Characterization of a Lactiplantibacillus plantarum R23 Isolated from Arugula by Whole-Genome Sequencing and Its Bacteriocin Production Ability
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin, Growth, and Storage Conditions of Lactiplantibacillus plantarum R23
2.2. Whole-Genome Sequencing and Bioinformatic Analyses
2.2.1. Virulence and Resistance Profiles of Lpb. plantarum R23
2.2.2. Identification of Lpb. plantarum R23-Produced Bacteriocin
2.3. Characterization of Lpb. plantarum R23-Produced Bacteriocin
2.3.1. Kinetics of Growth and Bacteriocin Production
2.3.2. Effect of Enzymes, Temperature, pH, and Surfactants on Bacteriocin Activity
2.3.3. Cell Lysis
2.3.4. Adsorption Studies
2.3.5. Partial Purification and Molecular Size
3. Results
3.1. Characterization of Lpb. plantarum R23 by Whole-Genome Sequencing
3.1.1. Presence of Antibiotic Resistances
3.1.2. Presence of Virulence Factors
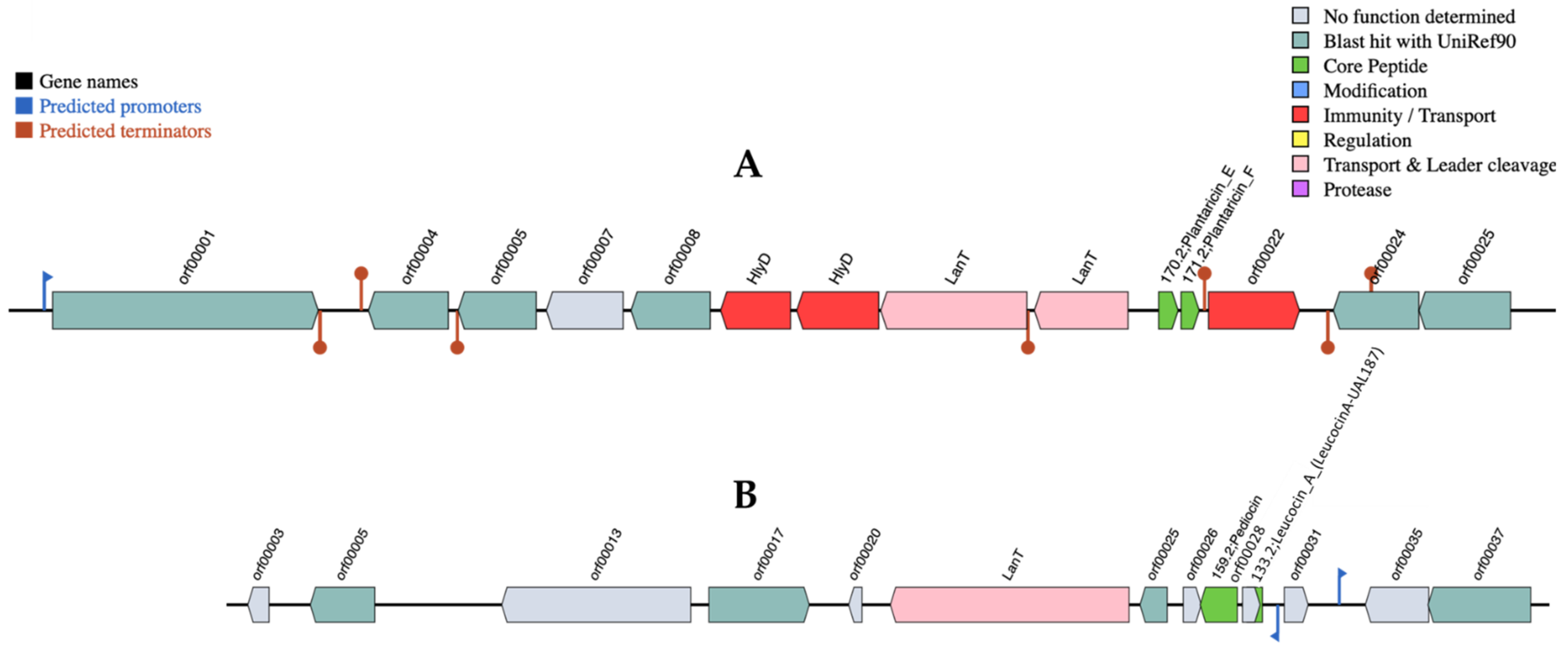
3.2. Identification of Lpb. plantarum R23 Bacteriocin by Whole-Genome Sequencing
3.3. Characterization of Lpb. plantarum R23-Produced Bacteriocin
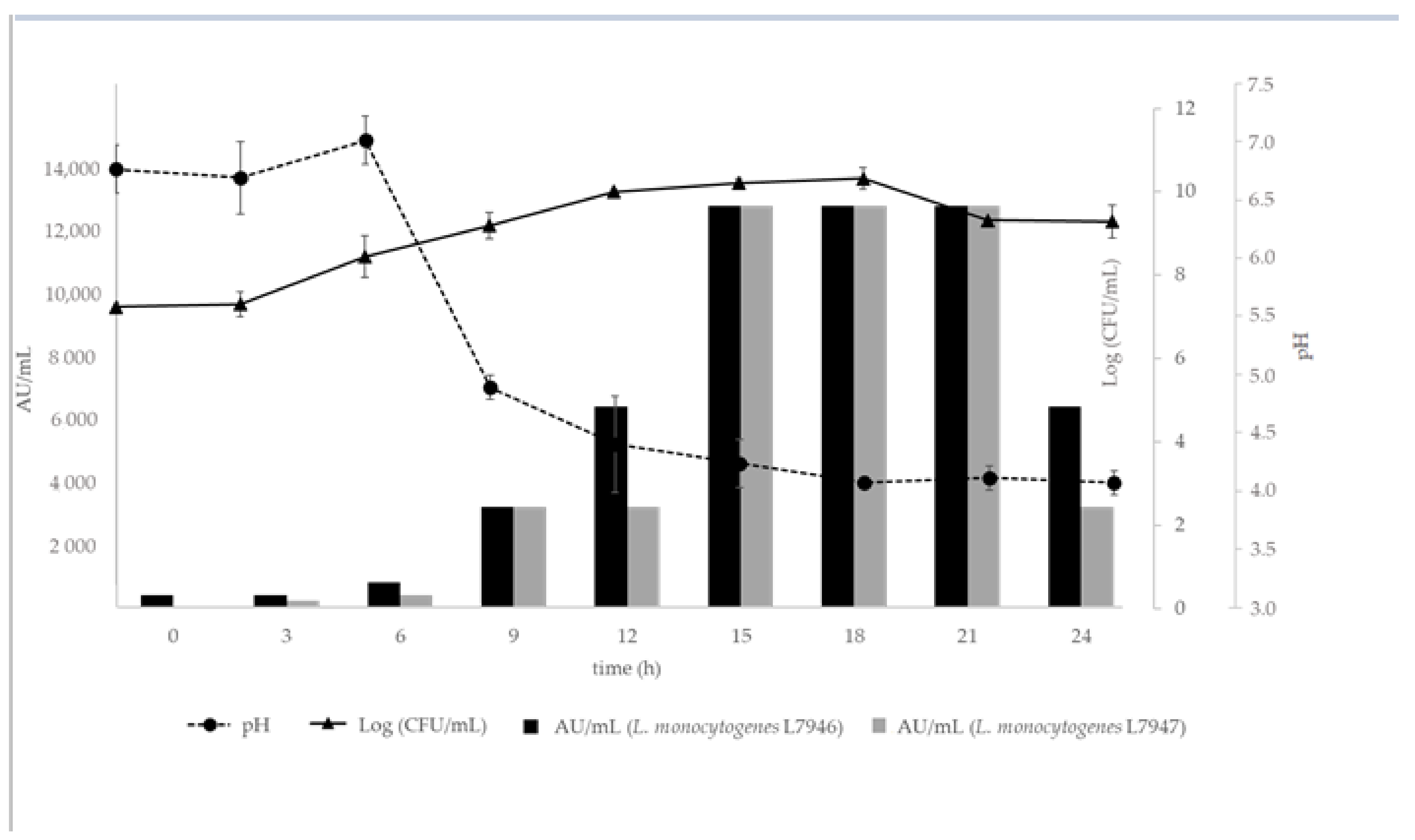
3.3.1. Kinetics of Growth and Bacteriocin Production
3.3.2. Effect of Enzymes, Temperature, pH, and Surfactants on Bacteriocin(s) Activity
3.3.3. Cell Lysis
3.3.4. Adsorption Studies and Molecular Size
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization. Fruit and Vegetables—Your Dietary Essentials; The International Year of Fruits and Vegetables, 2021, background paper; Food and Agriculture Organization: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Emilse, P.V.; Matías, V.; Cecilia, M.L.; Oscar, G.M.; Gisela, M.; Guadalupe, D.; Elizabeth, R.V.; Victorio, P.J.; Rodney, C.; Viviana, N.S.; et al. Enteric virus presence in green vegetables and associated irrigation waters in a rural area from Argentina. A quantitative microbial risk assessment. LWT 2021, 144, 111201. [Google Scholar] [CrossRef]
- Xylia, P.; Botsaris, G.; Chrysargyris, A.; Skandamis, P.; Tzortzakis, N. Variation of microbial load and biochemical activity of ready-to-eat salads in Cyprus as affected by vegetable type, season, and producer. Food Microbiol. 2019, 83, 200–210. [Google Scholar] [CrossRef]
- Park, S.; Szonyi, B.; Gautam, R.; Nightingale, K.; Anciso, J.; Ivanek, R. Risk factors for microbial contamination in fruits and vegetables at the preharvest level: A systematic review. J. Food Prot. 2012, 75, 2055–2081. [Google Scholar] [CrossRef]
