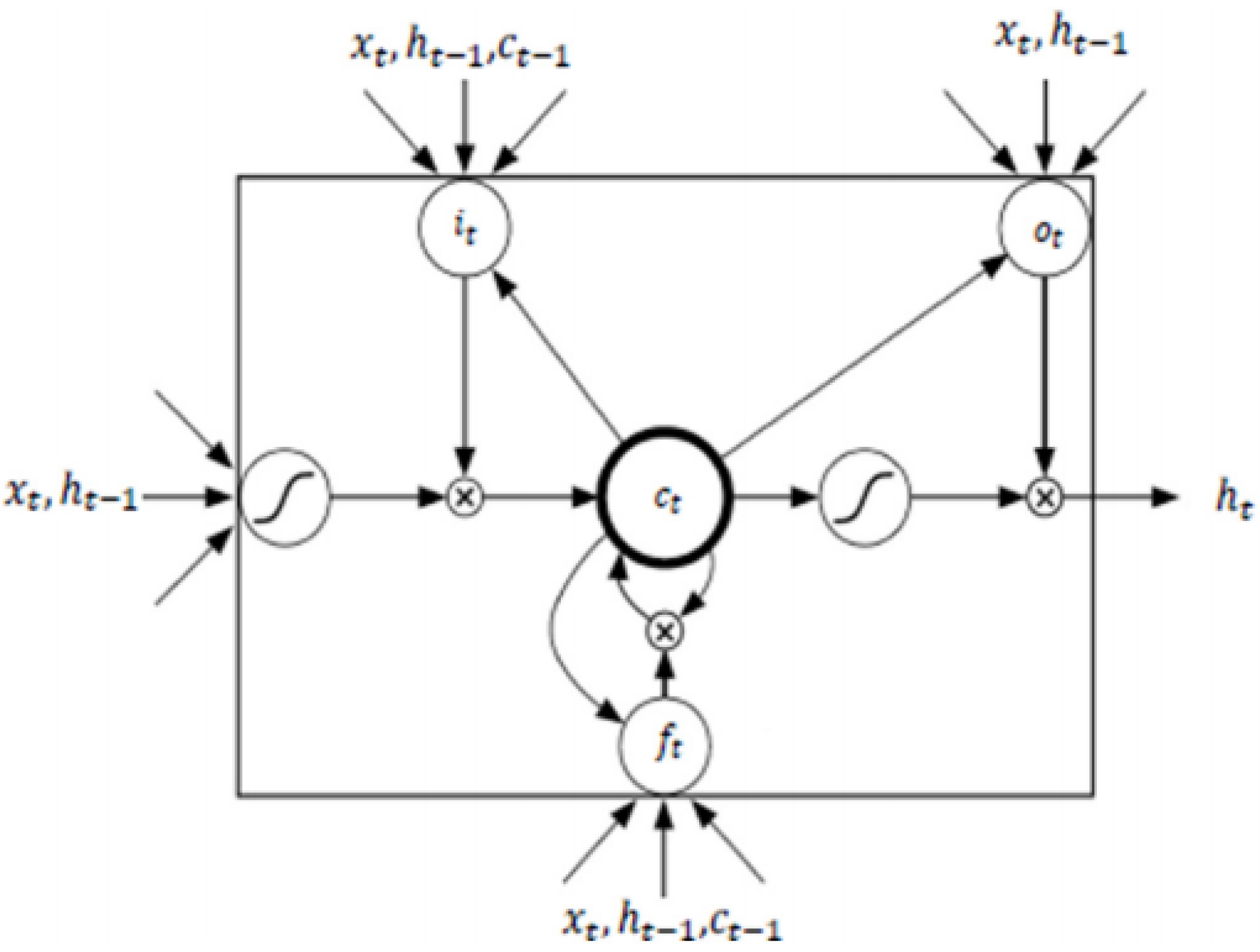
- Recurrent Neural Networks and Long-Short-Term Memory model: Recurrent Neural Networks (RNN) are a family of neural networks used to process sequential data [27], which are well-known and widely used to process time series data and natural language processing [40,41]. These networks are built upon the idea of using the output of the previous neuron in the network along with the next input of the sequence as input to the next. This ability gives the network the opportunity to model sequences. It facilitates modelling cases in which the relationships between variables are not simply parallel, but rather sequential (the value of a given variable at one time may determine the value of another at a later or earlier time). Sequential data can be trained as: complete sequences, forward or backward sequences, or a set of them.Figure 4 illustrates the basic architecture of a recurrent neural network. Given an input vector sequence , passed through to bunch of N recurrently connected hidden layers. The first hidden vector sequences are calculated as and the output vector sequence . Where N = 1, the architecture is simply reduced to a single layer. Hidden layer connections are calculated as follows:where H corresponds to the hidden layer functions, W equals the weight matrices (e.g., is the recurrent connection at the first hidden layer), b denotes bias vectors (e.g., is the output bias vector). Then the output is computed by:LSTM is one of the most famous types of RNN architecture [43]. It can memorize for long and short periods of time using a gating mechanism which makes it possible to control the information that has to be kept over time, the duration it has to be kept for and the time that it can be read through the memory cell [44]. The architecture of an LSTM cell, as described in [42], is shown in Figure 5, where is implemented by the following composite function:where is the logistic function , , , , correspond to the input gate, forget gate, output gate, memory cell and hidden state at time t respectively, and refers to the input of the system at time t.
- Support vector machine (SVM): this is an algorithm based on statistical learning and which has gained great popularity over the last decade. It is useful in several classification and regression problems [36,45,46]. SVM takes the structural risk minimization principle into account and attempts to find the locations of decision boundaries (also known as hyperplanes), which produce optimal separation among the classes [47,48].This paper used a support vector regression (SVR), which refers to a generalization of classification problems, where the model returns continuous values. SVM generalization to SVR is achieved by introducing an -insensitive region around the function, referred to as the -tube. The tube then reformulates the optimization problem in order to find a tube value which best fits the function, while balancing model complexity and prediction error [49]. SVR problem formulation derives from a geometrical representation, and its continuous-value functions could be approximately represented by:where w and b correspond to the weight and bias vectors, respectively. In spite of this, and in real applications, data tends to be non-linear and separable, and Kernel functions are therefore used to extend the concept of the optimal hyperplane.In multidimensional data, x augments by one, and b is included in the w vector for a simple mathematical notation (see Equation (17)). The multivariate regression in SVR then formulates the function approximation problem as an optimization which attempts to find the narrowest tube centred around the surface [49]. The objective function is shown below in Equation (18), where w equals the magnitude of the normal vector to the surface.The Grid search method was used to tune the hyperparameters, whereby three different kernel functions (i.e., radial basis function, polynomial kernel and sigmoid kernel) were used. These three kernel methods are defined by the Equations (19)–(21), respectively.where is the influence on classification outcomes; large values for leads to over-fitting and small values result in under-fitting [50].
