Subgingival Microbiota of Mexicans with Type 2 Diabetes with Different Periodontal and Metabolic Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subject Population
2.2. Clinical Evaluation and Sample Collection
2.3. Microbial Assessment
2.4. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Status Report on Noncommunicable Diseases; World Health Organization Library Cataloguing in Publication Data: Geneva, Switzerland, 2014; p. 280. [Google Scholar]
- IDF. Diabetes Atlas, 8th ed.; International Diabetes Federation: Brussels, Belgium, 2017; p. 150. [Google Scholar]
- Barquera, S.; Campos-Nonato, I.; Aguilar-Salinas, C.; Lopez-Ridaura, R.; Arredondo, A.; Rivera-Dommarco, J. Diabetes in mexico: Cost and management of diabetes and its complications and challenges for health policy. Glob. Health 2013, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.; Ceriello, A.; Buysschaert, M.; Chapple, I.; Demmer, R.T.; Graziani, F.; Herrera, D.; Jepsen, S.; Lione, L.; Madianos, P.; et al. Scientific evidence on the links between periodontal diseases and diabetes: Consensus report and guidelines of the joint workshop on periodontal diseases and diabetes by the international diabetes federation and the european federation of periodontology. J. Clin. Periodontol. 2018, 45, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Chapple, I.L.C.; Genco, R.; on behalf of working group 2 of the joint EFP/AAP workshop. Diabetes and periodontal diseases: Consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J. Clin. Periodontol. 2013, 84, S106–S112. [Google Scholar] [CrossRef] [PubMed]