- Trias, R.; Bañeras, L.; Badosa, E.; Montesinos, E. Bioprotection of Golden Delicious apples and Iceberg lettuce against foodborne bacterial pathogens by lactic acid bacteria. Int. J. Food Microbiol. 2008, 123, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Fessard, A.; Remize, F. Genetic and technological characterization of lactic acid bacteria isolated from tropically grown fruits and vegetables. Int. J. Food Microbiol. 2019, 301, 61–72. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Tosukhowong, A.; Visessanguan, W.; Pumpuang, L.; Tepkasikul, P.; Panya, A.; Valyasevi, R. Biogenic amine formation in Nham, a Thai fermented sausage, and the reduction by commercial starter culture, Lactobacillus plantarum BCC 9546. Food Chem. 2011, 129, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G. Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef]
- European Food Safety Authority. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 10: Suitability of taxonomic units notified to EFSA until March 2019. EFSA J. 2019, 17, 5753. [Google Scholar] [CrossRef]
- Soccol, C.R.; Vandenberghe, L.P.S.; Spier, M.R.; Medeiros, A.B.P.; Yamaguishi, C.T.; Lindner, J.D.D.; Pandey, A.; Thomaz-Soccol, V. The potential of probiotics: A review. Food Technol. Biotechnol. 2010, 48, 413–434. [Google Scholar]
- Kim, S.-K.; Guevarra, R.B.; Kim, Y.-T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.-H. Role of probiotics in human gut microbiome-associated diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Galvez, A.; Abriouel, H.; Lopez, R.L.; Ben Omar, N. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [PubMed]
- Kavitha, S.; Harikrishnan, A.; Jeevaratnam, K. Characterization and evaluation of antibacterial efficacy of a novel antibiotic-type compound from a probiotic strain Lactobacillus plantarum KJB23 against food-borne pathogens. LWT 2020, 118, 108759. [Google Scholar] [CrossRef]
- Pinto, A.; Barbosa, J.; Albano, H.; Isidro, J.; Teixeira, P. Screening of bacteriocinogenic lactic acid bacteria cultures and their characterization as potential probiotics. Microorganisms 2020, 8, 393. [Google Scholar] [CrossRef] [PubMed]
- Daba, G.M.; Elkhateeb, W.A. Bacteriocins of lactic acid bacteria as biotechnological tools in food and pharmaceuticals: Current applications and future prospects. Biocatal. Agric. Biotechnol. 2020, 28, 101750. [Google Scholar] [CrossRef]
- Zhao, S.; Han, J.; Bie, X.; Lu, Z.; Zhang, C.; Lv, F. Purification and characterization of plantaricin JLA-9: A novel bacteriocin against Bacillus spp. produced by Lactobacillus plantarum JLA-9 from Suan-Tsai, a traditional Chinese fermented cabbage. J. Agric. Food Chem. 2016, 64, 2754–2764. [Google Scholar] [CrossRef]
