Response of the Intertidal Microbial Community Structure and Metabolic Profiles to Zinc Oxide Nanoparticle Exposure
Abstract
1. Introduction
2. Materials and Methods
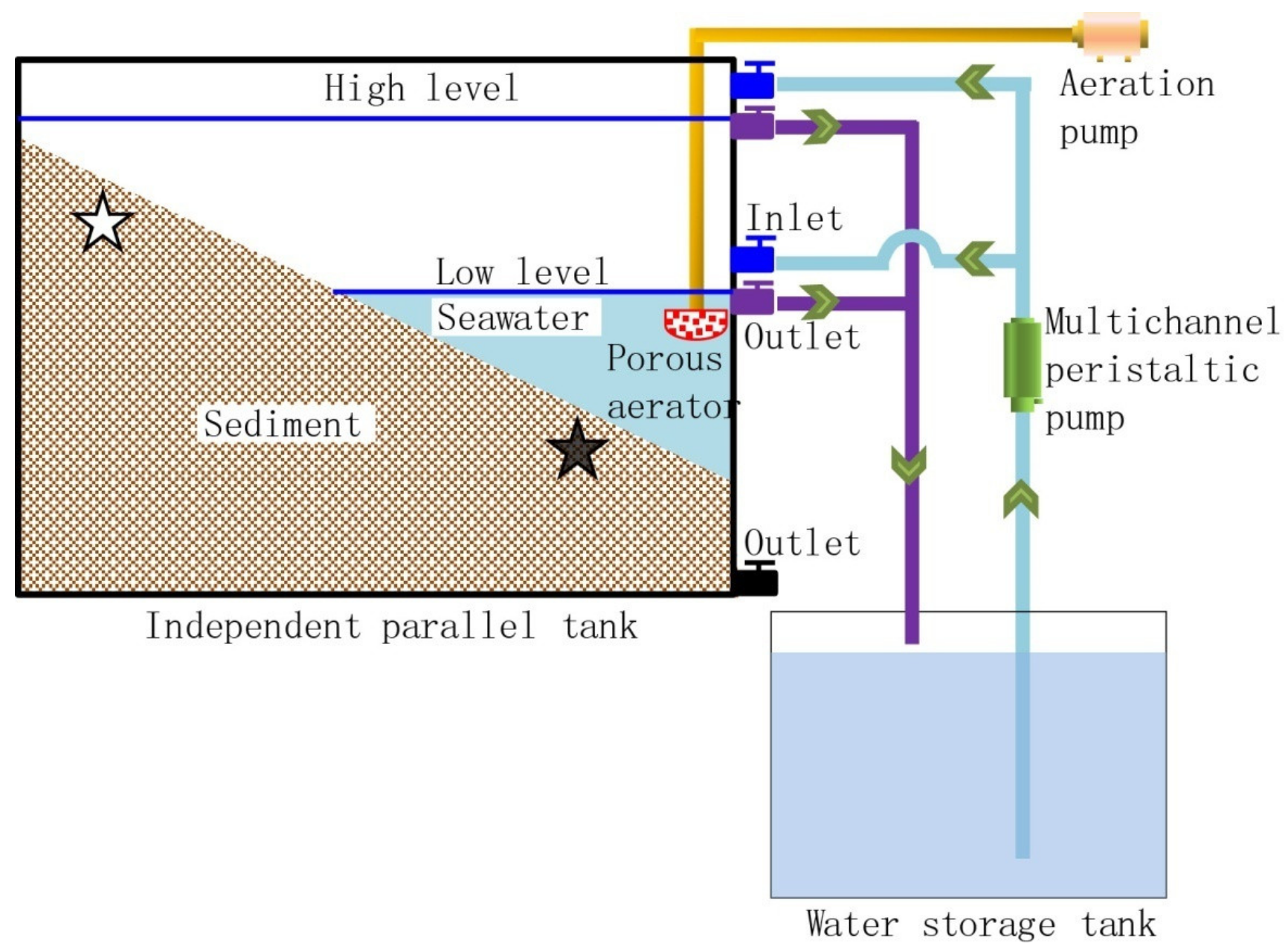
2.1. Reactor, Operation, and Sampling
2.2. DNA Extraction and High-Throughput Sequencing
2.3. High-Throughput Sequencing Data Analysis
2.4. Microbial Metabolic Activity Analysis
2.5. Physicochemical Analysis
2.6. Calculation and Statistical Analyses
3. Results
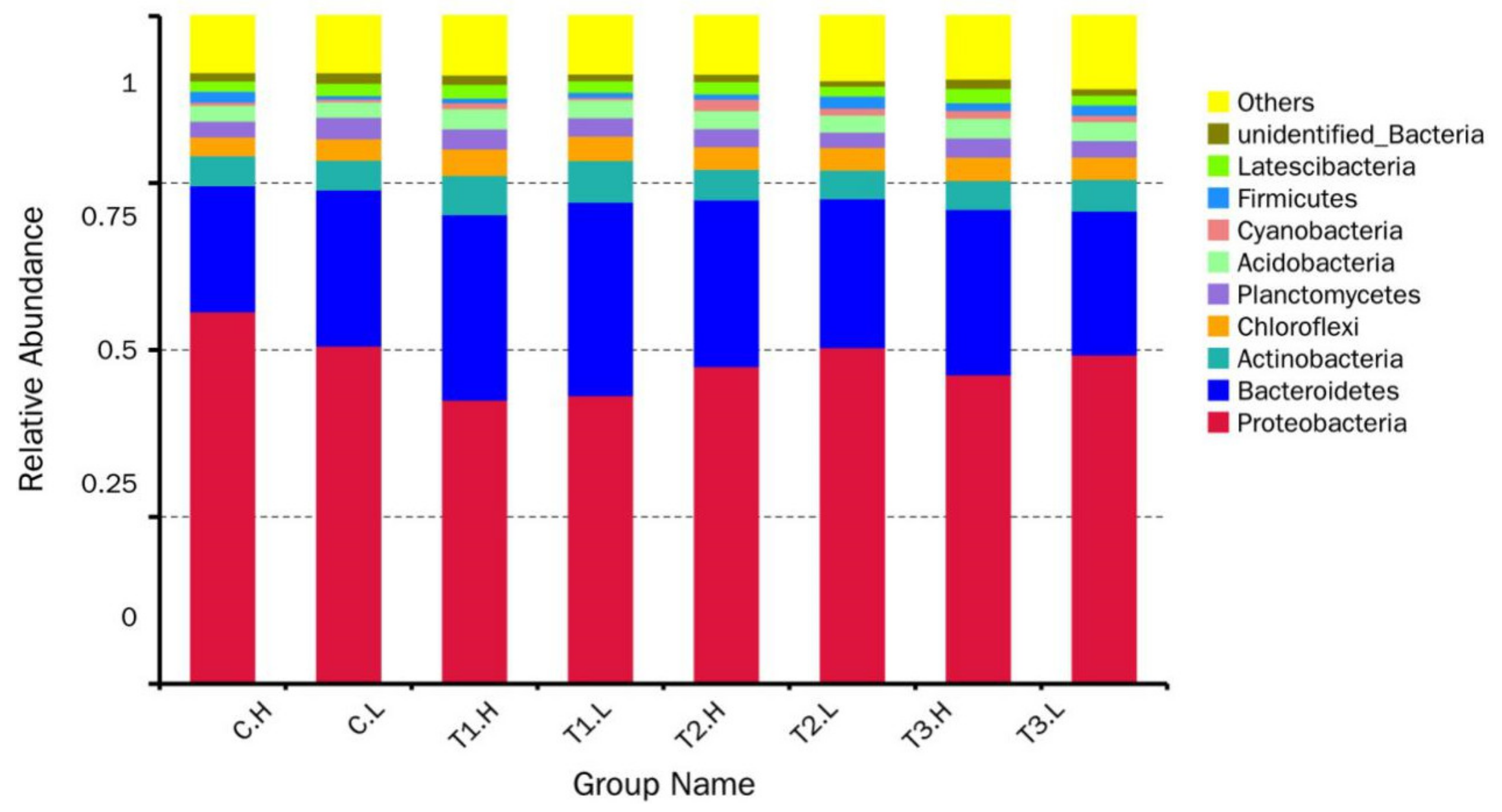
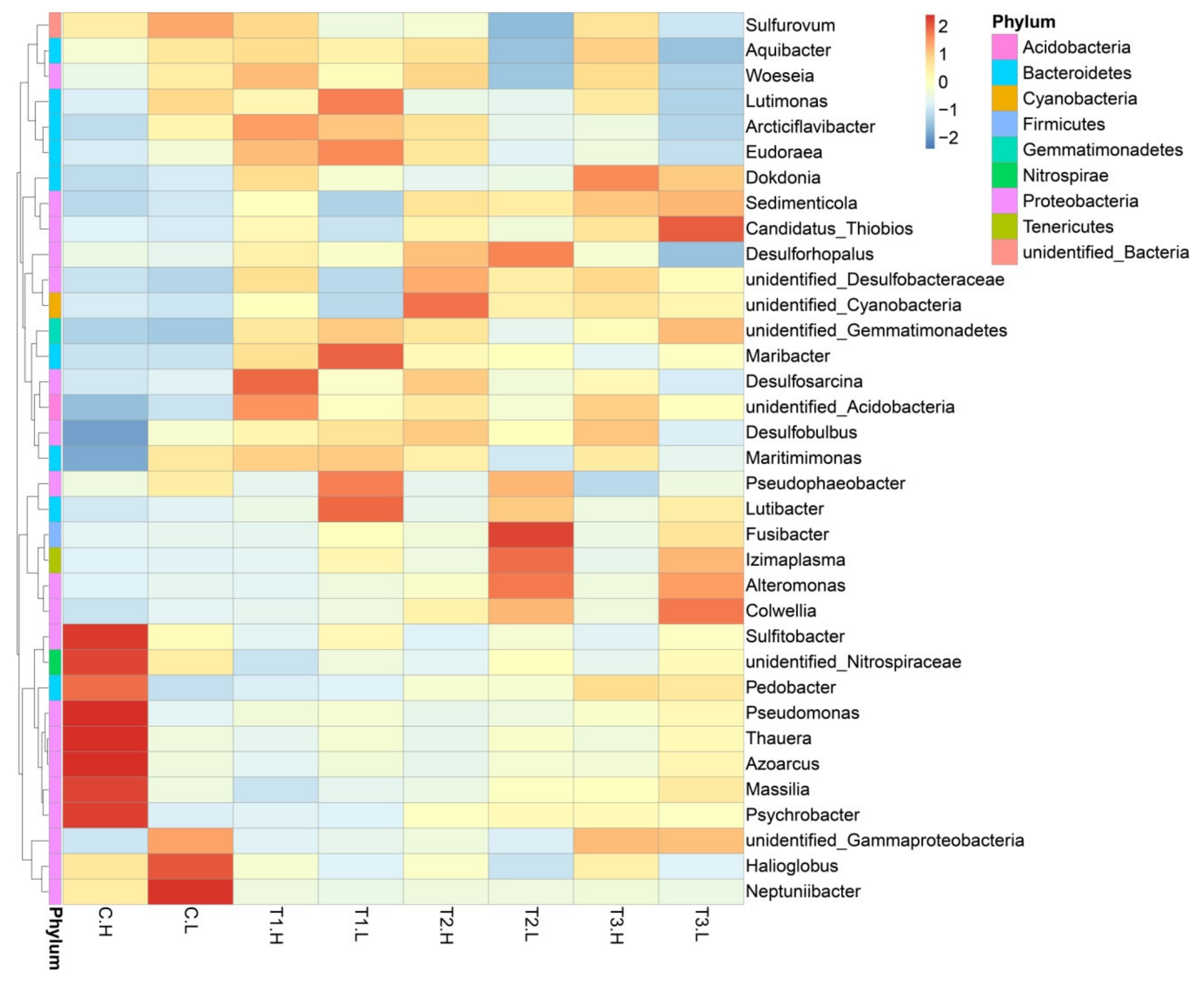
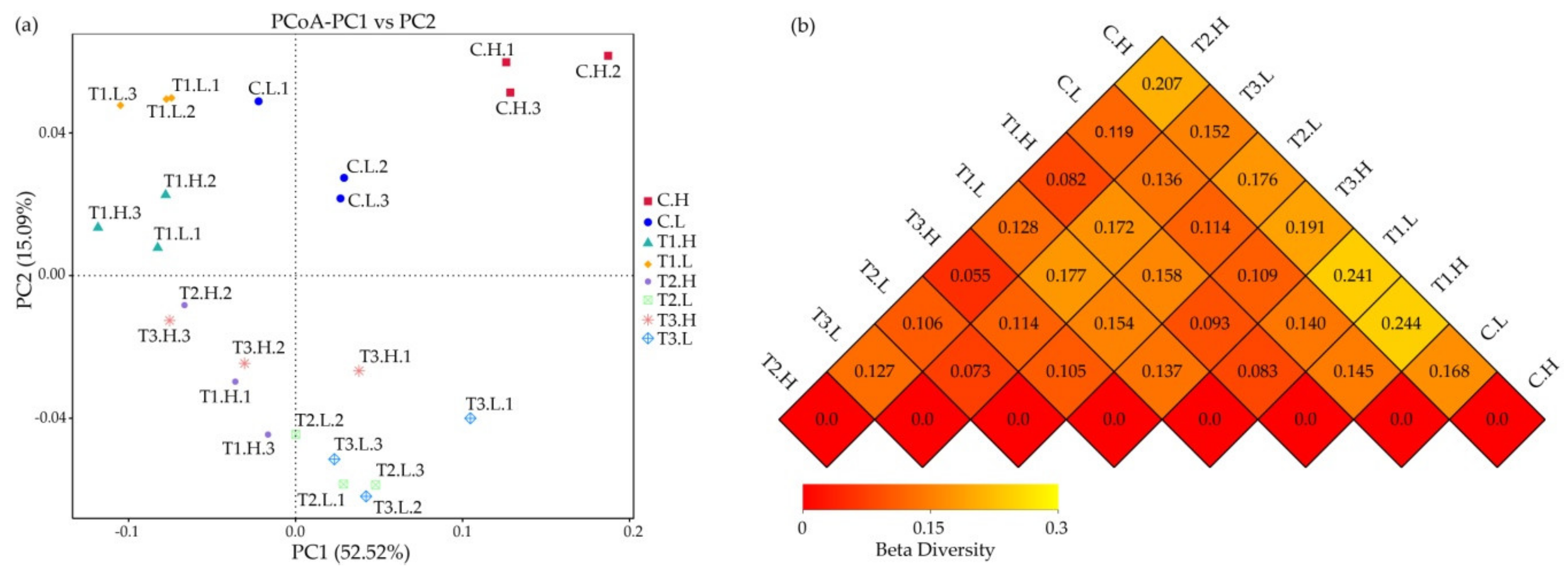
3.1. Microbial Community Composition
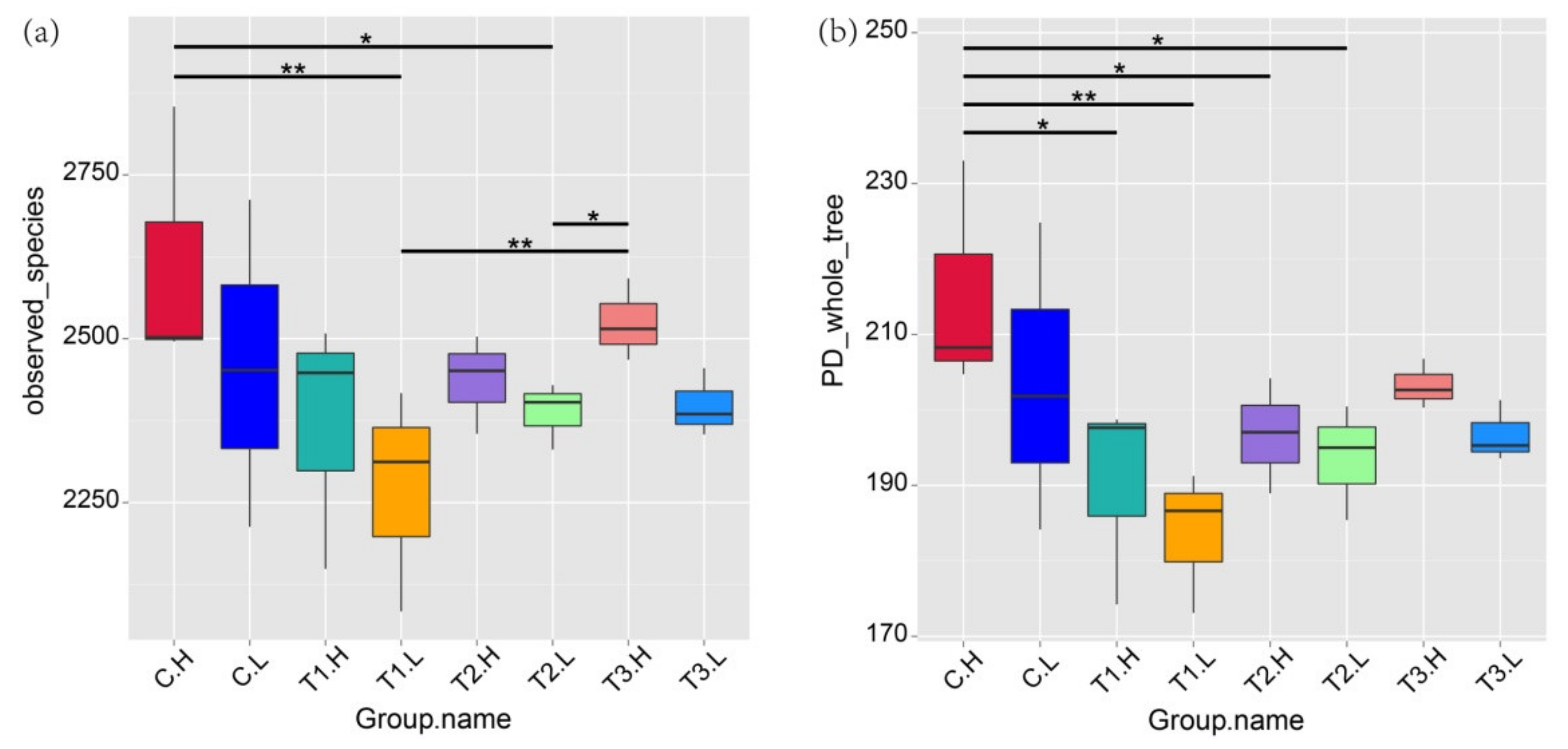
3.2. Microbial Community Diversity
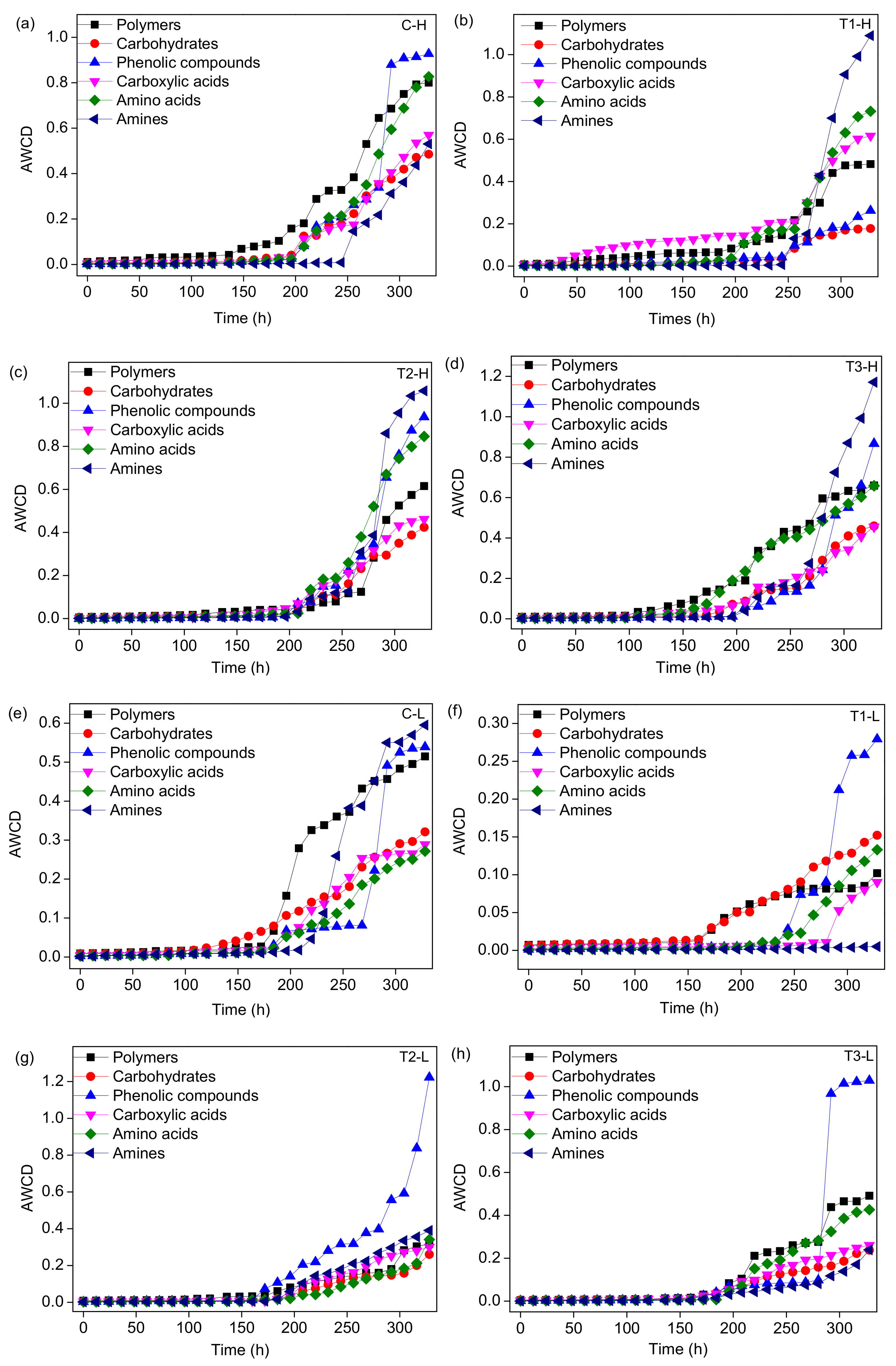
3.3. Microbial Metabolic Activity
3.4. Physicochemical Properties of the Sediment
4. Discussion
4.1. Microbial Community Shift with Concentrations of ZnO NPs
4.2. Shift of Microbial CLPP with Concentrations of ZnO NPs
4.3. The Effects of Environmental Factors on the Ecotoxicity of ZnO NPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Auffan, M.; Rose, J.; Bottero, J.Y.; Lowry, G.V.; Jolivet, J.P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Sharma, V.; Parmar, D. Nanomaterials: A challenge for toxicologists. Nanotoxicology 2009, 3, 1–9. [Google Scholar] [CrossRef]
- Farré, M.; Gajda-Schrantz, K.; Kantiani, L.; Barceló, D. Ecotoxicity and analysis of nanomaterials in the aquatic environment. Anal. Bioanal. Chem. 2009, 393, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, J.; Chen, M.; Liu, X.; Xiong, Y.; Wang, Y.; Feng, T.; Kang, S.; Wang, X. Recent advances in the biotoxicity of metal oxide nanoparticles: Impacts on plants, animals and microorganisms. Chemosphere 2019, 237, 124403. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Williams, P.L.; Diamond, S.A. Ecotoxicity of manufactured ZnO nanoparticles-a review. Environ. Pollut. 2013, 172, 76–85. [Google Scholar] [CrossRef]
- Ge, Y.; Schimel, J.P.; Holden, P.A. Evidence for negative effects of TiO2 and ZnO nanoparticles on soil bacterial communities. Environ. Sci. Technol. 2011, 45, 1659–1664. [Google Scholar] [CrossRef]
- Gottschalk, F.; Sun, T.; Nowack, B. Environmental concentrations of engineered nanomaterials: Review of modeling and analytical studies. Environ. Pollut. 2013, 181, 287–300. [Google Scholar] [CrossRef]
- Aravantinou, A.F.; Tsarpali, V.; Dailianis, S.; Manariotis, I.D. Effect of cultivation media on the toxicity of ZnO nanoparticles to freshwater and marine microalgae. Ecotox. Environ. Saf. 2015, 114, 109–116. [Google Scholar] [CrossRef]
- Sarkheil, M.; Johari, S.A.; An, H.J.; Asghari, S.; Park, H.S.; Sohn, E.K.; Yu, I.J. Acute toxicity, uptake, and elimination of zinc oxide nanoparticles (ZnO NPs) using saltwater microcrustacean, artemia franciscana. Environ. Toxicol. Phar. 2018, 57, 181–188. [Google Scholar] [CrossRef]
- Wong, S.W.; Leung, P.T.; Djurisic, A.B.; Leung, K.M. Toxicities of nano zinc oxide to five marine organisms: Influences of aggregate size and ion solubility. Anal. Bioanal. Chem. 2010, 396, 609–618. [Google Scholar] [CrossRef]