- Castrillon, C.A.; Hincapie, J.P.; Yepes, F.L.; Roldán, N.; Moreno, S.M.; Contreras, A.; Botero, J.E. Occurrence of red complex microorganisms and Aggregatibacter actinomycetemcomitans in patients with diabetes. J. Investig. Clin. Dent. 2013, 6, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Casarin, R.C.; Barbagallo, A.; Meulman, T.; Santos, V.R.; Sallum, E.A.; Nociti, F.H.; Duarte, P.M.; Casati, M.Z.; Goncalves, R.B. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal Res. 2013, 48, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Haffajee, A.; Bogren, A.; Hasturk, H.; Feres, M.; Lopez, N.; Socransky, S. Subgingival microbiota of chronic periodontitis subjects from different geographic locations [in process citation]. J. Clin. Periodontol. 2004, 31, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Haffajee, A.; Socransky, S. Microbiology of periodontal diseases: Introduction. Periodontology 2000 2005, 38, 9–12. [Google Scholar] [CrossRef]
- Hong, B.Y.; Furtado Araujo, M.V.; Strausbaugh, L.D.; Terzi, E.; Ioannidou, E.; Diaz, P.I. Microbiome profiles in periodontitis in relation to host and disease characteristics. PLoS ONE 2015, 10, e0127077. [Google Scholar] [CrossRef]
- Polak, D.; Shapira, L. An update on the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Clin. Periodontol. 2018, 45, 150–166. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef]
- Haffajee, A.D.; Socransky, S.S.; Goodson, J.M. Clinical parameters as predictors of destructive periodontal disease activity. J. Clin. Periodontol. 1983, 10, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Vogelstein, B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983, 132, 6–13. [Google Scholar] [CrossRef]
- Socransky, S.S.; Smith, C.; Martin, L.; Paster, B.J.; Dewhirst, F.E.; Levin, A.E. “Checkerboard” DNA-DNA hybridization. Biotechniques 1994, 17, 788–792. [Google Scholar]