2.2.5. Ensemble Models
2.2.6. Output
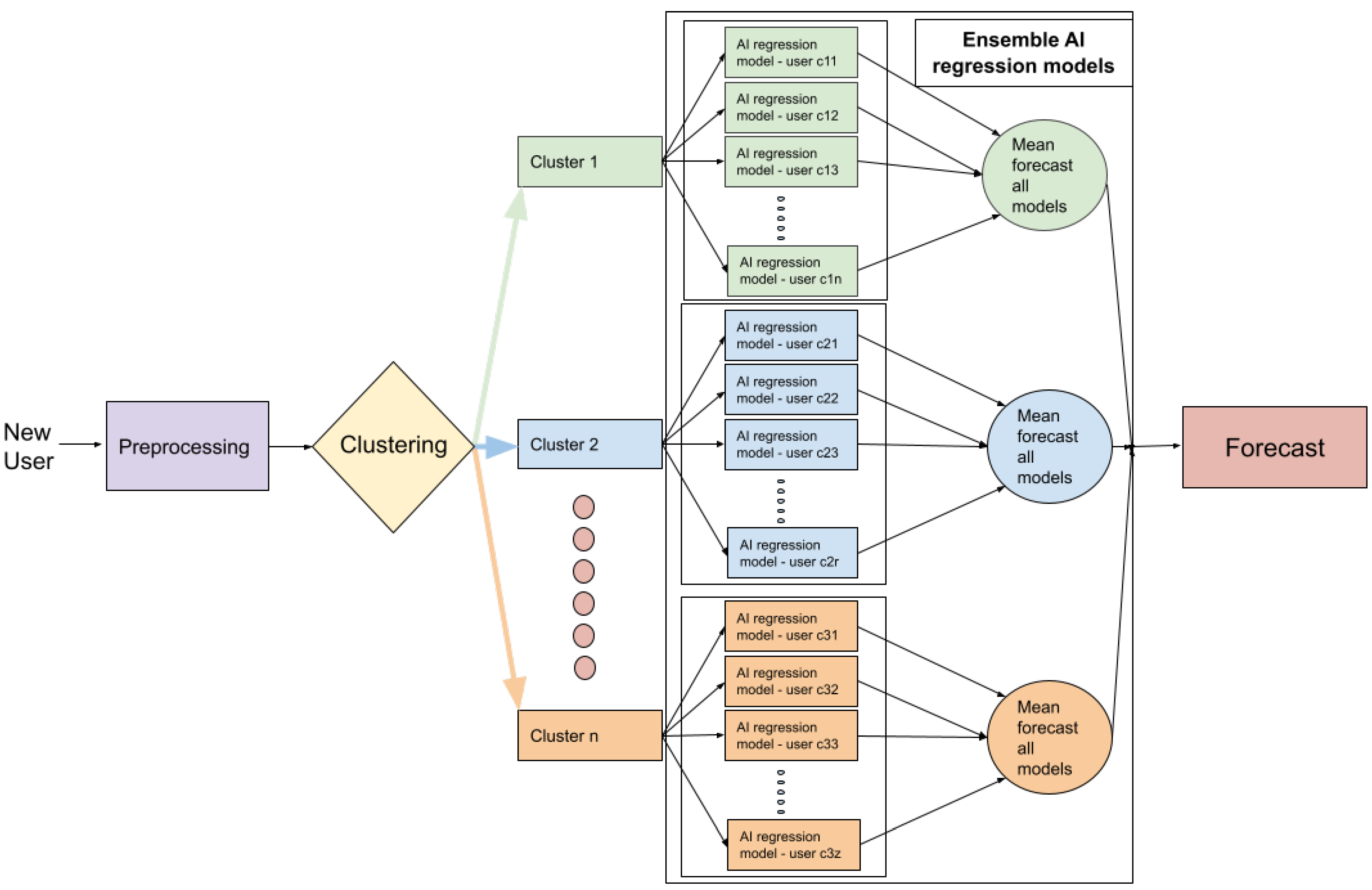
- Step 1 (Input data): Raw data from the three different users is given as input to the system. Users can belong to one of the three aforementioned categories (depicted in different colours).
- Step 2 (Pre-processing): In the pre-possessing step, all data cleaning procedures, as well as other operations, are applied in order to obtain the data from workout sessions.
- Step 3 (Clustering): When using the K-means algorithm, if three clusters are selected and the characteristics are the mean of accumulated seconds per day over three months, clusters categorize the users into three groups: people with high PA (orange colour), people with medium PA (blue colour), and people with low PA (green colour).
- Step 4 (Ensemble models): Assuming we are using the LSTM as the regression method, data corresponding to the first three months of use is given to the corresponding LSTM ensembles (orange, blue or green). The ensembles then use pre-trained models to calculate all regressions and the output will be the mean of the corresponding ensemble; Ē1: mean ensemble 1 (green), Ē2: mean ensemble 2 (blue), Ē3: mean ensemble 3 (orange).
- The system output corresponds to the average regression of the models within a given cluster. Since our aim was to determine adherence to training using a fitness app, we turned again to literature in order to follow a rule that defined user adherence. Previous researchers have established that exercise-derived health benefits taper off after 4–5 weeks of training cessation [53,54,55,56,57]. Taking this into account, we determined that a user would be considered non-adherent if he/she showed no training activity over a full month (the fourth month).
3. Experiments and Results
3.1. Implementation
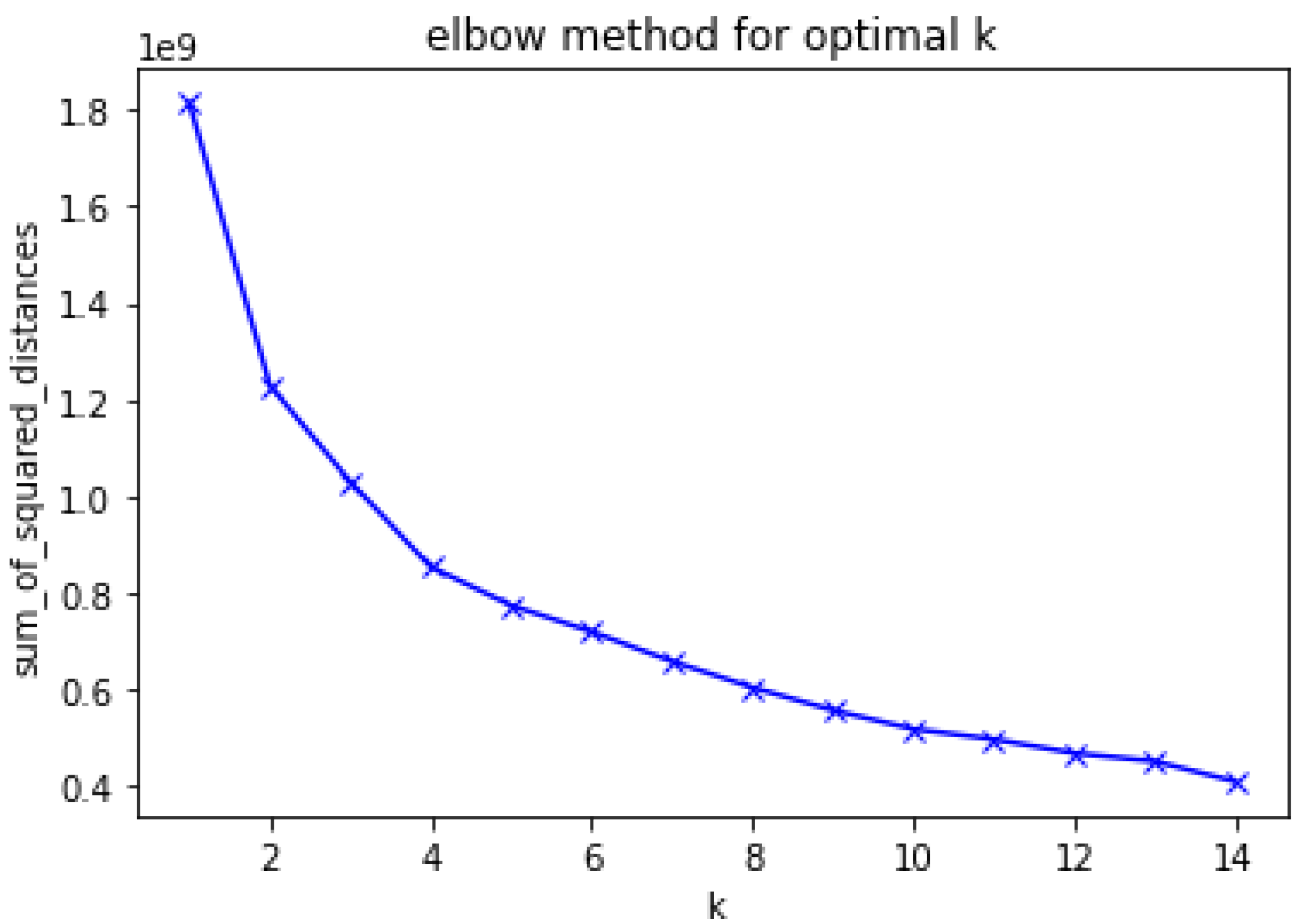
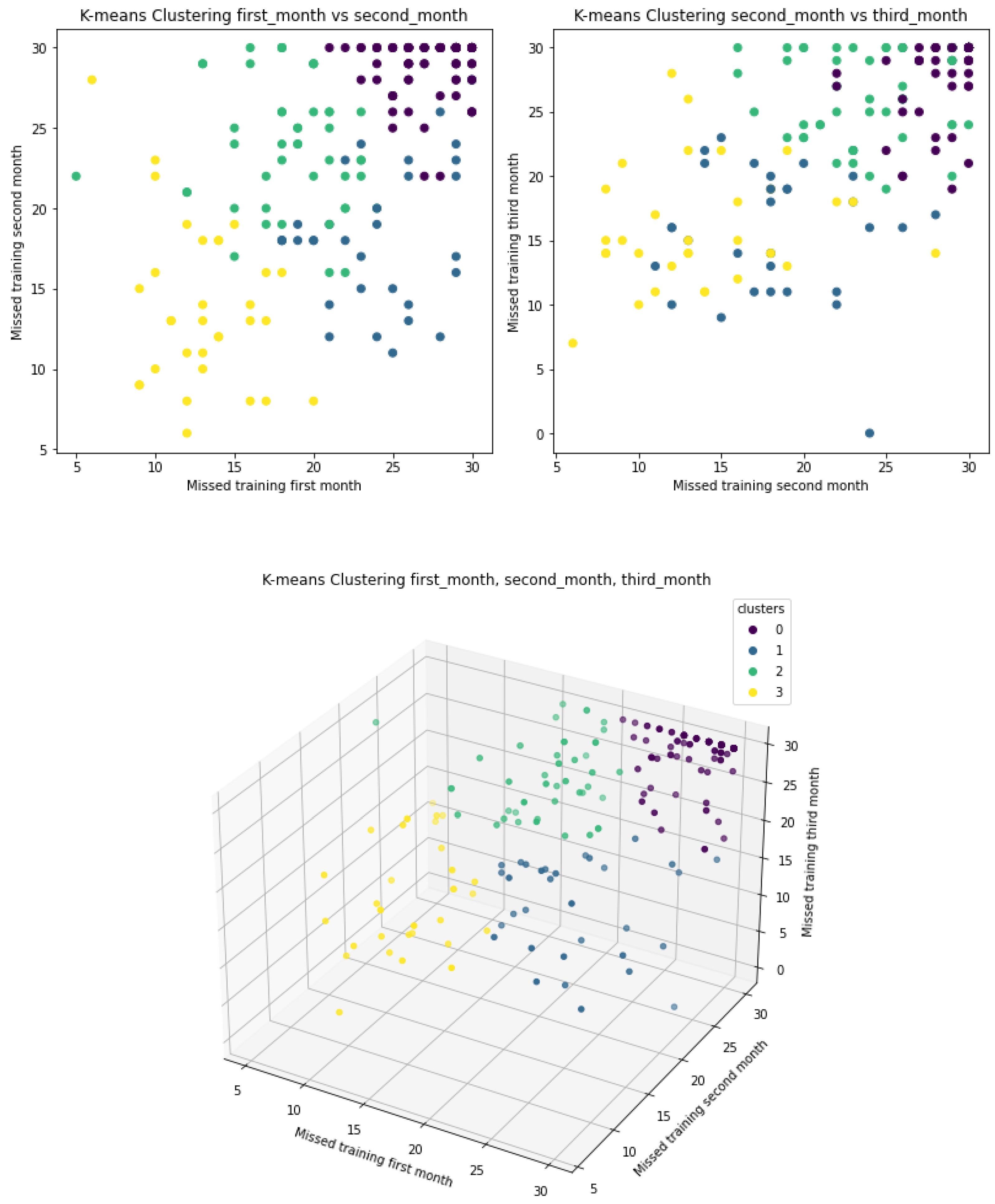
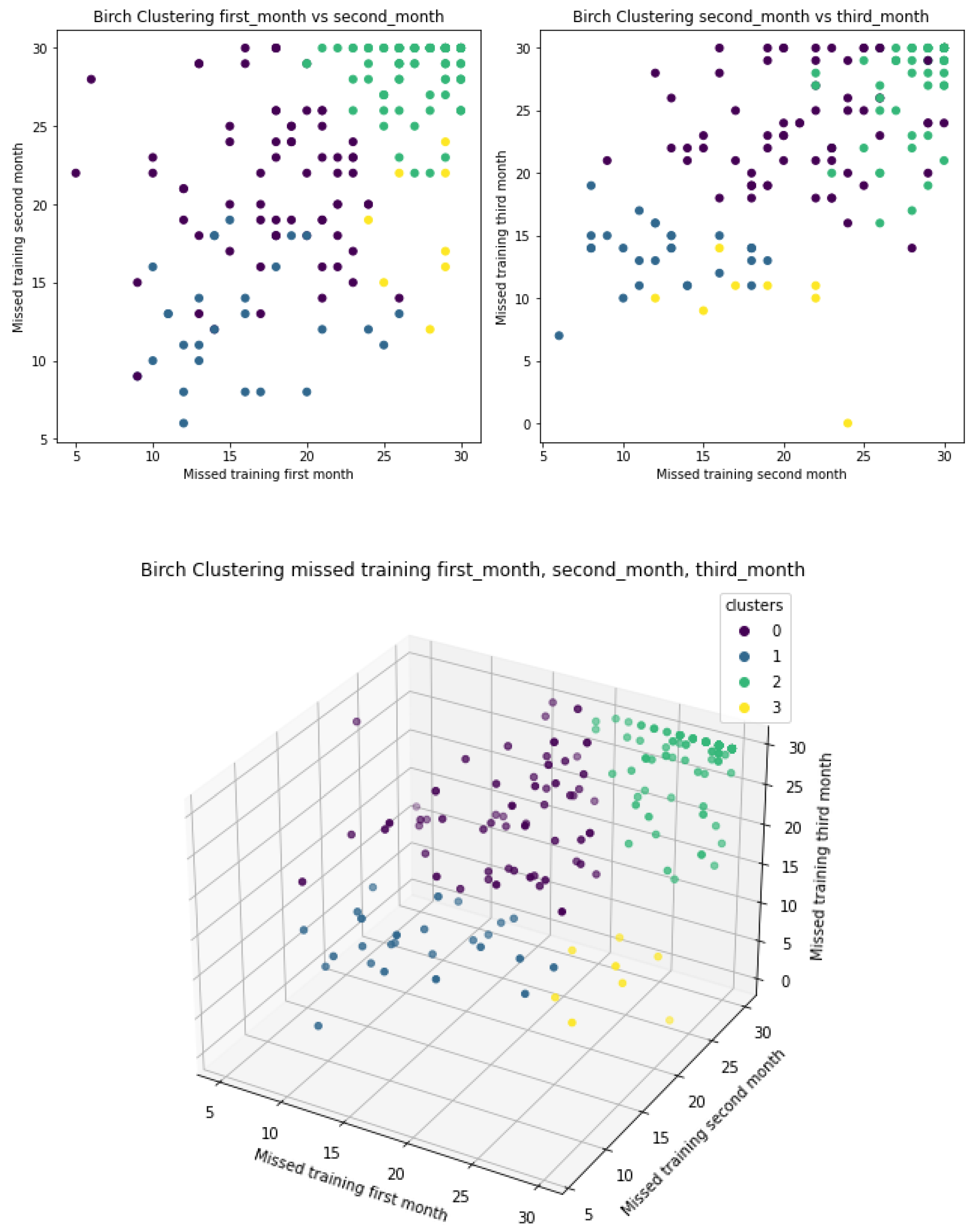
3.2. Clustering
3.2.1. K-Means
3.2.2. BIRCH
3.2.3. Affinity Propagation
3.3. Regression Results
Hyperparameter Tuning
- LSTM—grid search: following a large number of tests with different users, a wide range of hyperparameters was chosen for which the models generally adjusted the regression curve better. The same hyperparameters and number of neurons were then used for all users. Specifically, three dropout values after the first and second dense layers were applied. Similarly, three batch sizes of values 1,2,4 were used, taking