The Mediation Effects of Aluminum in Plasma and Dipeptidyl Peptidase Like Protein 6 (DPP6) Polymorphism on Renal Function via Genome-Wide Typing Association
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Questionnaires
2.3. Laboratory Analysis
2.4. Genotyping
2.5. Statistical Analysis
2.5.1. Descriptive Analysis
2.5.2. Genome-Wide Association (GWA)
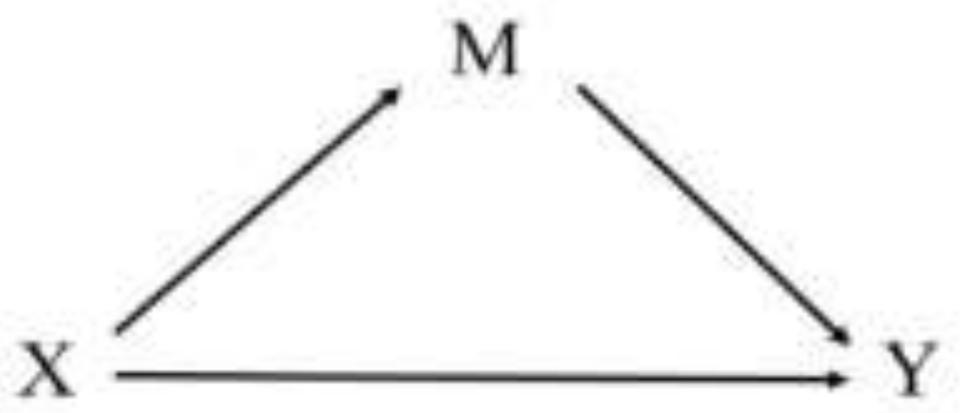
2.5.3. Mediation Analysis
3. Results
3.1. Study Population and SNPs Typing
