Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood
Abstract
1. Introduction
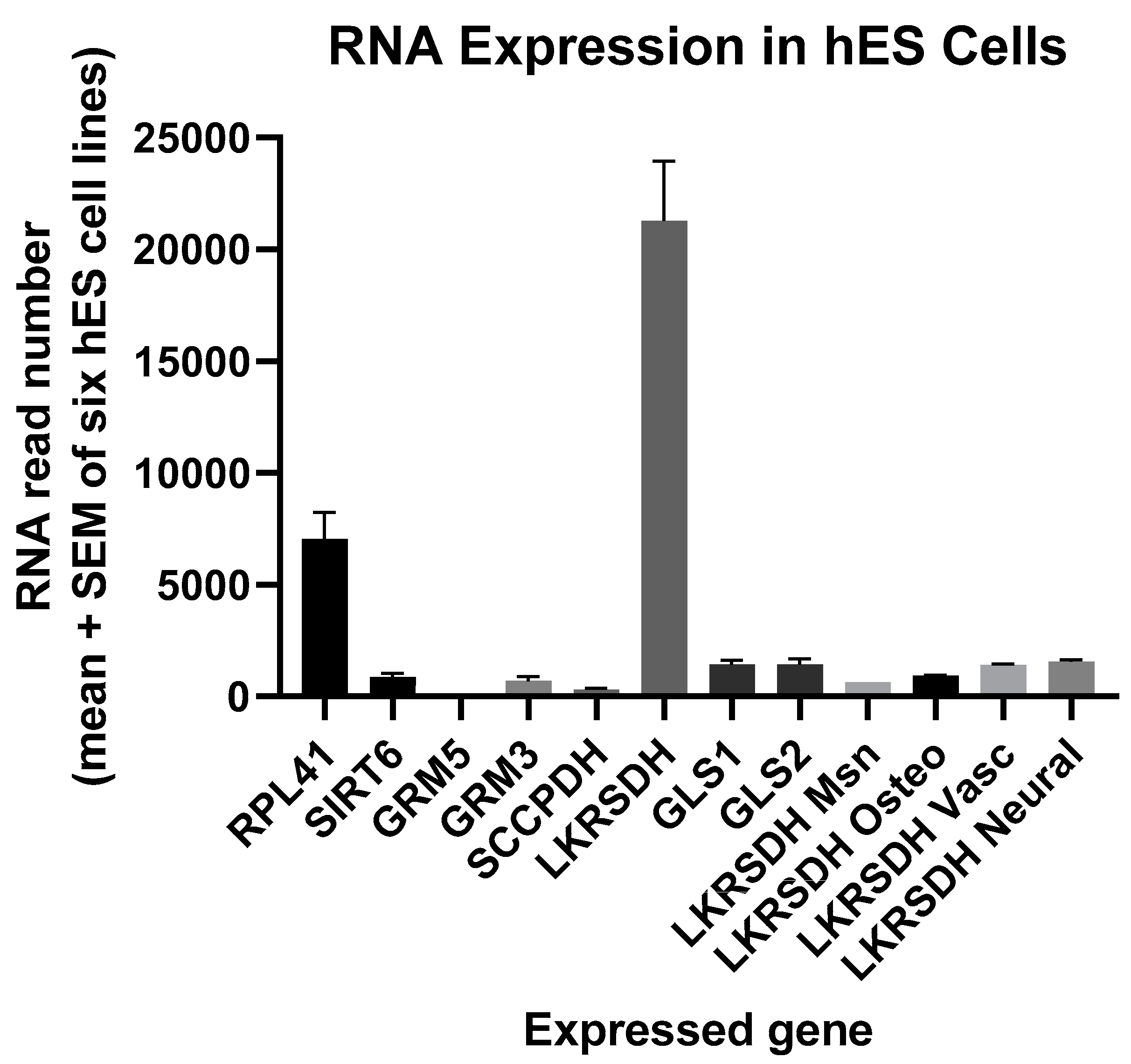
- Is RNA, encoding the protein that regulates glutamate synthesis from lysine (alpha-aminoadipic semialdehyde synthase), relatively abundant in hES cells?
- Is the amount of lysine consumed by hES cells equal to or greater than the amount of glutamate the cells produce?
- Is RNA encoding one or more metabotropic glutamate receptors expressed in hES cells?