into consideration the number of days in a week. Finally, five neuron values were applied to the first layer (LSTM layer). The range of aforementioned values are highlighted in Table 12.Remaining hyperparameters were selected from existing literature (previous work on prediction, even if aimed at different types of application), with high performance in their proposed architectures. Hence, in accordance with the previous explanation, we pursued a hyperparameter tuning strategy in the relevant literature [22,65], with a detailed explanation of all the values in Table 12. The lookback is a parameter which was selected and agreed with the MH team, as it was considered more appropriate for the purpose of analysing the evolution of training over the weeks, as people generally change their routines or lose their motivation within a period of one week [66]. Additionally, following some experiments, we also verified that curves were fitting better with a value of 7 days. The number of epochs was then selected to be 50 after observing in experiments that overfitting was occurring after 50 epochs. Next, based on [67], we selected the number of hidden layers to be 4 after performing several experiments. The activation function selected was Relu, since it resolved the problem of negative values, and had performed well in previous research [68]. The Relu activation function was applied to all layers (including LSTM and dense), except the final one, while early stopping with patience of 15 epochs, was configured in our architecture, in order to avoid over-fitting.
- SVR-grid search: The hyperparameters modified in the case of SVR were kernel, with the choices ‘poly’, ‘rbf’, and ’sigmoid’. Similarly, for the hyperparameter c, a range of 0 to 500 was chosen since the MAE error was not reduced beyond this number with any combination of kernel or other hyperparameters setup. Finally, the hyperparameter gamma, which assigns the scale option, and epsilon, which has a value of 10, were left unchanged.
3.4. Classification Results
3.4.1. Validation Metrics
- True Positives (TP): Users who were correctly predicted to exercise in the fourth month.
- True Negatives (TN): Users who were correctly predicted to not exercise in the fourth month.
- False Positives (FP): Users who were predicted to exercise but actually didn’t exercise in the fourth month.
- False Negatives (FN): Users who were predicted to not exercise in the fourth month, but who actually did.