3.2. Characteristics of Study Population
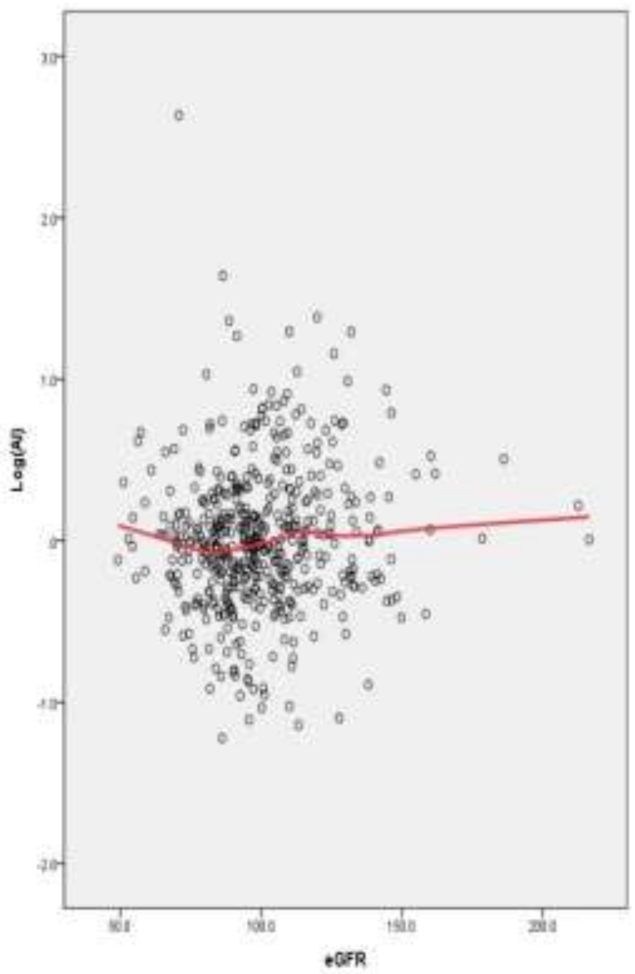
3.3. Association between eGFR and Al
3.4. Genome-Wide Association Test
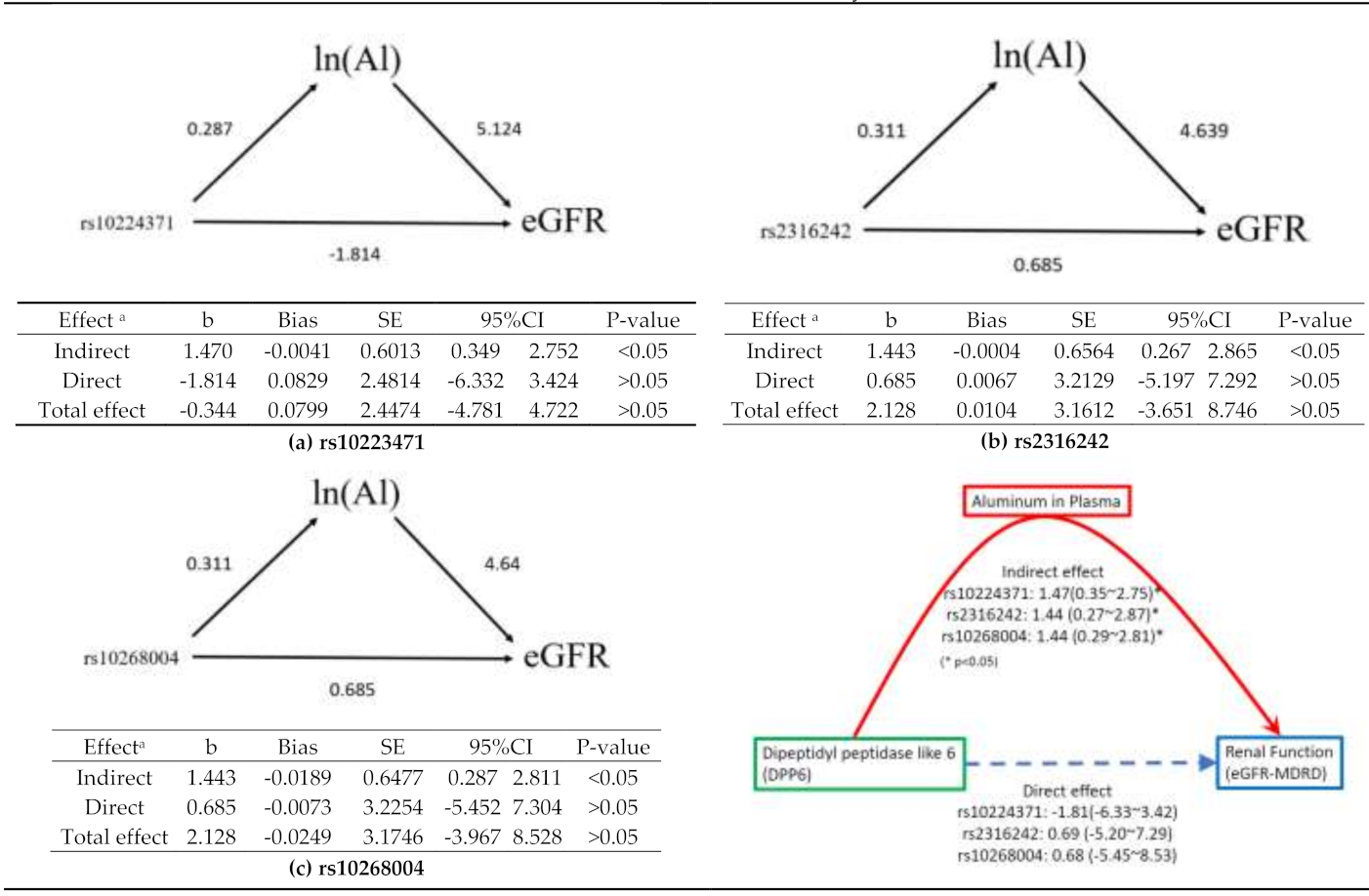
3.5. Mediation Analysis
3.5.1. rs10224371→ln(Al)→eGFR
3.5.2. rs2316242→ln(Al)→eGFR
3.5.3. rs10268004→ln(Al)→eGFR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Information
References
- Aluminum. Available online: https://en.wikipedia.org/wiki/Aluminium (accessed on 20 August 2021).