- Hwanhlem, N.; Ivanova, T.; Biscola, V.; Choiset, Y.; Haertlé, T. Bacteriocin producing Enterococcus faecalis isolated from chicken gastrointestinal tract originating from Phitsanulok, Thailand: Isolation, screening, safety evaluation and probiotic properties. Food Control 2017, 78, 187–195. [Google Scholar] [CrossRef]
- Qiao, Z.; Sun, H.; Zhou, Q.; Yi, L.; Wang, X.; Shan, Y.; Yi, Y.; Liu, B.; Zhou, Y.; Lü, X. Characterization and antibacterial action mode of bacteriocin BMP32r and its application as antimicrobial agent for the therapy of multidrug-resistant bacterial infection. Int. J. Biol. Macromol. 2020, 164, 845–854. [Google Scholar] [CrossRef]
- Balmeh, N.; Mahmoudi, S.; Fard, N.A. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inform. Med. Unlocked 2021, 23, 100515. [Google Scholar] [CrossRef] [PubMed]
- Abrams, D.; Barbosa, J.; Albano, H.; Silva, J.; Gibbs, P.A.; Teixeira, P. Characterization of bacPPK34 a bacteriocin produced by Pediococcus pentosaceus strain K34 isolated from “Alheira”. Food Control 2011, 22, 940–946. [Google Scholar] [CrossRef]
- Martinez, R.C.R.; Staliano, C.D.; Vieira, A.D.S.; Villarreal, M.L.M.; Todorov, S.D.; Saad, S.M.I.; Franco, B.D.G.M. Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp sakei 2a in a potentially synbiotic cheese spread. Food Microbiol. 2015, 48, 43–152. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.; Kim, J.; Kim, W.J. Isolation and characterization of bacteriocin-producing Pediococcus acidilactici HW01 from malt and its potential to control beer spoilage lactic acid bacteria. Food Control 2017, 80, 59–66. [Google Scholar] [CrossRef]
- Oliveira, M.; Barbosa, J.; Albano, H.; Teixeira, P. Bacteriocinogenic activity of Leuconostoc lactis RK18 isolated from fermented food. In Fermented Foods: Nutrition and Role in Health and Disease; Kovalyov, O., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2020; Chapter 3; pp. 159–181. [Google Scholar]
- Gomes, J.; Teixeira, P.; Barbosa, J. Preservation of meat products: Natural antimicrobial agents as an alternative to chemical antimicrobials. In Meat Products: Chemistry, Consumption and Health Aspects; Castro, M.P., Cayré, M.E., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2021; Chapter 3; pp. 49–87. [Google Scholar]