- Sulu-Gambari, F.; Seitaj, D.; Meysman, F.J.R.; Schauer, R.; Polerecky, L.; Slomp, C.P. Cable bacteria control iron-phosphorus dynamics in sediments of a coastal hypoxic basin. Environ. Sci. Technol. 2016, 50, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.K.; Lyon, D.Y.; Alvarez, P.J.J. Comparative eco-toxicity of nanoscale TiO2, SiO2, and ZnO water suspensions. Water Res. 2006, 40, 3532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, Y.; Ding, Y.; Povey, M.; York, D. Investigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids). J. Nanopart. Res. 2007, 9, 479–489. [Google Scholar] [CrossRef]
- Applerot, G.; Lipovsky, A.; Dror, R.; Perkas, N.; Nitzan, Y.; Lubart, R.; Gedanken, A. Enhanced antibacterial activity of nanocrystalline ZnO due to increased ros-mediated cell injury. Adv. Funct. Mater. 2009, 19, 842–852. [Google Scholar] [CrossRef]
- Zheng, X.; Wu, R.; Chen, Y. Effects of ZnO nanoparticles on wastewater biological nitrogen and phosphorus removal. Environ. Sci. Technol. 2011, 45, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pokhrel, S.; Jin, X.; Mädler, L.; Damoiseaux, R.; Hoek, E.M.V. Stability, bioavailability, and bacterial toxicity of ZnO and iron-doped ZnO nanoparticles in aquatic media. Environ. Sci. Technol. 2011, 45, 755–761. [Google Scholar] [CrossRef]
- Park, S.J.; Park, Y.C.; Lee, S.W.; Jeong, M.S.; Yu, K.N.; Jung, H.; Lee, J.K.; Kim, J.S.; Cho, M.H. Comparing the toxic mechanism of synthesized zinc oxide nanomaterials by physicochemical characterization and reactive oxygen species properties. Toxcol. Lett. 2011, 207, 0–203. [Google Scholar] [CrossRef]
- Han, Y.; Hwang, G.; Kim, D.; Bradford, S.A.; Lee, B.; Eom, L.; Kim, P.J.; Choi, S.Q.; Kim, H. Transport, retention, and long-term release behavior of ZnO nanoparticle aggregates in saturated quartz sand: Role of solution ph and biofilm coating. Water Res. 2016, 90, 247–257. [Google Scholar] [CrossRef]