3.4.2. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Affinity Propagation |
| BIRCH | Balanced Iterative Reducing and Clustering using Hierarchies |
| CF | Cluster Feature |
| DL | Deep Learning |
| FN | False negatives |
| FP | False positives |
| LSTM | Long Short Term Memory |
| MAE | Mean Absolute Error |
| MH | Mammoth Hunters |
| ML | Machine learning |
| PA | Physical activity |
| RNN | Recurrent Neural Networks |
| SSE | Sum of Squared Errors |
| SVM | Support vector machine |
| SVR | Support vector regression |
| TN | True negatives |
| TP | True positives |
| WHO | World Health Organization |
References
- Hall, G.; Laddu, D.R.; Phillips, S.A.; Lavie, C.J.; Arena, R. A tale of two pandemics: How will COVID-19 and global trends in physical inactivity and sedentary behavior affect one another? Prog. Cardiovasc. Dis. 2021, 64, 108. [Google Scholar] [CrossRef]
- Physical Activity. WHO Int. Available online: https://www.who.int/news-room/fact-sheets/detail/physical-activity (accessed on 26 November 2020).
- Booth, F.W.; Roberts, C.K.; Thyfault, J.P.; Ruegsegger, G.N.; Toedebusch, R.G. Role of inactivity in chronic diseases: Evolutionary insight and pathophysiological mechanisms. Physiol. Rev. 2017, 97, 1351–1402. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Action Plan on Physical Activity 2018–2030: More Active People for a Healthier World: At-a-Glance; Technical Report; World Health Organization: Geneva, Swizerland, 2018. [Google Scholar]
- Ding, D.; Lawson, K.D.; Kolbe-Alexander, T.L.; Finkelstein, E.A.; Katzmarzyk, P.T.; Van Mechelen, W.; Pratt, M.; Lancet Physical Activity Series 2 Executive Committee. The economic burden of physical inactivity: A global analysis of major non-communicable diseases. Lancet 2016, 388, 1311–1324. [Google Scholar] [CrossRef]
- Jason, A.; Bennie, K.D.C.; Tittlbach, S. The epidemiology of muscle-strengthening and aerobic physical activity guideline adherence among 24,016 German adults. Scand. J. Med. Sci. Sport. 2021, 31, 1096–1104. [Google Scholar] [CrossRef]
- Bull, F.; Saad, S.A.-A.; Biddle, S. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 2020, 54. Available online: https://bjsm.bmj.com/content/54/24/1451 (accessed on 26 November 2020). [CrossRef]
- Ekkekakis, P.; Vazou, S.; Bixby, W.; Georgiadis, E. The mysterious case of the public health guideline that is (almost) entirely ignored: Call for a research agenda on the causes of the extreme avoidance of physical activity in obesity. Obes. Rev. 2016, 17, 313–329. [Google Scholar] [CrossRef]
- Van Tuyckom, C.; Scheerder, J.; Bracke, P. Gender and age inequalities in regular sports participation: A cross-national study of 25 European countries. J. Sport Sci. 2010, 28, 1077–1084. [Google Scholar] [CrossRef]
- Direito, A.; Jiang, Y.; Whittaker, R.; Maddison, R. Smartphone apps to improve fitness and increase physical activity among young people: Protocol of the Apps for IMproving FITness (AIMFIT) randomized controlled trial. BMC Public Health 2015, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Ma, L.; Zhao, X.; Kankanhalli, A. Factors influencing user’s adherence to physical activity applications: A scoping literature review and future directions. Int. J. Med. Inform. 2020, 134, 104039. [Google Scholar] [CrossRef] [PubMed]
- Sieverink, F.; Kelders, S.M.; van Gemert-Pijnen, J.E. Clarifying the concept of adherence to eHealth technology: Systematic review on when usage becomes adherence. J. Med. Internet Res. 2017, 19, e8578. [Google Scholar] [CrossRef]
- Bailey, D.L.; Holden, M.A.; Foster, N.E.; Quicke, J.G.; Haywood, K.L.; Bishop, A. Defining adherence to therapeutic exercise for musculoskeletal pain: A systematic review. Br. J. Sport Med. 2020, 54, 326–331. [Google Scholar]
- Guertler, D.; Vandelanotte, C.; Kirwan, M.; Duncan, M.J. Engagement and nonusage attrition with a free physical activity promotion program: The case of 10,000 steps Australia. J. Med. Internet Res. 2015, 17, e4339. [Google Scholar] [CrossRef] [Green Version]
- Cugelman, B.; Thelwall, M.; Dawes, P. Online interventions for social marketing health behavior change campaigns: A meta-analysis of psychological architectures and adherence factors. J. Med. Internet Res. 2011, 13, e17. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Venkatakrishnan, A.; Youngblood, G.M.; Ram, A.; Pirolli, P. A group-based mobile application to increase adherence in exercise and nutrition programs: A factorial design feasibility study. JMIR MHealth UHealth 2016, 4, e4900. [Google Scholar] [CrossRef] [Green Version]
- Pratap, A.; Neto, E.C.; Snyder, P.; Stepnowsky, C.; Elhadad, N.; Grant, D.; Mohebbi, M.H.; Mooney, S.; Suver, C.; Wilbanks, J.; et al. Indicators of retention in remote digital health studies: A cross-study evaluation of 100,000 participants. NPJ Digit. Med. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.A.; Adjeroh, D.A. Deep learning using convolutional LSTM estimates biological age from physical activity. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Kassabi, H.T.; Khalil, K.; Serhani, M.A. Deep Learning Approach for Forecasting Athletes’ Performance in Sports Tournaments. In Proceedings of the 13th International Conference on Intelligent Systems: Theories and Applications, Rabat, Morocco, 23–24 September 2020; pp. 1–6. [Google Scholar]
- Zebin, T.; Sperrin, M.; Peek, N.; Casson, A.J. Human activity recognition from inertial sensor time-series using batch normalized deep LSTM recurrent networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 1–4. [Google Scholar]
- Kim, J.C.; Chung, K. Prediction model of user physical activity using data characteristics-based long short-term memory recurrent neural networks. KSII Trans. Internet Inf. Syst. (TIIS) 2019, 13, 2060–2077. [Google Scholar]
- Zahia, S.; Garcia-Zapirain, B.; Saralegui, I.; Fernandez-Ruanova, B. Dyslexia detection using 3D convolutional neural networks and functional magnetic resonance imaging. Comput. Methods Programs Biomed. 2020, 197, 105726. [Google Scholar] [CrossRef]
- Hameed, Z.; Garcia-Zapirain, B. Sentiment classification using a single-layered BiLSTM model. IEEE Access 2020, 8, 73992–74001. [Google Scholar] [CrossRef]
- Zahia, S.; Zapirain, M.B.G.; Sevillano, X.; González, A.; Kim, P.J.; Elmaghraby, A. Pressure injury image analysis with machine learning techniques: A systematic review on previous and possible future methods. Artif. Intell. Med. 2020, 102, 101742. [Google Scholar] [CrossRef]
- Acosta, M.F.J.; Tovar, L.Y.C.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 1–11. [Google Scholar]
- Rodríguez-Esparza, E.; Zanella-Calzada, L.A.; Oliva, D.; Pérez-Cisneros, M. Automatic detection and classification of abnormal tissues on digital mammograms based on a bag-of-visual-words approach. Med. Imaging 2020 Comput.-Aided Diagn. Int. Soc. Opt. Photonics 2020, 11314, 1131424. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Campos, L.C.; Campos, F.A.; Bezerra, T.A.; Pellegrinotti, Í.L. Effects of 12 weeks of physical training on body composition and physical fitness in military recruits. Int. J. Exerc. Sci. 2017, 10, 560. [Google Scholar] [PubMed]
- Hurst, C.; Weston, K.L.; Weston, M. The effect of 12 weeks of combined upper-and lower-body high-intensity interval training on muscular and cardiorespiratory fitness in older adults. Aging Clin. Exp. Res. 2019, 31, 661–671. [Google Scholar] [CrossRef] [Green Version]
- Oertzen-Hagemann, V.; Kirmse, M.; Eggers, B.; Pfeiffer, K.; Marcus, K.; de Marées, M.; Platen, P. Effects of 12 weeks of hypertrophy resistance exercise training combined with collagen peptide supplementation on the skeletal muscle proteome in recreationally active men. Nutrients 2019, 11, 1072. [Google Scholar] [CrossRef] [Green Version]
- Barranco-Ruiz, Y.; Villa-González, E. Health-related physical fitness benefits in sedentary women employees after an exercise intervention with Zumba Fitness®. Int. J. Environ. Res. Public Health 2020, 17, 2632. [Google Scholar] [CrossRef] [Green Version]
- Feito, Y.; Hoffstetter, W.; Serafini, P.; Mangine, G. Changes in body composition, bone metabolism, strength, and skill-specific performance resulting from 16-weeks of HIFT. PLoS ONE 2018, 13, e0198324. [Google Scholar] [CrossRef]
- Brownlee, J. Data Preparation for Machine Learning: Data Cleaning, Feature Selection, and Data Transforms in Python; Machine Learning Mastery, 2020. Available online: https://pubmed.ncbi.nlm.nih.gov/26806460/ (accessed on 26 November 2020).