- Woraprayote, W.; Malila, Y.; Sorapukdee, S.; Swetwiwathana, A.; Benjakul, S.; Visessanguan, W. Bacteriocins from lactic acid bacteria and their applications in meat and meat products. Meat Sci. 2016, 120, 118–132. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology, Proceedings of the 17th Annual International Conference, RECOMB 2013, Beijing, China, 7–10 April 2013; Lecture Notes in Computer Science; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7821, pp. 158–170. [Google Scholar]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Van Reenen, C.A.; Dicks, L.M.T.; Chikindas, M.L. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J. Appl. Microbiol. 1998, 84, 1131–1137. [Google Scholar] [CrossRef]
- McLauchlin, J.; Hampton, M.D.; Shah, S.; Threlfall, E.J.; Wieneke, A.A.; Curtis, G.D.W. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 1997, 83, 381–388. [Google Scholar] [CrossRef]
- Yang, R.; Monty, J.; Ray, B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl. Environ. Microbiol. 1992, 58, 3355–3359. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Malke, H., Ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; pp. 1538–1546. [Google Scholar]
- Schägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulphate polyacrylamide gel electrophoresis for the separation of protein in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- European Food Safety Authority. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance by EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). EFSA J. 2012, 10, 2740–2749. [Google Scholar] [CrossRef]
- Saelim, K.; Jampaphaeng, K.; Maneerat, S. Functional properties of Lactobacillus plantarum S0/7 isolated fermented stinky bean (Sa Taw Dong) and its use as a starter culture. J. Funct. Foods 2017, 38, 370–377. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Y.; Sun, M.; Zhang, H.; Mu, G.; Tuo, Y. Physiological function analysis of Lactobacillus plantarum Y44 based on genotypic and phenotypic characteristics. J. Dairy Sci. 2020, 103, 5916–5930. [Google Scholar] [CrossRef]
- Feriala, A.A.; Saif, A.; Sakr, E.A.E. Characterization and bioactivities of exopolysaccharide produced from probiotic Lactobacillus plantarum 47FE and Lactobacillus pentosus 68FE. Bioact. Carbohydr. Diet. Fibre 2020, 24, 100231. [Google Scholar] [CrossRef]
- European Food Safety Authority. EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021, 1–15. Available online: https://www.efsa.europa.eu/sites/default/files/2021-03/outcome-public-consultation-for-EFSA-statement-EFSA-Q-2019-00434.pdf (accessed on 17 May 2021).
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Diep, D.B.; Myhre, R.; Johnsborg, O.; Aakra, Å.; Nes, I.F. Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol. Microbiol. 2003, 47, 483–494. [Google Scholar] [CrossRef]