- Du, W.; Sun, Y.; Ji, R.; Zhu, J.; Wu, J.; Guo, H. TiO2 and ZnO nanoparticles negatively affect wheat growth and soil enzyme activities in agricultural soil. J. Environ. Monit. 2011, 13, 822–828. [Google Scholar] [CrossRef]
- Wu, Y.; He, T.; Chen, C.; Fang, X.; Wei, D.; Yang, J.; Zhang, R.; Han, R. Impacting microbial communities and absorbing pollutants by canna indica and cyperus alternifolius in a full-scale constructed wetland system. Int. J. Environ. Res. Public Health 2019, 16, 802. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef]
- Kachurina, O.M.; Zhang, H.; Raun, W.R.; Krenzer, E.G. Simultaneous determination of soil aluminum, ammonium- and nitrate-nitrogen using 1 M potassium chloride extraction. Commun. Soil. Sci. Plan. 2000, 31, 893–903. [Google Scholar] [CrossRef]
- Ren, J.; Wei, C.; Ma, H.; Dai, M.; Fan, J.; Liu, Y.; Wu, Y.; Han, R. The nitrogen-removal efficiency of a novel high-efficiency salt-tolerant aerobic denitrifier, halomonas alkaliphile hrl-9, isolated from a seawater biofilter. Int. J. Environ. Res. Public Health 2019, 16, 4451. [Google Scholar] [CrossRef]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microb. 1991, 57, 2351–2359. [Google Scholar] [CrossRef]
- Garland, J.L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 1997, 24, 289–300. [Google Scholar] [CrossRef]
- Classen, A.T.; Boyle, S.I.; Haskins, K.E.; Overby, S.T.; Hart, S.C. Community-level physiological profiles of bacteria and fungi: Plate type and incubation temperature influences on contrasting soils. FEMS Microbiol. Ecol. 2003, 44, 319–328. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, H.; Liu, J.; Wang, Q.; Shen, T.; Guo, W.; Wang, R. Shifts in microbial community function and structure along the successional gradient of coastal wetlands in yellow river estuary. Eur. J. Soil Biol. 2012, 49, 0–21. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Badireddy, A.R.; Hotze, E.M.; Chellam, S.; Alvarez, P.; Wiesner, M.R. Inactivation of bacteriophages via photosensitization of fullerol nanoparticles. Environ. Sci. Technol. 2007, 41, 6627–6632. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zhang, Y.; Hu, Z. Bacteria and bacteriophage inactivation by silver and zinc oxide nanoparticles. Colloid Surf. B 2011, 85, 161–167. [Google Scholar] [CrossRef]
- Wu, Y.; Han, R.; Yang, X.; Fang, X.; Chen, X.; Yang, D.; Zhang, R. Correlating microbial community with physicochemical indices and structures of a full-scale integrated constructed wetland system. Appl. Microbiol. Biot. 2016, 100, 6917–6926. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, L.; Lin, D. Toxicity of ZnO nanoparticles to Escherichia coli: Mechanism and the influence of medium components. Environ. Sci. Technol. 2011, 45, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lin, D.; Zhu, L. Effects of water chemistry on the dissolution of ZnO nanoparticles and their toxicity to Escherichia coli. Environ. Pollut. 2013, 173, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Batley, G.E.; Kirby, J.K.; McLaughlin, M.J. Fate and risks of nanomaterials in aquatic and terrestrial environments. Acc. Chem. Res. 2013, 46, 854–862. [Google Scholar] [CrossRef]
- Tony, J.B.; Charles, R.T.; Tamara, S.G. Impacts of metal and metal oxide nanoparticles on marine organisms. Environ. Pollut. 2014, 186, 257–271. [Google Scholar] [CrossRef]