- Li, B.; Liu, B.; Lin, W.; Zhang, Y. Performance analysis of clustering algorithm under two kinds of big data architecture. J. High Speed Netw. 2017, 23, 49–57. [Google Scholar] [CrossRef]
- Tan, P.N.; Steinbach, M.; Karpatne, A.; Kumar, V. Introduction to Data Mining, 2nd ed.; Pearson: London, UK, 2018. [Google Scholar]
- Chaves, A.; Jossa, O.; Jojoa, M. Classification of Hosts in a WLAN Based on Support Vector Machine. In Proceedings of the 2018 Congreso Internacional de Innovación y Tendencias en Ingeniería (CONIITI), Bogotá, Colombia, 3–5 October 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Larose, D. Data Mining and Predictive Analytics; Wiley Series on Methods and Applications in Data Mining; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [Green Version]
- Thavikulwat, P. Affinity propagation: A clustering algorithm for computer-assisted business simulations and experiential exercises. Dev. Bus. Simul. Exp. Learn. 2008, 35, 220–224. [Google Scholar]
- Basiri, M.E.; Nemati, S.; Abdar, M.; Cambria, E.; Acharya, U.R. ABCDM: An attention-based bidirectional CNN-RNN deep model for sentiment analysis. Future Gener. Comput. Syst. 2021, 115, 279–294. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, C.; Yang, L.; Chen, Y. DSTP-RNN: A dual-stage two-phase attention-based recurrent neural network for long-term and multivariate time series prediction. Expert Syst. Appl. 2020, 143, 113082. [Google Scholar] [CrossRef]
- Graves, A. Generating sequences with recurrent neural networks. arXiv 2013, arXiv:1308.0850. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Karevan, Z.; Suykens, J.A. Transductive LSTM for time-series prediction: An application to weather forecasting. Neural Netw. 2020, 125, 1–9. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Jojoa, M.; Garcia-Zapirain, B. Forecasting COVID 19 Confirmed Cases Using Machine Learning: The Case of America. Preprints 2020, 2020090228. [Google Scholar] [CrossRef]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support vector regression. In Efficient Learning Machines; Apress: Berkeley, CA, USA, 2015; pp. 67–80. [Google Scholar]
- Rahmadani, F.; Lee, H. ODE-based epidemic network simulation of viral Hepatitis A and kernel support vector machine based vaccination effect analysis. J. Korean Inst. Intell. Syst. 2020, 30, 106–112. [Google Scholar] [CrossRef]
- Opitz, D.; Maclin, R. Popular ensemble methods: An empirical study. J. Artif. Intell. Res. 1999, 11, 169–198. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Mujika, I.; Padilla, S. Detraining: Loss of training-induced physiological and performance adaptations. Part I. Sports Med. 2000, 30, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.C.; Neiva, H.P.; Izquierdo, M.; Cadore, E.L.; Alves, A.R.; Marinho, D.A. Concurrent training and detraining: Brief review on the effect of exercise intensities. Int. J. Sports Med. 2019, 40, 747–755. [Google Scholar] [CrossRef]
- Maldonado-Martín, S.; Cámara, J.; James, D.V.; Fernández-López, J.R.; Artetxe-Gezuraga, X. Effects of long-term training cessation in young top-level road cyclists. J. Sport Sci. 2017, 35, 1396–1401. [Google Scholar] [CrossRef]
- Sousa, A.C.; Marinho, D.A.; Gil, M.H.; Izquierdo, M.; Rodríguez-Rosell, D.; Neiva, H.P.; Marques, M.C. Concurrent training followed by detraining: Does the resistance training intensity matter? J. Strength Cond. Res. 2018, 32, 632–642. [Google Scholar] [CrossRef]
- Zacca, R.; Toubekis, A.; Freitas, L.; Silva, A.F.; Azevedo, R.; Vilas-Boas, J.P.; Pyne, D.B.; Castro, F.A.D.S.; Fernandes, R.J. Effects of detraining in age-group swimmers performance, energetics and kinematics. J. Sports Sci. 2019, 37, 1490–1498. [Google Scholar] [CrossRef]
- Buitinck, L.; Louppe, G.; Blondel, M.; Pedregosa, F.; Mueller, A.; Grisel, O.; Niculae, V.; Prettenhofer, P.; Gramfort, A.; Grobler, J.; et al. API design for machine learning software: Experiences from the scikit-learn project. arXiv 2013, arXiv:1309.0238. [Google Scholar]
- Nasir, V.; Sassani, F. A review on deep learning in machining and tool monitoring: Methods, opportunities, and challenges. Int. J. Adv. Manuf. Technol. 2021, 115, 2683–2709. [Google Scholar] [CrossRef]
- Victoria, A.H.; Maragatham, G. Automatic tuning of hyperparameters using Bayesian optimization. Evol. Syst. 2021, 12, 217–223. [Google Scholar] [CrossRef]
- Wu, J.; Toscano-Palmerin, S.; Frazier, P.I.; Wilson, A.G. Practical multi-fidelity Bayesian optimization for hyperparameter tuning. In Proceedings of the 35th Uncertainty in Artificial Intelligence Conference, PMLR, Tel Aviv, Israel, 22–25 July 2020; Volume 115, pp. 788–798. [Google Scholar]
- Koller, D.; Friedman, N. Probabilistic Graphical Models: Principles and Techniques; MIT Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Caballero, L.; Jojoa, M.; Percybrooks, W.S. Optimized neural networks in industrial data analysis. SN Appl. Sci. 2020, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Amirabadi, M.A.; Kahaei, M.H.; Nezamalhosseini, S.A. Novel suboptimal approaches for hyperparameter tuning of deep neural network [under the shelf of optical communication]. Phys. Commun. 2020, 41, 101057. [Google Scholar] [CrossRef]
- Hameed, Z.; Zahia, S.; Garcia-Zapirain, B.; Javier Aguirre, J.; María Vanegas, A. Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors 2020, 20, 4373. [Google Scholar] [CrossRef]
- Pilloni, P.; Piras, L.; Carta, S.; Fenu, G.; Mulas, F.; Boratto, L. Recommender system lets coaches identify and help athletes who begin losing motivation. Computer 2018, 51, 36–42. [Google Scholar] [CrossRef]
- Sunny, M.A.I.; Maswood, M.M.S.; Alharbi, A.G. Deep Learning-Based Stock Price Prediction Using LSTM and Bi-Directional LSTM Model. In Proceedings of the 2020 2nd Novel Intelligent and Leading Emerging Sciences Conference (NILES), Giza, Egypt, 24–26 October 2020; pp. 87–92. [Google Scholar]
- Gokulan, S.; Narmadha, S.; Pavithra, M.; Rajmohan, R.; Ananthkumar, T. Determination of Various Deep Learning Parameter for Sleep Disorder. In Proceedings of the 2020 International Conference on System, Computation, Automation and Networking (ICSCAN), Puducherry, India, 3–4 July 2020; pp. 1–6. [Google Scholar]
- Basso, J.C.; Suzuki, W.A. The effects of acute exercise on mood, cognition, neurophysiology, and neurochemical pathways: A review. Brain Plast. 2017, 2, 127–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.H.; Emerson, J.A.; Williams, D.M. The exercise–affect–adherence pathway: An evolutionary perspective. Front. Psychol. 2016, 7, 1285. [Google Scholar] [CrossRef] [PubMed]
- Milne-Ives, M.; Lam, C.; De Cock, C.; Van Velthoven, M.H.; Meinert, E. Mobile apps for health behavior change in physical activity, diet, drug and alcohol use, and mental health: Systematic review. JMIR MHealth UHealth 2020, 8, e17046. [Google Scholar] [CrossRef]