- Smith, G.L.; Socransky, S.S.; Sansone, C. “Reverse” DNA hybridization method for the rapid identification of subgingival microorganisms. Oral Microbiol. Immunol. 1989, 4, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Smith, C.; Dibart, S. Relation of counts of microbial species to clinical status at the sampled site. J. Clin. Periodontol. 1991, 18, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.W.; Machen, R.L.; Jotwani, R.; Iacopino, A.M. Heightened gingival inflammation and attachment loss in type 2 diabetics with hyperlipidemia. J. Periodontol. 1999, 70, 1313–1321. [Google Scholar] [CrossRef]
- Armitage, G.C. The complete periodontal examination. Periodontology 2000 2004, 34, 22–33. [Google Scholar] [CrossRef]
- Tanner, A.; Goodson, J. Sampling of microorganisms associated with periodontal disease. Oral Microbiol. Immunol. 1986, 1, 15–20. [Google Scholar] [CrossRef]
- Liu, L.S.; Gkranias, N.; Farias, B.; Spratt, D.; Donos, N. Differences in the subgingival microbial population of chronic periodontitis in subjects with and without type 2 diabetes mellitus—A systematic review. Clin. Oral Investig. 2018, 22, 2743–2762. [Google Scholar] [CrossRef] [PubMed]
- Zambon, J.J.; Reynolds, H.; Fisher, J.G.; Shlossman, M.; Dunford, R.; Genco, R.J. Microbiological and immunological studies of adult periodontitis in patients with noninsulin-dependent diabetes mellitus. J. Periodontol. 1988, 59, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Collin, H.L.; Uusitupa, M.; Niskanen, L.; Kontturi-Narhi, V.; Markkanen, H.; Koivisto, A.M.; Meurman, J.H. Periodontal findings in elderly patients with non-insulin dependent diabetes mellitus. J. Periodontol. 1998, 69, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Chang, C.J.; Hsu, P.C.; Sun, H.S.; Tseng, C.C.; Wang, J.R. Detection of putative periodontal pathogens in non-insulin-dependent diabetes mellitus and non-diabetes mellitus by polymerase chain reaction. J. Periodontal Res. 2001, 36, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Hintao, J.; Teanpaisan, R.; Chongsuvivatwong, V.; Ratarasan, C.; Dahlen, G. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol. Immunol. 2007, 22, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.; Hoffmann, T.; Fischer, S.; Bornstein, S.; Grassler, J.; Noack, B. Obesity alters composition and diversity of the oral microbiota in patients with type 2 diabetes mellitus independently of glycemic control. PLoS ONE 2018, 13, e0204724. [Google Scholar] [CrossRef]
- Demmer, R.T.; Breskin, A.; Rosenbaum, M.; Zuk, A.; LeDuc, C.; Leibel, R.; Paster, B.; Desvarieux, M.; Jacobs, D.R., Jr.; Papapanou, P.N. The subgingival microbiome, systemic inflammation and insulin resistance: The oral infections, glucose intolerance and insulin resistance study. J. Clin. Periodontol. 2017, 44, 255–265. [Google Scholar] [CrossRef]
- Sardi, J.C.; Duque, C.; Camargo, G.A.; Hofling, J.F.; Goncalves, R.B. Periodontal conditions and prevalence of putative periodontopathogens and candida spp. In insulin-dependent type 2 diabetic and non-diabetic patients with chronic periodontitis—A pilot study. Arch. Oral Biol. 2011, 56, 1098–1105. [Google Scholar] [CrossRef]
- Zhou, M.; Rong, R.; Munro, D.; Zhu, C.; Gao, X.; Zhang, Q.; Dong, Q. Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16s rdna pyrosequencing. PLoS ONE 2013, 8, e61516. [Google Scholar] [CrossRef]
- Ximenez-Fyvie, L.A.; Almaguer-Flores, A.; Jacobo-Soto, V.; Lara-Cordoba, M.; Sanchez-Vargas, L.O.; Alcantara-Maruri, E. Description of the subgingival microbiota of periodontally untreated mexican subjects: Chronic periodontitis and periodontal health. J. Periodontol. 2006, 77, 460–471. [Google Scholar] [CrossRef]