2. Materials and Methods
2.1. hES Cell Culture
2.2. Mesenchymal Differentiation
2.3. Osteogenic Differentiation
2.4. Vascular Differentiation
2.5. Neural Differentiation
2.6. RNA Isolation
2.7. Microarray Data Preparation
2.8. RNA Sequencing Analysis
2.9. Amino Acid Production/Utilization
2.10. Statistical Analyses
3. Results
3.1. Expression of RNA Encoding Alpha-Aminoadipic Semialdehyde Synthase (LKRSDH)
3.2. Production/Utilization of Glutamate and Lysine by hES Cells (H9 Cell Line)
3.3. hES Cells Express at Least Two Metabotropic Glutamate Receptors
4. Discussion
5. Limitations/Future Directions
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Winkle, L.J. Amino acid transport regulation and early embryo development. Biol. Reprod. 2001, 64, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.K.; Lane, M. Alleviation of the ‘2-cell block’and development to the blastocyst of CF1 mouse embryos: Role of amino acids, EDTA and physical parameters. Hum. Reprod. 1996, 11, 2703–2712. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.; Gardner, D.K. Differential regulation of mouse embryo development and viability by amino acids. Reproduction 1997, 109, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.M.; Sutherland, A.E.; Van Winkle, L.J. Amino acid transport regulates blastocyst implantation. Biol. Reprod. 2003, 69, 1101–1108. [Google Scholar] [CrossRef]
- Van Winkle, L.J.; Ryznar, R. Amino acid transporters: Roles for nutrition, signaling and epigenetic modifications in embryonic stem cells and their progenitors. In eLS 2019; John Wiley & Sons, Ltd.: Chichester, UK, 2019; pp. 1–3. [Google Scholar]
- Van Winkle, L.J.; Ryznar, R. One-carbon metabolism regulates embryonic stem cell fate through epigenetic DNA and histone modifications: Implications for transgenerational metabolic disorders in adults. Front. Cell Dev. Biol. 2019, 7, 300. [Google Scholar] [CrossRef]
- Kaye, P.L.; Schultz, G.A.; Johnson, M.H.; Pratt, H.P.; Church, R.B. Amino acid transport and exchange in preimplantation mouse embyros. Reproduction 1982, 65, 367–380. [Google Scholar] [CrossRef]
- Van Winkle, L.J.; Campione, A.L.; Gorman, J.M.; Weimer, B.D. Changes in the activities of amino acid transport systems b0,+ and L during development of preimplantation mouse conceptuses. Biochim. Biophys. Acta (BBA)-Biomembr. 1990, 1021, 77–84. [Google Scholar] [CrossRef]
- Van Winkle, L.J. Biomembrane Transport, 1st ed.; Academic Press: San Diego, CA, USA, 1999; pp. 120–124. [Google Scholar]
- Eckert, J.J.; Porter, R.; Watkins, A.J.; Burt, E.; Brooks, S.; Leese, H.J.; Humpherson, P.G.; Cameron, I.T.; Fleming, T.P. Metabolic induction and early responses of mouse blastocyst developmental programming following maternal low protein diet affecting life-long health. PLoS ONE 2012, 7, e52791. [Google Scholar] [CrossRef]
- Fleming, T.P.; Watkins, A.J.; Velazquez, M.A.; Mathers, J.C.; Prentice, A.M.; Stephenson, J.; Barker, M.; Saffery, R.; Yajnik, C.S.; Eckert, J.J.; et al. Origins of lifetime health around the time of conception: Causes and consequences. Lancet 2018, 391, 1842–1852. [Google Scholar] [CrossRef]
- Wang, J.; Alexander, P.; Wu, L.; Hammer, R.; Cleaver, O.; McKnight, S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009, 325, 435–439. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Papes, F.; Surpili, M.J.; Langone, F.; Trigo, J.R.; Arruda, P. The essential amino acid lysine acts as precursor of glutamate in the mammalian central nervous system. FEBS Lett. 2001, 488, 34–38. [Google Scholar] [CrossRef]
- Sacksteder, K.A.; Biery, B.J.; Morrell, J.C.; Goodman, B.K.; Geisbrecht, B.V.; Cox, R.P.; Gould, S.J.; Geraghty, M.T. Identification of the α-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am. J. Hum. Genet. 2000, 66, 1736–1743. [Google Scholar] [CrossRef] [PubMed]
- Crowther, L.M.; Mathis, D.; Poms, M.; Plecko, B. New insights into human lysine degradation pathways with relevance to pyridoxine-dependent epilepsy due to antiquitin deficiency. J. Inherit. Metab. Dis. 2019, 42, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.; Maussion, G.; Jefri, M.; Peng, H.; Theroux, J.F.; Silveira, H.; Soubannier, V.; Wu, H.; Hu, P.; Galat, E.; et al. Disruption of GRIN2B impairs differentiation in human neurons. Stem Cell Rep. 2018, 11, 183–196. [Google Scholar] [CrossRef]