- Lasi, H.; Fettke, P.; Kemper, H.G.; Feld, T.; Hoffmann, M. Industry 4.0. Bus. Inf. Syst. Eng. 2014, 6, 239–242. [Google Scholar] [CrossRef]




| Personal Data | |
|---|---|
| Age | Date of Birth |
| Gender | Male or Female |
| city | City and country |
| Fitness data | |
| Weight | Kg |
| Height | Cm |
| Body fat | Individual body fat estimation (%) |
| Body type | Thin, normal or overweight |
| User goals | |
| Body fat goal | Desired body fat goal (%) |
| User goal | Lose weight Stay healthy Gain strength |
| Information application data | |
| Profile creation | Date |
| App downloads | Date/s and number of downloads |
| App visits | Number of visits (total, per day, per week, per month) |
| Training plan purchased | Plan type |
| Training programme used | Programme type |
| Total sessions performed | Number of sessions Degree of difficulty of the sessions |
| Total sessions completed | Number of sessions |
| Completed sessions | Type of the most completed session Type of the most discarded session Number of sessions per week Date and hour of session completion Duration Finished YES/NO |
| Features | Age | Weight (Kgs) | Height (cm) | BMI | Body Fat | Objective of Body Fat |
|---|---|---|---|---|---|---|
| Mean | 40 | 71.4 | 170.6 | 24.5 | 24.1 | 13.0 |
| Standard Deviation | 8 | 13.7 | 9.3 | 4.0 | 8.5 | 3.5 |
| min | 19 | 37.0 | 150.0 | 16.2 | 6.0 | 5.0 |
| 25% | 34 | 62.0 | 164.0 | 22.0 | 20.0 | 10.0 |
| 50% | 41 | 70.0 | 170.0 | 23.8 | 25.0 | 15.0 |
| 75% | 47 | 79.9 | 177.5 | 25.9 | 30.0 | 15.0 |
| Max | 66 | 129.0 | 201.0 | 50.4 | 60.0 | 25.0 |
| Features | First Month | Second Month | Third Month | Fourth Month |
|---|---|---|---|---|
| Mean [s] | 398.95 | 393.74 | 377.83 | 318.73 |
| Standard Deviation [s] | 594.89 | 632.84 | 631.76 | 620.22 |
| min [s] | 0.00 | 0.00 | 0.00 | 0.00 |
| 25% | 22.03 | 0.00 | 0.00 | 0.00 |
| 50% | 161.38 | 60.45 | 14.23 | 0.00 |
| 75% | 514.50 | 604.14 | 553.71 | 466.95 |
| Max [s] | 4106.47 | 3643.43 | 3522.57 | 4524.93 |
| Features | Number of Users First Month | Number of Users Second Month | Number of Users Third Month | Number of Users Fourth Month |
|---|---|---|---|---|
| Time = 0 s | 41 | 106 | 121 | 132 |
| Time = [0 s–300 s] | 102 | 49 | 40 | 45 |
| Time = [300 s–1800 s] | 96 | 81 | 74 | 61 |
| Time = [1800 s–3600 s] | 4 | 9 | 11 | 6 |
| Time = [3600 s–7200 s] | 3 | 1 | 0 | 2 |
| 1. Prepare the data: pre-processing, cleaning, scaling, splitting |
| 2. Train clustering algorithms with all users with workout sessions over first three months |
| 3. For each user in all users: |
| 3.1. Configure architecture and set parameters to tune |
| 3.2. For each parameters combination in the grid: |
| 3.2.1. Train regression algorithms (LSTM, SVR) |
| 3.2.2. Select the best model based on Mean absolute error (MAE) |
| 3.2.3. Save the best model |
| 4. Build ensemble of clusters with the best regression algorithms for each user, and with the corresponding clusters built in step 2 |
| 1. Prepare new user data: pre-processing, cleaning, scaling, splitting |
| 2. Obtain user features from workouts sessions of first three months |
| 3. Select the corresponding ensemble of models according to clustering result |
| 4. For each model in ensemble: |
| 4.1. Predict data of fourth month with the trained models |
| 4.2. Save prediction |
| 5. Obtain the result of workout in fourth month by calculating the mean of all predictions |
| 6. Apply rule to determine adherence |
| 7. Classify user adherence to MH fitness app |
| Value | Description | |
|---|---|---|
| Total users | 246 | Total number of users, with at least 4 months of data |
| Longitudinal Period selected | 120 [days] | Longitudinal period of training sessions based on state of the art |
| Days selected for training | 90 [days] | Days selected to train the artificial intelligence system |
| Days selected for testing | 30 [days] | Days selected to test the artificial intelligence system |
| Scores | Sessions [s] | Array with training session data for users over 4 months |
| Output class 1 | 112 | Users who are adherent during the fourth month |
| Output class 0 | 134 | Users who are not adherent during the fourth month |
| 1. Select K points as initial centroids |
| 2. while |
| 2.1. K clusters by assigning each point to its closest centroid |
| 2.2. Recompute the centroid of each cluster |
| 3. until centroids do not change |
| 1. For each record in set of elements D: |
| 1.1. Determine correct leaf node for insertion |
| 1.2. If threshold condition is not violated then: |
| 1.2.1. Add to cluster and update CF |
| 1.3. else if threshold condition is violated: |
| 1.3.1. Insert as single cluster and update CF |
| 2. Apply an existing clustering algorithm to the sub-clusters, with a view to combining these sub-clusters into clusters |
| 1. Set “availabilities” to zero i.e., ∀ i,k: a(i,k) = 0 |
| 1.2. While responsibility and availability matrices are updated until they converge: |
| 1.3. Calculate similarity matrix |
| 1.4. Calculate responsibility matrix |
| 1.5. Calculate availability matrix |
| 2. Cluster assignments corresponding to the highest criterion values of each row is designated as the exemplar i.e., argmax_k [a(i,k) + r(i,k)] |
| Variable | Description |
|---|---|
| mean_first_month | Mean of completed sessions in the first month of training (seconds) |
| mean_second_month | Mean of completed sessions in the second month of training (seconds) |
| mean_third_month | Mean of completed sessions in the three month of training (seconds) |
| missed_first_month | The number of skipped sessions in the first month |
| missed_second_month | The number of skipped sessions in the second month |
| missed_third_month | The number of skipped sessions in the third month |
| mean_week_1 | Mean of completed sessions in the first month of training (seconds) |
| mean_week_2 | Mean of completed sessions in the first week of training (seconds) |
| mean_week_3 | Mean of completed sessions in the second week of training (seconds) |
| ... | ... |
| mean_week_12 | Mean of completed sessions in the 12th month of training (seconds) |
| Hyperparameters | Values |
|---|---|
| lookback | 7 |
| Number of epochs | 50 |
| Number of hidden layers | 4 |
| Number of LSTM layers | 1 |
| Number of dense layers | 3 |
| Activation function | relu |
| Optimizer | adam |
| Loss | mse |
| Early stopping | Patience:15 Monitor: loss |
| Dropout | [ 0.2, 0.4, 0.6] |
| Batch size | [1, 2, 4] |
| Number of Neurons | [50, 75, 100, 125, 150] |
| Accuracy | Recall | Precision | Specificity | F1_score |
|---|---|---|---|---|
| 0.7276 | 0.4196 | 0.9592 | 0.9851 | 0.5839 |
| Features Selected | |||||