- Letzel, S.; Lang, C.; Schaller, K.; Angerer, J.; Fuchs, S.; Neundörfer, B.; Lehnert, G. Longitudinal study of neurotoxicity with occupational exposure to aluminum dust. Neurology 2000, 54, 997–1000. [Google Scholar] [CrossRef]
- Sinczuk-Walczak, H.; Szymczak, M.; Razniewska, G.; Matczak, W.; Szymczak, W. Effects of occupational exposure to aluminum on nervous system: Clinical and electroencephalographic findings. Int J Occup Med Environ Health 2003, 16, 301–310. [Google Scholar] [PubMed]
- Rønneberg, A.; Haldorsen, T.; Romundstad, P.; Andersen, A. Occupational exposure and cancer incidence among workers from an aluminum smelter in western Norway. J Work Environ Health 1999, 25, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Alfrey, A.; Hegg, A.; Craswell, P. Metabolism and toxicity of aluminum in renal failure. Am. J. Clin. Nutr. 1980, 33, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Recker, R.; Blotcky, A.; Leffler, J.; Rack, E. Evidence of aluminum absorption from the gastrointestinal tract and bone deposition by aluminum carbonate ingestion with normal renal function. J Lab Clin Med 1977, 90, 810–815. [Google Scholar]
- Jeffery, E.; Abreo, K.; Burgess, E.; Cannata, J.; Greger, J. Systemic aluminum toxicity: Effects on bone, hematopoietic tissue, and kidney. J. Toxicol. Environ. Health Part A 1996, 48, 649–666. [Google Scholar]
- Hewitt, C.; Savory, J.; Wills, M. Aspects of aluminum toxicity. Clin. Lab. Med. 1990, 10, 403–422. [Google Scholar] [CrossRef]
- Nayak, P. Aluminum: Impacts and disease. Environ. Res. 2002, 89, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Darbre, P. Aluminium, antiperspirants and breast cancer. J. Inorg. Biochem. 2005, 99, 1912–1919. [Google Scholar] [CrossRef]
- Ng, E.; Lind, P.; Lindgren, C.; Ingelsson, E.; Mahajan, A.; Morris, A.; Lind, L. Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum Mol Genet 2015, 24, 4739–4745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Zhang, X.; Tarantini, L.; Nordio, F.; Bonzini, M.; Angelici, L.; Marinelli, B.; Rizzo, G.; Cantone, L.; Apostoli, P. Ambient PM exposure and DNA methylation in tumor suppressor genes: A cross-sectional study. Part. Fibre Toxicol. 2011, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Tajuddin, S.M.; Amaral, A.F.S.; Fernández, A.F.; Rodríguez-Rodero, S.; Rodríguez, R.M.; Moore, L.E.; Tardón, A.; Carrato, A.; García-Closas, M.; Silverman, D.T.; et al. Genetic and Non-genetic Predictors of LINE-1 Methylation in Leukocyte DNA. Environ. Health Perspect. 2013, 121, 650–656. [Google Scholar] [CrossRef] [Green Version]
- Kile, M.L.; Fang, S.; Baccarelli, A.A.; Tarantini, L.; Cavallari, J.; Christiani, D.C. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. J. Environ. Health 2013, 12, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Yuan, Y.; Lu, X.; Yang, J.; Wang, L.; Song, J.; Nie, J.; Zhang, Q.; Niu, Q. The Relationship Between Cognitive Impairment and Global DNA Methylation Decrease Among Aluminum Potroom Workers. J. Occup. Environ. 2015, 57, 713–717. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.; Kochian, L. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 6503–6508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Famoso, A.; Zhao, K.; Clark, R.; Tung, C.; Wright, M.; Bustamante, C.; Kochian, L.; McCouch, S. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011, 7, e1002221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, C.; Lin, J.; Lee, C. Taiwan Biobank: A project aiming to aid Taiwan’s transition into a biomedical island. Pharmacogenomics 2008, 9, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, J.; Chiang, C.; Hsiung, C.; Wu, P.; Chang, L.; Chu, H.; Chang, J.; Song, I.; Yang, S. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum Mol Genet 2016, 25, 5321–5331. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, R.; Belmont, J.; Hardenbol, P.; Willis, T.; Yu, F.; Yang, H.; Ch’’ang, L.; Huang, W.; Liu, B.; Shen, Y. The international HapMap project. Nature 2003, 426, 789–796. [Google Scholar]
- Siva, N. 1000 Genomes project. Nat. Biotechnol. 2008, 26, 256. [Google Scholar] [CrossRef]
- Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.; Daly, M. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J Comput Graph Stat 1996, 5, 299–314. [Google Scholar]
- Ma, Y.C.; Zuo, L.; Chen, J.H.; Luo, Q.; Yu, X.Q.; Li, Y.; Xu, J.S.; Huang, S.M.; Wang, L.N.; Huang, W.; et al. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J. Am. Soc. Nephrol. 2006, 17, 2937–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bland, J.; Altman, D. Multiple significance tests: The Bonferroni method. Br. Med. J. 1995, 310, 170. [Google Scholar] [CrossRef] [Green Version]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g: Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Barrett, J.; Fry, B.; Maller, J.; Daly, M. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2004, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. J. Open Source Softw. 2018, 3, 0731. [Google Scholar] [CrossRef] [Green Version]
- Baron, R.; Kenny, D. The moderator–mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J Pers Soc Psychol 1986, 51, 1173. [Google Scholar] [CrossRef]
- MacKinnon, D.; Fairchild, A.; Fritz, M. Mediation analysis. Annu. Rev. Psychol. 2007, 58, 593–614. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, D.P.; Lockwood, C.M.; Williams, J. Confidence Limits for the Indirect Effect: Distribution of the Product and Resampling Methods. Multivariate Behav Res 2004, 39, 99. [Google Scholar] [CrossRef] [Green Version]
- Preacher, K.; Hayes, A. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav. Res. Methods 2008, 40, 879–891. [Google Scholar] [CrossRef]
- Thakkinstian, A.; Anothaisintawee, T.; Chailurkit, L.; Ratanachaiwong, W.; Yamwong, S.; Sritara, P.; Ongphiphadhanakul, B. Potential causal associations between vitamin D and uric acid: Bidirectional mediation analysis. Sci. Rep. 2015, 5, 14528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preacher, K.; Selig, J. Advantages of Monte Carlo confidence intervals for indirect effects. Commun Methods Meas 2012, 6, 77–98. [Google Scholar] [CrossRef] [Green Version]
- Hayes, A.; Scharkow, M. The relative trustworthiness of inferential tests of the indirect effect in statistical mediation analysis: Does method really matter? Psychol. Sci. 2013, 24, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.; Chibi, M.; Xie, P.; Koundrioukoff, S.; Moolman-Smook, H.; Pugh, D.; Debatisse, M.; He, F.; Zhang, L.; Defossez, P.A. The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability. Cell Rep. 2014, 7, 575–587. [Google Scholar] [CrossRef]
- Jones, M.H.; Hamana, N.; Nezu, J.; Shimane, M. A novel family of bromodomain genes. Genomics 2000, 63, 40–45. [Google Scholar] [CrossRef]