- Fairbairn, E.A.; Kellern, A.A.; Madler, L.; Zhou, D.; Pokhrel, S.; Cherr, G.N. Metal oxide nanomaterials in seawater: Linking physicochemical characteristics with biological response in sea urchin development. J. Hazard Mater. 2011, 192, 1565–1571. [Google Scholar] [CrossRef]
- Miller, R.J.; Lenihan, H.S.; Muller, E.B.; Tseng, N.; Hanna, S.K.; Keller, A.A. Impacts of metal oxide nanoparticles on marine phytoplankton. Environ. Sci. Technol. 2010, 44, 7329–7334. [Google Scholar] [CrossRef]
- Kumar, A.; Pandey, A.K.; Singh, S.S.; Shanker, R.; Dhawan, A. Cellular uptake and mutagenic potential of metal oxide nanoparticles in bacterial cells. Chemosphere 2011, 83, 1124–1132. [Google Scholar] [CrossRef]
- Mariappan, P.P.; Krishnamoorthy, K.M.; Kadarkaraithangam, J.P.; Govindasamy, M.M. Selective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidation. Nanomedcine 2011, 7, 184–192. [Google Scholar] [CrossRef]
- Ge, Z.; Du, H.; Gao, Y.; Qiu, W. Analysis on metabolic functions of stored rice microbial communities by BIOLOG ECO microplates. Front. Microbiol. 2018, 9, 1375. [Google Scholar] [CrossRef]
- Nair, A.; Ngouajio, M. Soil microbial biomass, functional microbial diversity, and nematode community structure as affected by cover crops and compost in an organic vegetable production system. Appl. Soil Ecol. 2012, 58, 45–55. [Google Scholar] [CrossRef]
- Park, M.H.; Igarashi, K. Polyamines and their metabolites as diagnostic markers of human diseases. Biomol. Ther. 2013, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sala, M.M.; Arin, L.; Balagué, V.; Felipe, J.; Guadayol, Ò.; Vaqué, D. Functional diversity of bacterioplankton assemblages in western antarctic seawaters during late spring. Mar. Ecol. Prog. Ser. 2005, 292, 13–21. [Google Scholar] [CrossRef]