- Le Marrec, C.; Hyronimus, B.; Bressollier, P.; Verneuil, B.; Urdaci, M.C. Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the Pediocin family of bacteriocins, produced by Bacillus coagulans I4. Appl. Environ. Microbiol. 2000, 66, 5213–5220. [Google Scholar] [CrossRef]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Ennahar, S.; Aoude-Werner, D.; Sorokine, O.; van Dorsselaer, A.; Bringel, F.; Hubert, J.-C.; Hasselmann, C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl. Environ. Microbiol. 1996, 62, 4381–4387. [Google Scholar] [CrossRef]
- Loessner, M.; Guenther, S.; Steffan, S.; Scherer, S. A Pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl. Environ. Microbiol. 2003, 69, 1854–1857. [Google Scholar] [CrossRef]
- West, C.A.; Warner, P.J. Plantacin B, a bacteriocin produced by Lactobacillus plantarum NCDO 1193. FEMS Microbiol. Lett. 1988, 49, 163–165. [Google Scholar] [CrossRef]
- Remiger, A.; Eijsink, V.G.; Ehrmann, M.A.; Sletten, K.; Nes, I.F.; Vogel, R.F. Purification and partial amino acid sequence of plantaricin 1.25 alpha and 1.25 beta, two bacteriocins produced by Lactobacillus plantarum TMW1.25. J. Appl. Microbiol. 1999, 86, 1053–1058. [Google Scholar] [CrossRef]
- Holo, H.; Jeknic, Z.; Daeschel, M.; Stevanovic, S.; Nes, I.F. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology 2001, 147, 643–651. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Y.; Xie, Q.; Zhang, Y.; Hu, J.; Li, P. Purification and characterization of plantaricin LPL-1, a novel class IIa bacteriocin produced by Lactobacillus plantarum LPL-1 isolated from fermented fish. Front. Microbiol. 2018, 9, 2276. [Google Scholar] [CrossRef]
- Martinez, R.C.R.; Wachsman, M.; Torres, N.I.; LeBlanc, J.G.; Todorov, S.D.; Franco, B.D.G.M. Biochemical, antimicrobial and molecular characterization of a noncytotoxic bacteriocin produced by Lactobacillus plantarum ST71KS. Food Microbiol. 2013, 34, 376–381. [Google Scholar] [CrossRef]
- Peng, S.; Song, J.; Zeng, W.; Wang, H.; Zhang, Y.; Xin, J.; Suo, H. A broad-spectrum novel bacteriocin produced by Lactobacillus plantarum SHY 21–2 from yak yogurt: Purification, antimicrobial characteristics and antibacterial mechanism. LWT 2021, 142, 110955. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, N.; Qin, Y.; Zhang, Y.; Zhang, J.; Li, P. The complete genome sequence of Lactobacillus plantarum LPL-1, a novel antibacterial probiotic producing class IIa bacteriocin. J. Biotechnol. 2018, 266, 84–88. [Google Scholar] [CrossRef]
- Pei, J.; Jin, W.; Abd El-Aty, A.M.; Baranenko, D.A.; Gou, X.; Zhang, H.; Geng, J.; Jiang, L.; Chen, D.; Yue, T. Isolation, purification, and structural identification of a new bacteriocin made by Lactobacillus plantarum found in conventional kombucha. Food Control 2020, 110, 106923. [Google Scholar] [CrossRef]