- McKenna, M.C. The glutamate-glutamine cycle is not stoichiometric: Fates of glutamate in brain. J. Neurosci. Res. 2007, 85, 3347–3358. [Google Scholar] [CrossRef]
- Cappuccio, I.; Spinsanti, P.; Porcellini, A.; Desiderati, F.; De Vita, T.; Storto, M.; Capobianco, L.; Battaglia, G.; Nicoletti, F.; Melchiorri, D. Endogenous activation of mGlu5 metabotropic glutamate receptors supports self-renewal of cultured mouse embryonic stem cells. Neuropharmacology 2005, 49, 196–205. [Google Scholar] [CrossRef]
- Spinsanti, P.; De Vita, T.; Di Castro, S.; Storto, M.; Formisano, P.; Nicoletti, F.; Melchiorri, D. Endogenously activated mGlu5 metabotropic glutamate receptors sustain the increase in c-Myc expression induced by leukaemia inhibitory factor in cultured mouse embryonic stem cells. J. Neurochem. 2006, 99, 299–307. [Google Scholar] [CrossRef]
- Hashimoto, S.; Nishihara, T.; Murata, Y.; Oku, H.; Fukuda, A.; Morimoto, Y. Novel stable glutamine derivative achieved dramatic improvement of developmental competence of human embryos. Fertil. Steril. 2007, 88, S66–S67. [Google Scholar] [CrossRef]
- Hashimoto, S.; Nishihara, T.; Murata, Y.; Oku, H.; Nakaoka, Y.; Fukuda, A.; Morimoto, Y. Medium without ammonium accumulation supports the developmental competence of human embryos. J. Reprod. Dev. 2008, 54, 370–374. [Google Scholar] [CrossRef][Green Version]
- Galat, V.; Malchenko, S.; Galat, Y.; Ishkin, A.; Nikolsky, Y.; Kosak, S.T.; Soares, B.M.; Iannaccone, P.; Crispino, J.D.; Hendrix, M.J. A model of early human embryonic stem cell differentiation reveals inter-and intracellular changes on transition to squamous epithelium. Stem Cells Dev. 2012, 21, 1250–1263. [Google Scholar] [CrossRef] [PubMed]
- Galat, V.; Galat, Y.; Perepitchka, M.; Jennings, L.J.; Iannaccone, P.M.; Hendrix, M.J. Transgene reactivation in induced pluripotent stem cell derivatives and reversion to pluripotency of induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cells Dev. 2016, 25, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Galat, Y.; Dambaeva, S.; Elcheva, I.; Khanolkar, A.; Beaman, K.; Iannaccone, P.M.; Galat, V. Cytokine-free directed differentiation of human pluripotent stem cells efficiently produces hemogenic endothelium with lymphoid potential. Stem Cell Res. Ther. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali-Ellis, Z.; Galat, V.; Galat, Y.; Gilgur, A.; Seftor, E.A.; Hendrix, M.J. Lefty glycoproteins in human embryonic stem cells: Extracellular delivery route and posttranslational modification in differentiation. Stem Cells Dev. 2016, 25, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Van Winkle, L.J.; Dickinson, H.R. Differences in amino acid content of preimplantation mouse embryos that develop in vitro versus in vivo: In vitro effects of five amino acids that are abundant in oviductal secretions. Biol. Reprod. 1995, 52, 96–104. [Google Scholar] [CrossRef] [PubMed]
- De La Barca, J.M.C.; Arrázola, M.S.; Bocca, C.; Arnauné-Pelloquin, L.; Iuliano, O.; Tcherkez, G.; Lenaers, G.; Simard, G.; Belenguer, P.; Reynier, P. The metabolomic signature of Opa1 deficiency in rat primary cortical neurons shows aspartate/glutamate depletion and phospholipids remodeling. Sci. Rep. 2019, 9, 6107. [Google Scholar] [CrossRef]
- Galili, G. New insights into the regulation and functional significance of lysine metabolism in plants. Annu. Rev. Plant Biol. 2002, 53, 27–43. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Winkle, L.J.; Galat, V.; Iannaccone, P.M. Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood. Int. J. Environ. Res. Public Health 2020, 17, 5462. https://doi.org/10.3390/ijerph17155462
Van Winkle LJ, Galat V, Iannaccone PM. Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood. International Journal of Environmental Research and Public Health. 2020; 17(15):5462. https://doi.org/10.3390/ijerph17155462
Chicago/Turabian StyleVan Winkle, Lon J., Vasiliy Galat, and Philip M. Iannaccone. 2020. "Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood" International Journal of Environmental Research and Public Health 17, no. 15: 5462. https://doi.org/10.3390/ijerph17155462
APA StyleVan Winkle, L. J., Galat, V., & Iannaccone, P. M. (2020). Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood. International Journal of Environmental Research and Public Health, 17(15), 5462. https://doi.org/10.3390/ijerph17155462