|---|---|---|---|---|---|
| Parameters | Metrics | mean_fm, mean_sm, mean_tm | missed_first_month, missed_second_month, missed_third_month | week_0, week_1, week_2, ..., week_11 | missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9 week10, week 11 |
| k = 3 | Accuracy | 0.7073 | 0.7764 | 0.7398 | 0.7195 |
| Recall | 0.375 | 0.5268 | 0.4464 | 0.4018 | |
| Precision | 0.9545 | 0.9672 | 0.9615 | 0.9574 | |
| Specificity | 0.9851 | 0.9851 | 0.9851 | 0.9851 | |
| F1_score | 0.5385 | 0.6821 | 0.6098 | 0.566 | |
| k = 4 | Accuracy | 0.7073 | 0.7358 | 0.7398 | 0.7236 |
| Recall | 0.3661 | 0.4643 | 0.4375 | 0.4107 | |
| Precision | 0.9762 | 0.9123 | 0.98 | 0.9583 | |
| Specificity | 0.9925 | 0.9627 | 0.9925 | 0.9851 | |
| F1_score | 0.5325 | 0.6154 | 0.6049 | 0.575 | |
| Features Selected | |||||
|---|---|---|---|---|---|
| Parameters | Metrics | mean_fm, mean_sm, mean_tm | missed_first_month, missed_second_month, missed_third_month | week_0, week_1, week_2, ..., week_11 | missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9 week10, week 11 |
| k = 4, threshold = 0.001 | Accuracy | 0.687 | 0.7764 | 0.7764 | 0.7236 |
| Recall | 0.3214 | 0.5268 | 0.5268 | 0.4018 | |
| Precision | 0.973 | 0.9672 | 0.9672 | 0.9783 | |
| Specificity | 0.9925 | 0.9851 | 0.9851 | 0.9925 | |
| F1_score | 0.4832 | 0.6821 | 0.6821 | 0.5696 | |
| k = 4, threshold = 0.1 | Accuracy | 0.687 | 0.7764 | 0.752 | 0.7154 |
| Recall | 0.3214 | 0.5268 | 0.4732 | 0.3929 | |
| Precision | 0.973 | 0.9672 | 0.9636 | 0.9565 | |
| Specificity | 0.9925 | 0.9851 | 0.9851 | 0.9851 | |
| F1_score | 0.4832 | 0.6821 | 0.6347 | 0.557 | |
| k = 4, threshold = 100 | Accuracy | 0.7602 | 0.752 | 0.7114 | 0.752 |
| Recall | 0.4911 | 0.4732 | 0.375 | 0.4732 | |
| Precision | 0.9649 | 0.9636 | 0.9767 | 0.9636 | |
| Specificity | 0.9851 | 0.9851 | 0.9925 | 0.9851 | |
| F1_score | 0.6509 | 0.6347 | 0.5419 | 0.6347 | |
| Parameters: Damping | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Features selected | Metrics | 0.66 | 0.68 | 0.7 | 0.72 | 0.74 | 0.76 | 0.78 | 0.8 |
| mean_fm, mean_sm, mean_tm, | Accuracy | 0.752 | 0.7114 | 0.7114 | 0.7154 | 0.7154 | 0.7195 | 0.7154 | 0.7195 |
| Recall | 0.4732 | 0.375 | 0.375 | 0.3839 | 0.3839 | 0.3929 | 0.3839 | 0.3929 | |
| Precision | 0.9636 | 0.9767 | 0.9767 | 0.9773 | 0.9773 | 0.9778 | 0.9773 | 0.9778 | |
| Specificity | 0.9851 | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9552 | 0.9925 | 0.9925 | |
| F1_score | 0.6347 | 0.5419 | 0.5419 | 0.5513 | 0.5513 | 0.5605 | 0.5513 | 0.5605 | |
| missed_first_month, missed_second_month, missed_third_month | Accuracy | 0.6992 | 0.6911 | 0.687 | 0.687 | 0.687 | 0.687 | 0.687 | 0.687 |
| Recall | 0.3393 | 0.3214 | 0.3125 | 0.3125 | 0.3125 | 0.3125 | 0.3125 | 0.3125 | |
| Precision | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Specificity | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| F1_score | 0.5067 | 0.4865 | 0.4762 | 0.4762 | 0.4762 | 0.4762 | 0.4762 | 0.4762 | |
| week_0, week_1 week_2,...,week_11 | Accuracy | 0.752 | 0.7317 | 0.7317 | 0.7317 | 0.7358 | 0.752 | 0.7317 | 0.7317 |
| Recall | 0.4732 | 0.4196 | 0.4196 | 0.4196 | 0.4196 | 0.4732 | 0.4196 | 0.4196 | |
| Precision | 0.9636 | 0.9792 | 0.9792 | 0.9792 | 1.0 | 0.9636 | 0.9792 | 0.9792 | |
| Specificity | 0.9851 | 0.9925 | 0.9925 | 0.9925 | 1.0 | 0.9851 | 0.9925 | 0.9925 | |
| F1_score | 0.6347 | 0.5875 | 0.5875 | 0.5875 | 0.5912 | 0.6347 | 0.5875 | 0.5875 | |
| missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9, week10, week 11 | Accuracy | 0.752 | 0.7317 | 0.7317 | 0.7317 | 0.7358 | 0.7317 | 0.7317 | 0.7317 |
| Recall | 0.4732 | 0.4107 | 0.4107 | 0.4107 | 0.4196 | 0.4107 | 0.4107 | 0.4107 | |
| Precision | 0.9636 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Specificity | 0.9851 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| F1_score | 0.6347 | 0.5823 | 0.5823 | 0.5823 | 0.5912 | 0.5823 | 0.5823 | 0.5823 | |
| Features selected | Metrics | 0.82 | 0.84 | 0.86 | 0.88 | 0.9 | 0.92 | 0.94 | 0.96 |
| mean_fm, mean_sm, mean_tm, | Accuracy | 0.7033 | 0.7033 | 0.7033 | 0.7033 | 0.7398 | 0.7398 | 0.7398 | 0.7398 |
| Recall | 0.3571 | 0.3571 | 0.3571 | 0.3571 | 0.4464 | 0.4464 | 0.4464 | 0.4464 | |
| Precision | 0.9756 | 0.9756 | 0.9756 | 0.9756 | 0.9615 | 0.9615 | 0.9615 | 0.9615 | |
| Specificity | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9851 | 0.9851 | 0.9851 | 0.9851 | |
| F1_score | 0.5229 | 0.5229 | 0.5229 | 0.5229 | 0.6098 | 0.6098 | 0.6098 | 0.6098 | |
| missed_first_month, missed_second_month, missed_third_month | Accuracy | 0.6911 | 0.687 | 0.687 | 0.6911 | 0.6911 | 0.6911 | 0.6992 | 0.7195 |
| Recall | 0.3214 | 0.3125 | 0.3125 | 0.3214 | 0.3214 | 0.3214 | 0.3393 | 0.3929 | |
| Precision | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.9778 | |
| Specificity | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.9925 | |
| F1_score | 0.4865 | 0.4762 | 0.4762 | 0.4865 | 0.4865 | 0.4865 | 0.5067 | 0.5605 | |
| week_0, week_1 week_2,...,week_11 | Accuracy | 0.7073 | 0.7317 | 0.5875 | 0.7073 | 0.7073 | 0.7317 | 0.7317 | 0.7195 |
| Recall | 0.3661 | 0.4196 | 0.4196 | 0.3661 | 0.3661 | 0.4196 | 0.4196 | 0.4018 | |
| Precision | 0.9762 | 0.9792 | 0.9792 | 0.9762 | 0.9762 | 0.9792 | 0.9792 | 0.9574 | |
| Specificity | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9925 | 0.9851 | |
| F1_score | 0.5325 | 0.5875 | 0.5875 | 0.5325 | 0.5325 | 0.5875 | 0.5875 | 0.566 | |
| missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9, week10, week 11 | Accuracy | 0.7317 | 0.7317 | 0.7317 | 0.7195 | 0.752 | 0.752 | 0.6951 | 0.6992 |
| Recall | 0.4107 | 0.4107 | 0.4107 | 0.3839 | 0.4732 | 0.4732 | 0.3393 | 0.3482 | |
| Precision | 1.0 | 1.0 | 1.0 | 1.0 | 0.9636 | 0.9636 | 0.9744 | 0.975 | |
| Specificity | 1.0 | 1.0 | 1.0 | 1.0 | 0.9851 | 0.9851 | 0.9925 | 0.9925 | |
| F1_score | 0.5823 | 0.5823 | 0.5823 | 0.5548 | 0.6347 | 0.6347 | 0.5033 | 0.5132 | |
| Features Selected | |||||
|---|---|---|---|---|---|
| Parameters | Metrics | mean_fm, mean_sm, mean_tm, | missed_first_month, missed_second_month, missed_third_month | week_0, week_1 week_2,...,week_11 | missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9 week10, week 11 |
| k = 3 | Accuracy | 0.7967 | 0.8496 | 0.7805 | 0.7967 |
| Recall | 0.5714 | 0.7054 | 0.5268 | 0.5625 | |
| Precision | 0.9697 | 0.9518 | 0.9833 | 0.9844 | |
| Specificity | 0.9851 | 0.9701 | 0.9925 | 0.9925 | |
| F1_score | 0.7191 | 0.8103 | 0.686 | 0.7159 | |
| k = 4 | Accuracy | 0.8089 | 0.7967 | 0.7764 | 0.8089 |
| Recall | 0.5714 | 0.6161 | 0.5179 | 0.5982 | |
| Precision | 0.9697 | 0.9452 | 0.9831 | 0.971 | |
| Specificity | 0.9851 | 0.9701 | 0.9925 | 0.9851 | |
| F1_score | 0.7191 | 0.7459 | 0.6784 | 0.7403 | |
| Features Selected | ||||||