- Manzoor, A.; Tayyeb, A. Functional probiotic attributes and gene encoding plantaracin among variant Lactobacillus plantarum strains. Microb. Pathog. 2019, 131, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Handa, S.; Sharma, N. In vitro study of probiotic properties of Lactobacillus plantarum F22 isolated from chhang—A traditional fermented beverage of Himachal Pradesh, India. J. Genet. Eng. Biotechnol. 2016, 14, 91–97. [Google Scholar] [CrossRef]
- Todorov, S.D.; Prévost, H.; Lebois, M.; Dousset, X.; LeBlanc, J.G.; Franco, B.D.G.M. Bacteriocinogenic Lactobacillus plantarum ST16Pa isolated from papaya (Carica papaya)—From isolation to application: Characterization of a bacteriocin. Food Res. Int. 2011, 44, 1351–1363. [Google Scholar] [CrossRef]
- Wen, L.S.; Philip, K.; Ajam, N. Purification, characterization and mode of action of plantaricin K25 produced by Lactobacillus plantarum. Food Control 2016, 60, 430–439. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Lactobacillus plantarum isolated from molasses produces bacteriocins active against Gram-negative bacteria. Enzyme Microb. Technol. 2005, 36, 318–326. [Google Scholar] [CrossRef]
- Powell, J.E.; Witthuhn, R.C.; Todorov, S.D.; Dicks, L.M.T. Characterization of bacteriocin ST8KF produced by a kefir isolate Lactobacillus plantarum ST8KF. Int. Dairy J. 2007, 17, 190–198. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Rehman, M.U.; Mehmood, K.; Jiang, X.; Iqbal, M.; Tong, X.; Gao, X.; Li, J. Antibacterial activity of Lactobacillus plantarum isolated from Tibetan yaks. Microbial Pathogenesis. Microb. Pathog. 2018, 115, 293–298. [Google Scholar] [CrossRef]
- Todorov, S.; Onno, B.; Sorokine, O.; Chobert, J.M.; Ivanova, I.; Dousset, X. Detection and characterization of a novel antibacterial substance produced by Lactobacillus plantarum ST31 isolated from sourdough. Int. J. Food Microbiol. 1999, 48, 167–177. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Effect of growth medium on bacteriocin production by Lactobacillus plantarum ST194BZ, a strain isolated from boza. Food Technol. Biotechnol. 2005, 43, 165–173. [Google Scholar]
- Hu, M.; Zhao, H.; Zhang, C.; Yu, J.; Lu, Z. Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. J. Agric. Food Chem. 2013, 61, 11676–11682. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, P.K.; Nehra, K. Purification and characterization of bacteriocin Bac23 extracted from Lactobacillus plantarum PKLP5 and its interaction with silver nanoparticles for enhanced antimicrobial spectrum against food-borne pathogens. LWT 2021, 139, 110546. [Google Scholar] [CrossRef]