- He, Z.; Deng, Y.; Van Nostrand, J.D.; Tu, Q.; Xu, M.; Hemme, C.L.; Li, X.; Wu, L.; Gentry, T.J.; Yin, Y.; et al. Geochip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010, 4, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298. [Google Scholar] [CrossRef]
- Bakhtchadjian, R.; Manucharova, L.; Tavadyan, L. Selective oxidation of ddt by dioxygen on the dioxo-mo (vi) complex anchored on a TiO2 under uv-irradiation. Catal. Commun. 2015, 69, 193–195. [Google Scholar] [CrossRef]
- Majedi, S.M.; Kelly, B.C.; Lee, H.K. Combined effects of water temperature and chemistry on the environmental fate and behavior of nanosized zinc oxide. J. Hazard. Mater. 2014, 264, 370–379. [Google Scholar] [CrossRef]
- Majedi, S.M.; Kelly, B.C.; Lee, H.K. Role of combinatorial environmental factors in the behavior and fate of ZnO nanoparticles in aqueous systems: A multiparametric analysis. J. Hazard. Mater. 2014, 264, 370–379. [Google Scholar] [CrossRef]
- Murphy, R.R.; Kemp, W.M.; Ball, W.P. Long-term trends in chesapeake bay seasonal hypoxia, stratification, and nutrient loading. Estuar. Coast. 2011, 34, 1293–1309. [Google Scholar] [CrossRef]
- Costa, A.L.; Paixão, S.M.; Caçador, I.; Carolino, M. ClPP and EEA profiles of microbial communities in salt marsh sediments. J. Soils Sediments 2007, 7, 418–425. [Google Scholar] [CrossRef]

| Group Name | Temperature (°C) | DO (mg·L−1) | pH | Salinity (PPT) | NH4-N (mg·L−1) |
|---|---|---|---|---|---|
| C-L | 11.1 | 0.22 | 8.29 | 5.77 | 0.11 |