- Saeb, A.T.M.; Al-Rubeaan, K.A.; Aldosary, K.; Udaya Raja, G.K.; Mani, B.; Abouelhoda, M.; Tayeb, H.T. Relative reduction of biological and phylogenetic diversity of the oral microbiota of diabetes and pre-diabetes patients. Microb. Pathog. 2019, 128, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Maciel, S.S.; Feres, M.; Goncalves, T.E.; Zimmermann, G.S.; da Silva, H.D.; Figueiredo, L.C.; Duarte, P.M. Does obesity influence the subgingival microbiota composition in periodontal health and disease? J. Clin. Periodontol. 2016, 43, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Silva-Boghossian, C.M.; Cesario, P.C.; Leao, A.T.T.; Colombo, A.P.V. Subgingival microbial profile of obese women with periodontal disease. J. Periodontol. 2018, 89, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.M.; Matthews, J.B.; DJ, O.H.; Griffiths, H.R.; Chapple, I.L. Oxidative and inflammatory status in type 2 diabetes patients with periodontitis. J. Clin. Periodontol. 2011, 38, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; Hayes, C.; Taylor, G.W. Glycemic control of type 2 diabetes and severe periodontal disease in the us adult population. Community Dent. Oral Epidemiol. 2002, 30, 182–192. [Google Scholar] [CrossRef]
- Karima, M.; Kantarci, A.; Ohira, T.; Hasturk, H.; Jones, V.; Nam, B.H.; Malabanan, A.; Trackman, P.; Badwey, J.; Van Dyke, T. Enhanced superoxide release and elevated protein kinase c activity in neutrophils from diabetic patients: Association with periodontitis. J. Leukoc. Biol. 2005, 78, 862–870. [Google Scholar] [CrossRef]
- Bastos, A.S.; Graves, D.T.; Loureiro, A.P.d.M.; Rossa Júnior, C.; Abdalla, D.S.P.; Faulin, T.d.E.S.; Olsen Câmara, N.; Andriankaja, O.M.; Orrico, S.R. Lipid peroxidation is associated with the severity of periodontal disease and local inflammatory markers in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2012, 97, E1353–E1362. [Google Scholar] [CrossRef]