| L. monocytogenes L7946 | L. monocytogenes L7947 | ||
|---|---|---|---|
| pH | 2 | 1.6% | 1.6% |
| 4 | 0.0% | 0.0% | |
| 6 | 0.0% | 0.0% | |
| 8 | 1.6% | 1.6% | |
| 10 | 12.5% | 12.5% | |
| 12 | 100.0% | 100.0% | |
| Temperature (°C) | 4 | 3.1% | 3.1% |
| 25 | 1.6% | 1.6% | |
| 30 and 37 | 0.0% | 0.0% | |
| 45 | 3.1% | 6.3% | |
| 60 | 3.1% | 6.3% | |
| 80 | 6.3% | 6.3% | |
| 100 | 12.5% | 12.5% | |
| 121 | 100.0% | 100.0% | |
| Enzymes (mg/mL) | Proteinase K1.0and0.1 | 100.0% | 100.0% |
| Papain1.0 | 50.0% | 50.0% | |
| Papain0.1 | 50.0% | 50.0% | |
| Pepsin1.0 | 100.0% | 100.0% | |
| Pepsin0.1 | 50.0% | 25.0% | |
| Trypsin1.0and0.1 | 100.0% | 100.0% | |
| α-amylase1.0 | 6.3% | 25.0% | |
| α-amylase0.1 | 0.0% | 0.0% | |
| Catalase1.0and0.1 | 0.0% | 0.0% | |
| Detergents | Tween 20 and Tween 800.01g/mL | 3.1% | 3.1% |
| Triton X-114 and Triton X-1000.01g/mL | 50.0% | 50.0% | |
| SDS0.01g/mL | 0.0% | 1.6% | |
| EDTA0.1mM | 3.1% | 3.1% | |
| EDTA2.0mM | 3.1% | 3.1% | |
| EDTA5.0mM | 6.3% | 6.3% | |
| Ox-bile0.01g/mL | 1.6% | 1.6% | |
| Urea and NaCl0.01g/mL | 0.0% | 0.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, J.; Albano, H.; Silva, B.; Almeida, M.H.; Nogueira, T.; Teixeira, P. Characterization of a Lactiplantibacillus plantarum R23 Isolated from Arugula by Whole-Genome Sequencing and Its Bacteriocin Production Ability. Int. J. Environ. Res. Public Health 2021, 18, 5515. https://doi.org/10.3390/ijerph18115515
Barbosa J, Albano H, Silva B, Almeida MH, Nogueira T, Teixeira P. Characterization of a Lactiplantibacillus plantarum R23 Isolated from Arugula by Whole-Genome Sequencing and Its Bacteriocin Production Ability. International Journal of Environmental Research and Public Health. 2021; 18(11):5515. https://doi.org/10.3390/ijerph18115515
Chicago/Turabian StyleBarbosa, Joana, Helena Albano, Beatriz Silva, Maria Helena Almeida, Teresa Nogueira, and Paula Teixeira. 2021. "Characterization of a Lactiplantibacillus plantarum R23 Isolated from Arugula by Whole-Genome Sequencing and Its Bacteriocin Production Ability" International Journal of Environmental Research and Public Health 18, no. 11: 5515. https://doi.org/10.3390/ijerph18115515
APA StyleBarbosa, J., Albano, H., Silva, B., Almeida, M. H., Nogueira, T., & Teixeira, P. (2021). Characterization of a Lactiplantibacillus plantarum R23 Isolated from Arugula by Whole-Genome Sequencing and Its Bacteriocin Production Ability. International Journal of Environmental Research and Public Health, 18(11), 5515. https://doi.org/10.3390/ijerph18115515