| T1-L | 11.2 | 0.19 | 8.29 | 5.92 | 0.17 |
| T2-L | 11.1 | 0.20 | 8.28 | 5.96 | 0.11 |
| T3-L | 11.0 | 0.20 | 8.30 | 5.98 | 0.11 |
| C-H | 10.1 | 6.60 | 8.33 | 5.97 | 0.11 |
| T1-H | 10.0 | 6.52 | 8.34 | 5.80 | 0.21 |
| T2-H | 10.2 | 6.55 | 8.33 | 5.79 | 0.17 |
| T3-H | 10.1 | 6.56 | 8.34 | 5.83 | 0.08 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Rong, X.; Zhang, C.; Zhang, R.; He, T.; Yu, Y.; Zhao, Z.; Yang, J.; Han, R. Response of the Intertidal Microbial Community Structure and Metabolic Profiles to Zinc Oxide Nanoparticle Exposure. Int. J. Environ. Res. Public Health 2020, 17, 2253. https://doi.org/10.3390/ijerph17072253
Wu Y, Rong X, Zhang C, Zhang R, He T, Yu Y, Zhao Z, Yang J, Han R. Response of the Intertidal Microbial Community Structure and Metabolic Profiles to Zinc Oxide Nanoparticle Exposure. International Journal of Environmental Research and Public Health. 2020; 17(7):2253. https://doi.org/10.3390/ijerph17072253
Chicago/Turabian StyleWu, Yinghai, Xinyu Rong, Cuiya Zhang, Renduo Zhang, Tao He, Yunjun Yu, Zhuangming Zhao, Jing Yang, and Rui Han. 2020. "Response of the Intertidal Microbial Community Structure and Metabolic Profiles to Zinc Oxide Nanoparticle Exposure" International Journal of Environmental Research and Public Health 17, no. 7: 2253. https://doi.org/10.3390/ijerph17072253
APA StyleWu, Y., Rong, X., Zhang, C., Zhang, R., He, T., Yu, Y., Zhao, Z., Yang, J., & Han, R. (2020). Response of the Intertidal Microbial Community Structure and Metabolic Profiles to Zinc Oxide Nanoparticle Exposure. International Journal of Environmental Research and Public Health, 17(7), 2253. https://doi.org/10.3390/ijerph17072253