- Baldwin, D.; Marshall, W. Heavy metal poisoning and its laboratory investigation. Ann. Clin. Biochem 1999, 36, 267–300. [Google Scholar] [CrossRef] [Green Version]
- Goullé, J.; Mahieu, L.; Castermant, J.; Neveu, N.; Bonneau, L.; Lainé, G.; Bouige, D.; Lacroix, C. Metal and metalloid multi-elementary ICP-MS validation in whole blood, plasma, urine and hair: Reference values. Sci. Int. 2005, 153, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Cronin, S.; Tomik, B.; Bradley, D.G.; Slowik, A.; Hardiman, O. Screening for replication of genome-wide SNP associations in sporadic ALS. Eur. J. Hum. Genet. 2009, 17, 213–218. [Google Scholar] [CrossRef] [Green Version]
- Chio, A.; Schymick, J.C.; Restagno, G.; Scholz, S.W.; Lombardo, F.; Lai, S.L.; Mora, G.; Fung, H.C.; Britton, A.; Arepalli, S.; et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009, 18, 1524–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwee, L.C.; Liu, Y.; Haynes, C.; Gibson, J.R.; Stone, A.; Schichman, S.A.; Kamel, F.; Nelson, L.M.; Topol, B.; Van den Eeden, S.K.; et al. A high-density genome-wide association screen of sporadic ALS in US veterans. PloS ONE 2012, 7, e32768. [Google Scholar] [CrossRef] [Green Version]
- Maussion, G.; Cruceanu, C.; Rosenfeld, J.A.; Bell, S.C.; Jollant, F.; Szatkiewicz, J.; Collins, R.L.; Hanscom, C.; Kolobova, I.; de Champfleur, N.M.; et al. Implication of LRRC4C and DPP6 in neurodevelopmental disorders. Am. J. Med. Genet. Part A. 2017, 173, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Wolf, E.J.; Rasmusson, A.M.; Mitchell, K.S.; Logue, M.W.; Baldwin, C.T.; Miller, M.W. A genome-wide association study of clinical symptoms of dissociation in a trauma-exposed sample. Depress Anxiety. 2014, 31, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Long, L.K.; Hatch, M.M.; Hoffman, D.A. DPP6 Domains Responsible for Its Localization and Function. J. Biol. Chem. 2014, 289, 32153–32165. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Murphy, J.G.; Karlsson, R.M.; Petralia, R.S.; Gutzmann, J.J.; Abebe, D.; Wang, Y.X.; Cameron, H.A.; Hoffman, D.A. DPP6 Loss Impacts Hippocampal Synaptic Development and Induces Behavioral Impairments in Recognition, Learning and Memory. Front. Cell. Neurosci. 2018, 12, 84. [Google Scholar] [CrossRef] [Green Version]
- Keith, S.; Jones, D.; Rosemond, Z.; Ingerman, L.; Chappell, L. Toxicological profile for aluminum. DIANE Publishing: Collingdale, PA, USA, 2008. [Google Scholar]
- Tokar, E.; Boyd, W.; Freedman, J.; Waalkes, M. Toxic effects of metals. In Casarett & Doull’s Toxicology: The Basic Science of Poisons, 8th ed.; Klaassen, C., Ed.; McGraw-Hill Health Professions Division: New York, NY, USA, 2013. [Google Scholar]
- Mayor, G.H.; Burnatowska-Hledin, M.A. Impaired renal function and aluminum metabolism. FASEB J. 1983, 42, 2979–2983. [Google Scholar]
- Brambilla, P.; Esposito, F.; Lindstrom, E.; Sorosina, M.; Giacalone, G.; Clarelli, F.; Rodegher, M.; Colombo, B.; Moiola, L.; Ghezzi, A.; et al. Association between DPP6 polymorphism and the risk of progressive multiple sclerosis in Northern and Southern Europeans. Neurosci. Lett. 2012, 530, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Martyn, C.; Coggon, D.; Inskip, H.; Lacey, R.; Young, W. Aluminum concentrations in drinking water and risk of Alzheimer’’s disease. Epidemiology. 1997, 8, 281–286. [Google Scholar] [CrossRef]