| Parameters | Metrics | mean_fm, mean_sm, mean_tm, | missed_first_month, missed_second_month, missed_third_month | week_0, week_1 week_2,...,week_11 | missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9, week10, week 11 | |
| k = 4, threshold = 0.001 | Accuracy | 0.8089 | 0.7967 | 0.7967 | 0.8333 | |
| Recall | 0.5893 | 0.5625 | 0.5893 | 0.6518 | ||
| Precision | 0.9851 | 0.9844 | 0.9429 | 0.9733 | ||
| Specificity | 0.9925 | 0.9925 | 0.9701 | 0.9851 | ||
| F1_score | 0.7374 | 0.7159 | 0.7253 | 0.7807 | ||
| k = 4, threshold = 0.1 | Accuracy | 0.8089 | 0.7967 | 0.7967 | 0.8415 | |
| Recall | 0.5893 | 0.5625 | 0.5893 | 0.6607 | ||
| Precision | 0.9851 | 0.9844 | 0.9429 | 0.9867 | ||
| Specificity | 0.9925 | 0.9925 | 0.9701 | 0.9925 | ||
| F1_score | 0.7374 | 0.7159 | 0.7253 | 0.7914 | ||
| k = 4, threshold = 100 | Accuracy | 0.7398 | 0.7276 | 0.7805 | 0.7276 | |
| Recall | 0.4286 | 0.4196 | 0.5536 | 0.4196 | ||
| Precision | 1.0 | 0.9592 | 0.9394 | 0.9592 | ||
| Specificity | 1.0 | 0.9851 | 0.9701 | 0.9851 | ||
| F1_score | 0.6 | 0.5839 | 0.6966 | 0.5839 | ||
| Parameters: Damping | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Features selected | Metrics | 0.66 | 0.68 | 0.7 | 0.72 | 0.74 | 0.76 | 0.78 | 0.8 |
| mean_fm, mean_sm, mean_tm, | Accuracy | 0.7276 | 0.8293 | 0.8293 | 0.8252 | 0.8252 | 0.8252 | 0.8252 | 0.8089 |
| Recall | 0.4196 | 0.6786 | 0.6786 | 0.6696 | 0.6696 | 0.6696 | 0.6696 | 0.5893 | |
| Precision | 0.9592 | 0.9268 | 0.9268 | 0.9259 | 0.9259 | 0.9259 | 0.9259 | 0.9851 | |
| Specificity | 0.9851 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9925 | |
| F1_score | 0.5839 | 0.7835 | 0.7835 | 0.7772 | 0.7772 | 0.7772 | 0.7772 | 0.7374 | |
| missed_first_month, missed_second_month, missed_third_month | Accuracy | 0.8496 | 0.8496 | 0.7967 | 0.7967 | 0.7967 | 0.7967 | 0.7967 | 0.7967 |
| Recall | 0.6875 | 0.6964 | 0.5893 | 0.5893 | 0.5893 | 0.5893 | 0.5893 | 0.5893 | |
| Precision | 0.9747 | 0.963 | 0.9429 | 0.9429 | 0.9429 | 0.9429 | 0.9429 | 0.9429 | |
| Specificity | 0.9851 | 0.9776 | 0.9701 | 0.9701 | 0.9701 | 0.9701 | 0.9701 | 0.9701 | |
| F1_score | 0.8063 | 0.8083 | 0.7253 | 0.7253 | 0.7253 | 0.7253 | 0.7253 | 0.7253 | |
| week_0, week_1 week_2,...,week_11 | Accuracy | 0.8577 | 0.8577 | 0.8577 | 0.8577 | 0.8618 | 0.8577 | 0.8577 | 0.8577 |
| Recall | 0.7411 | 0.7411 | 0.7411 | 0.7411 | 0.75 | 0.7411 | 0.7411 | 0.7411 | |
| Precision | 0.9326 | 0.9326 | 0.9326 | 0.9326 | 0.9333 | 0.9326 | 0.9326 | 0.9326 | |
| Specificity | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9552 | |
| F1_score | 0.8259 | 0.8259 | 0.8259 | 0.8259 | 0.8317 | 0.8259 | 0.8259 | 0.8259 | |
| missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9, week10, week 11 | Accuracy | 0.7276 | 0.8775 | 0.8333 | 0.8333 | 0.8333 | 0.8333 | 0.8333 | 0.8333 |
| Recall | 0.4196 | 0.7748 | 0.6964 | 0.6964 | 0.6964 | 0.6964 | 0.6964 | 0.6964 | |
| Precision | 0.9592 | 0.9451 | 0.9176 | 0.9176 | 0.9176 | 0.9176 | 0.9176 | 0.9176 | |
| Specificity | 0.9851 | 0.9627 | 0.9478 | 0.9478 | 0.9478 | 0.9478 | 0.9478 | 0.9478 | |
| F1_score | 0.5839 | 0.8514 | 0.7919 | 0.7919 | 0.7919 | 0.7919 | 0.7919 | 0.7919 | |
| Features selected | Metrics | 0.82 | 0.84 | 0.86 | 0.88 | 0.9 | 0.92 | 0.94 | 0.96 |
| mean_fm, mean_sm, mean_tm, | Accuracy | 0.8252 | 0.8252 | 0.8252 | 0.8252 | 0.7561 | 0.7561 | 0.7561 | 0.7561 |
| Recall | 0.6696 | 0.6696 | 0.6696 | 0.6696 | 0.4732 | 0.4732 | 0.4732 | 0.4732 | |
| Precision | 0.9259 | 0.9259 | 0.9259 | 0.9259 | 0.9815 | 0.9815 | 0.9815 | 0.9815 | |
| Specificity | 0.9552 | 0.9552 | 0.9552 | 0.9552 | 0.9925 | 0.9925 | 0.9925 | 0.9925 | |
| F1_score | 0.7772 | 0.7772 | 0.7772 | 0.7772 | 0.6386 | 0.6386 | 0.6386 | 0.6386 | |
| missed_first_month, missed_second_month, missed_third_month | Accuracy | 0.7967 | 0.7967 | 0.7967 | 0.7967 | 0.7967 | 0.8577 | 0.813 | 0.813 |
| Recall | 0.5893 | 0.5893 | 0.5893 | 0.5893 | 0.5893 | 0.7143 | 0.6161 | 0.6161 | |
| Precision | 0.9429 | 0.9429 | 0.9429 | 0.9429 | 0.9429 | 0.9639 | 0.9583 | 0.9583 | |
| Specificity | 0.9701 | 0.9701 | 0.9701 | 0.9701 | 0.9701 | 0.9776 | 0.9776 | 0.9776 | |
| F1_score | 0.7253 | 0.7253 | 0.7253 | 0.7253 | 0.7253 | 0.8205 | 0.75 | 0.75 | |
| week_0, week_1 week_2,...,week_11 | Accuracy | 0.8577 | 0.8618 | 0.8618 | 0.8577 | 0.8577 | 0.8659 | 0.8659 | 0.8496 |
| Recall | 0.7411 | 0.7411 | 0.7411 | 0.7411 | 0.7411 | 0.7411 | 0.7411 | 0.6786 | |
| Precision | 0.9326 | 0.9432 | 0.9432 | 0.9326 | 0.9326 | 0.954 | 0.954 | 0.987 | |
| Specificity | 0.9552 | 0.9627 | 0.9627 | 0.9552 | 0.9552 | 0.9701 | 0.9701 | 0.9925 | |
| F1_score | 0.8259 | 0.83 | 0.83 | 0.8259 | 0.8259 | 0.8342 | 0.8342 | 0.8042 | |
| missed_first_month, missed_second_month, missed_third_month, mean_fm, mean_sm, mean_tm, week8, week9, week10, week 11 | Accuracy | 0.8333 | 0.8333 | 0.8374 | 0.8333 | 0.7276 | 0.7276 | 0.7927 | 0.7886 |
| Recall | 0.6964 | 0.6964 | 0.6607 | 0.6964 | 0.4196 | 0.4196 | 0.5536 | 0.5446 | |
| Precision | 0.9176 | 0.9176 | 0.9737 | 0.9176 | 0.9592 | 0.9592 | 0.9841 | 0.9839 | |
| Specificity | 0.9478 | 0.9478 | 0.9851 | 0.9478 | 0.9851 | 0.9851 | 0.9925 | 0.9925 | |
| F1_score | 0.7919 | 0.7919 | 0.7872 | 0.7919 | 0.5839 | 0.5839 | 0.7086 | 0.7011 | |
| Confusion Matrix | Predicted: No | Predicted: Yes |
|---|---|---|
| Actual: No | TN: 129 | FP: 5 |
| Actual: Yes | FN: 25 | TP: 86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jossa-Bastidas, O.; Zahia, S.; Fuente-Vidal, A.; Sánchez Férez, N.; Roda Noguera, O.; Montane, J.; Garcia-Zapirain, B. Predicting Physical Exercise Adherence in Fitness Apps Using a Deep Learning Approach. Int. J. Environ. Res. Public Health 2021, 18, 10769. https://doi.org/10.3390/ijerph182010769
Jossa-Bastidas O, Zahia S, Fuente-Vidal A, Sánchez Férez N, Roda Noguera O, Montane J, Garcia-Zapirain B. Predicting Physical Exercise Adherence in Fitness Apps Using a Deep Learning Approach. International Journal of Environmental Research and Public Health. 2021; 18(20):10769. https://doi.org/10.3390/ijerph182010769
Chicago/Turabian StyleJossa-Bastidas, Oscar, Sofia Zahia, Andrea Fuente-Vidal, Néstor Sánchez Férez, Oriol Roda Noguera, Joel Montane, and Begonya Garcia-Zapirain. 2021. "Predicting Physical Exercise Adherence in Fitness Apps Using a Deep Learning Approach" International Journal of Environmental Research and Public Health 18, no. 20: 10769. https://doi.org/10.3390/ijerph182010769
APA StyleJossa-Bastidas, O., Zahia, S., Fuente-Vidal, A., Sánchez Férez, N., Roda Noguera, O., Montane, J., & Garcia-Zapirain, B. (2021). Predicting Physical Exercise Adherence in Fitness Apps Using a Deep Learning Approach. International Journal of Environmental Research and Public Health, 18(20), 10769. https://doi.org/10.3390/ijerph182010769