- McLachlan, D. Aluminium and the risk for Alzheimer’’s disease. Environmetrics. 1995, 6, 233–275. [Google Scholar] [CrossRef]
- Virk, S.A.; Eslick, G.D. Occupational Exposure to Aluminum and Alzheimer Disease: A Meta-Analysis. J. Occup. Environ. Med. 2015, 57, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Krewski, D.; Yokel, R.; Nieboer, E.; Borchelt, D.; Cohen, J.; Harry, J.; Kacew, S.; Lindsay, J.; Mahfouz, A.; Rondeau, V. Human health risk assessment for aluminium, aluminium oxide, and aluminium hydroxide. J. Toxicol. Environ. Health Part B. 2007, 10, 1–269. [Google Scholar] [CrossRef] [PubMed]

| Term (n = 494) | Mean ± SD, N(%) | Min | Q1 | Q2 | Q3 | Max |
|---|---|---|---|---|---|---|
| Gender (Male) | 275(55.7%) | |||||
| Drink (Current drinking) a | 39(7.9%) | |||||
| Smoke experience (Yes) | 169(34.2%) | |||||
| Age (year) | 48.3 ± 10.9 | 30 | 40 | 47 | 54 | 70 |
| Height (cm) | 165.18 ± 8.63 | 142.0 | 188.5 | 158.5 | 165.5 | 171.5 |
| Weight (kg) | 66.95 ± 12.32 | 39.4 | 120.8 | 58.2 | 66.80 | 74.8 |
| γ-GT (U/L) | 26.25 ± 27.09 | 6.0 | 13.0 | 18.0 | 30.0 | 293.0 |
| BUN (mg/dL) | 13.06 ± 3.48 | 5.0 | 10.6 | 12.6 | 15.2 | 28.2 |
| Creatinine (mg/dL) | 0.78 ± 0.19 | 0.33 | 0.62 | 0.79 | 0.91 | 1.50 |
| eGFR b (mL/min/1.73 m2) | 99.52 ± 22.00 | 49.15 | 85.30 | 96.99 | 110.37 | 216.66 |
| Uric acid (mg/dL) | 5.73 ± 1.56 | 2.6 | 4.5 | 5.6 | 6.9 | 12.4 |
| HbA1C (%) | 5.66 ± 0.55 | 4.4 | 5.4 | 5.6 | 5.8 | 11.9 |
| Total cholesterol (mg/dL) | 197.7 ± 35.8 | 117 | 358 | 174 | 195 | 217 |
| TG (mg/dL) | 122.9 ± 85.7 | 30 | 798 | 68 | 101 | 145 |
| HDL (mg/dL) | 53.8 ± 13.2 | 26 | 108 | 44 | 52 | 62 |
| LDL (mg/dL) | 125.5 ± 32.4 | 47 | 258 | 105 | 124 | 146 |
| AST (U/L) | 23.38 ± 11.44 | 10 | 18 | 21 | 26 | 167 |
| ALT (U/L) | 25.6 ± 23.4 | 7 | 15 | 20 | 28 | 344 |
| Aluminum (μg/L) | 1.13 ± 0.80 | 0.29 | 0.78 | 0.99 | 1.27 | 13.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-H.; Yang, C.-C.; Luo, K.-H.; Dai, C.-Y.; Chuang, Y.-C.; Chuang, H.-Y. The Mediation Effects of Aluminum in Plasma and Dipeptidyl Peptidase Like Protein 6 (DPP6) Polymorphism on Renal Function via Genome-Wide Typing Association. Int. J. Environ. Res. Public Health 2021, 18, 10484. https://doi.org/10.3390/ijerph181910484
Chen T-H, Yang C-C, Luo K-H, Dai C-Y, Chuang Y-C, Chuang H-Y. The Mediation Effects of Aluminum in Plasma and Dipeptidyl Peptidase Like Protein 6 (DPP6) Polymorphism on Renal Function via Genome-Wide Typing Association. International Journal of Environmental Research and Public Health. 2021; 18(19):10484. https://doi.org/10.3390/ijerph181910484
Chicago/Turabian StyleChen, Ting-Hao, Chen-Cheng Yang, Kuei-Hau Luo, Chia-Yen Dai, Yao-Chung Chuang, and Hung-Yi Chuang. 2021. "The Mediation Effects of Aluminum in Plasma and Dipeptidyl Peptidase Like Protein 6 (DPP6) Polymorphism on Renal Function via Genome-Wide Typing Association" International Journal of Environmental Research and Public Health 18, no. 19: 10484. https://doi.org/10.3390/ijerph181910484
APA StyleChen, T.-H., Yang, C.-C., Luo, K.-H., Dai, C.-Y., Chuang, Y.-C., & Chuang, H.-Y. (2021). The Mediation Effects of Aluminum in Plasma and Dipeptidyl Peptidase Like Protein 6 (DPP6) Polymorphism on Renal Function via Genome-Wide Typing Association. International Journal of Environmental Research and Public Health, 18(19), 10484. https://doi.org/10.3390/ijerph181910484