- Dhir, S.; Wangnoo, S.; Kumar, V. Impact of glycemic levels in type 2 diabetes on periodontitis. Indian J. Endocrinol. Metab. 2018, 22, 672–677. [Google Scholar] [CrossRef]
- D’Aiuto, F.; Gkranias, N.; Bhowruth, D.; Khan, T.; Orlandi, M.; Suvan, J.; Masi, S.; Tsakos, G.; Hurel, S.; Hingorani, A.D.; et al. Systemic effects of periodontitis treatment in patients with type 2 diabetes: A 12 month, single-centre, investigator-masked, randomised trial. Lancet Diabetes Endocrinol. 2018, 6, 954–965. [Google Scholar] [CrossRef]
- Longo, P.L.; Dabdoub, S.; Kumar, P.; Artese, H.P.; Dib, S.A.; Romito, G.A.; Mayer, M.P.A. Glycaemic status affects the subgingival microbiome of diabetic patients. J. Clin. Periodontol. 2018, 45, 932–940. [Google Scholar] [CrossRef]
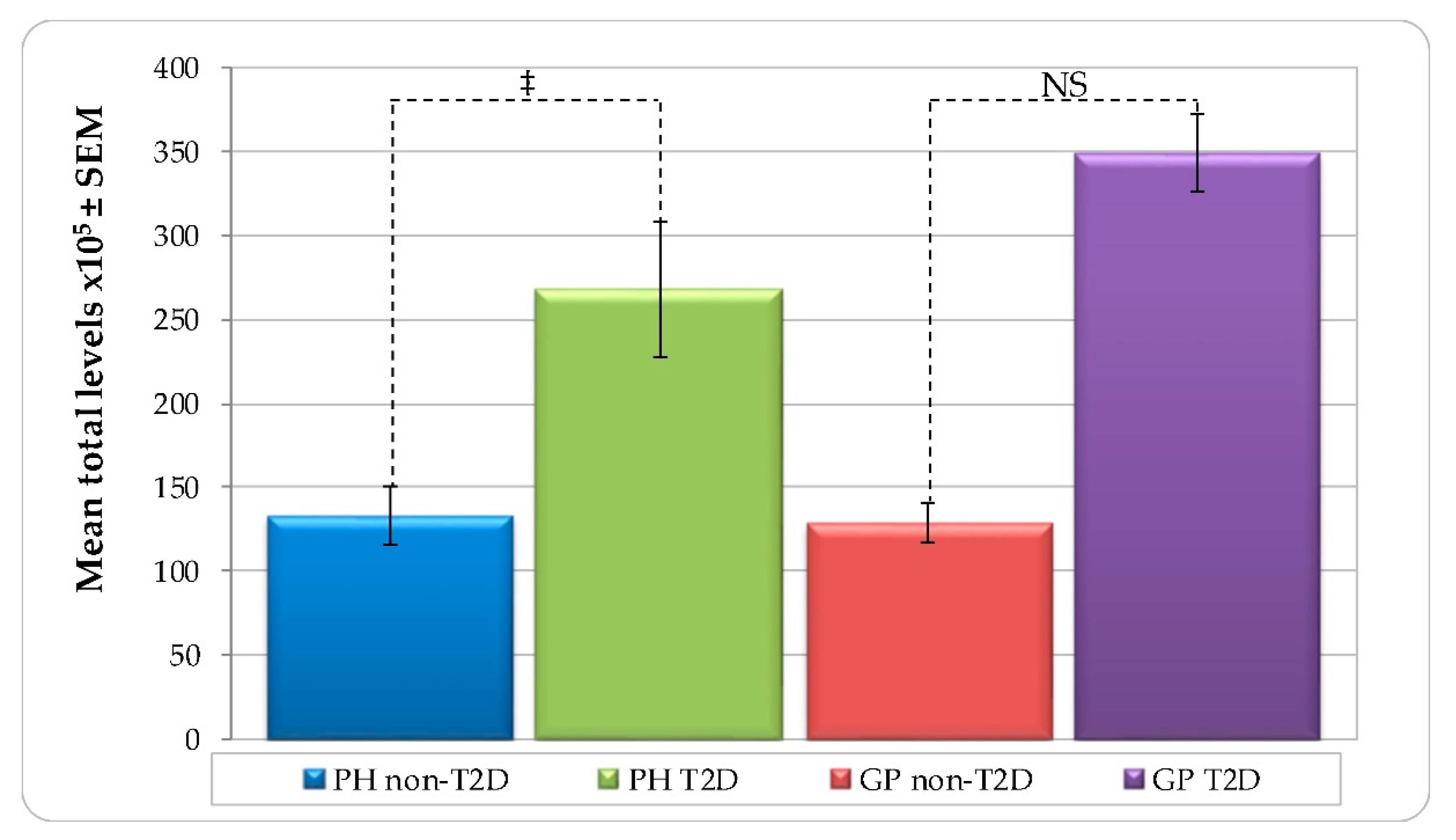
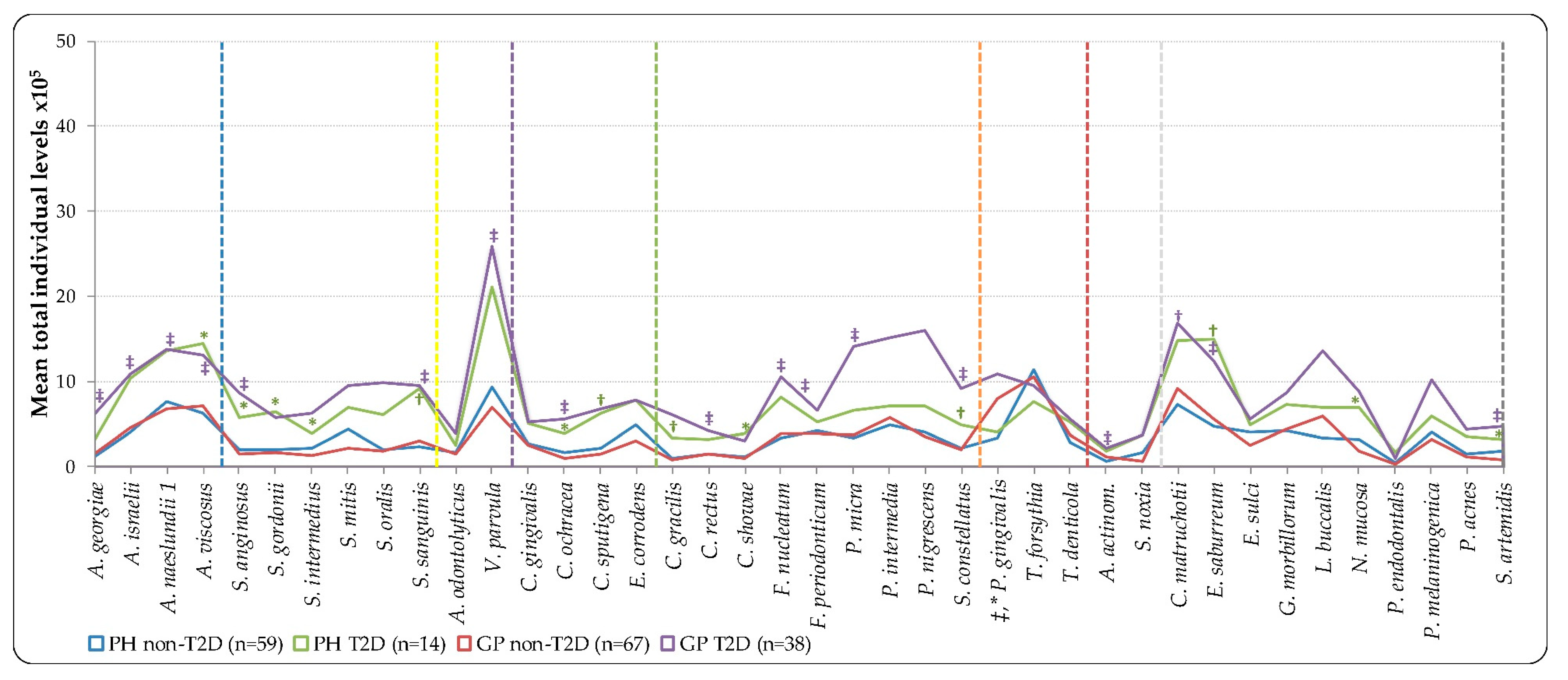
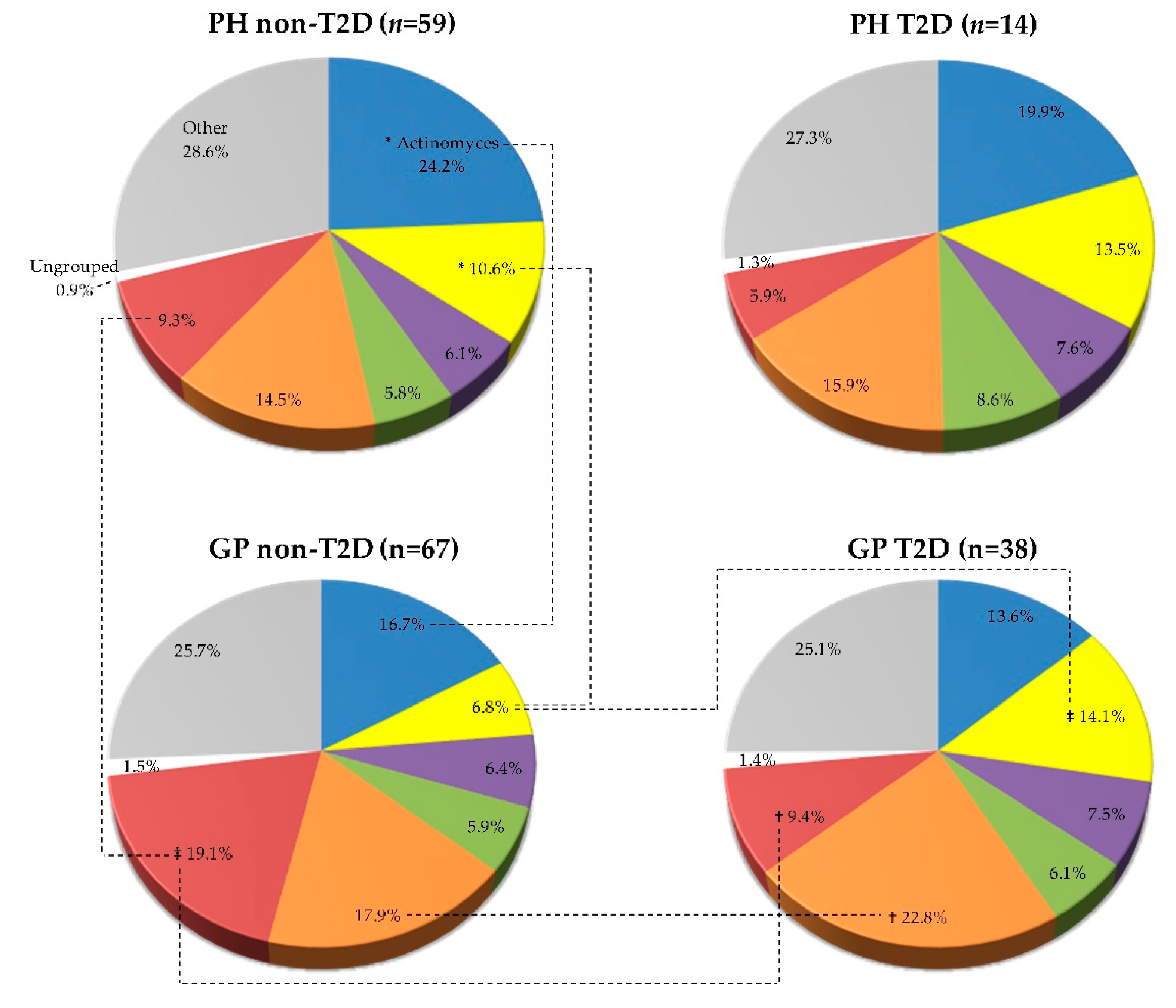
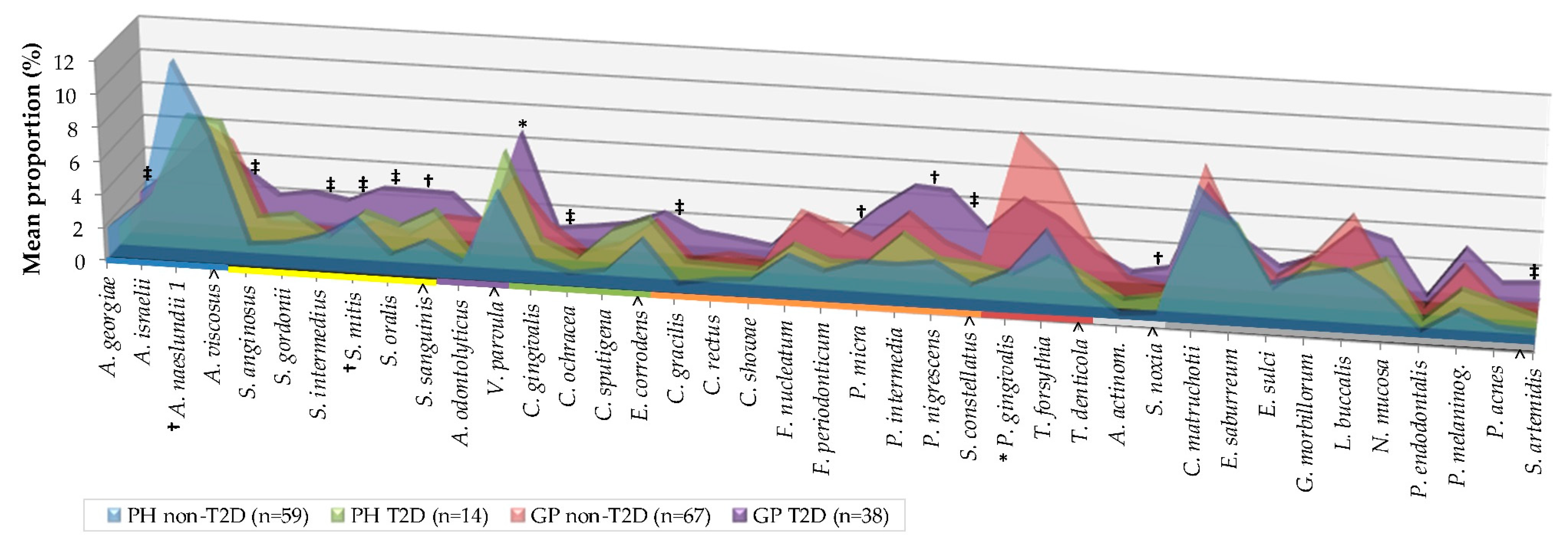
| Variable | PH Non-T2D (n = 59) | PH T2D (n = 14) | GP Non-T2D (n = 67) | GP T2D (n = 38) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Media | SEM | Media | SEM | MW | Media | SEM | Media | SEM | MW | |||||
| Age (years) ‡ † | 31.39 | ± | 1.20 | 46.00 | ± | 2.10 | ‡ | 49.10 | ± | 1.24 | 59.47 | ± | 1.77 | ‡ |
| Gender (% female) | 59.32 | ± | 57.14 | ± | 53.73 | ± | 68.42 | ± | ||||||
| Height (cm) | 160.86 | ± | 1.97 | 159.57 | ± | 2.69 | 160.50 | ± | 1.46 | 151.08 | ± | 1.50 | ||
| Weight (kg) | 68.53 | ± | 2.12 | 69.93 | ± | 3.11 | 70.36 | ± | 2.56 | 64.11 | ± | 2.33 | ||
| Body fat (%) * | 35.22 | ± | 2.09 | 31.11 | ± | 1.94 | 27.11 | ± | 1.41 | 30.69 | ± | 1.20 | ||
| Mean body mass index | 26.58 | ± | 0.76 | 27.45 | ± | 1.09 | 26.91 | ± | 0.74 | 27.98 | ± | 0.81 | ||
| Overweight and obesity (%) | 57.58 | ± | 64.29 | ± | 59.09 | ± | 71.05 | ± | ||||||
| Obesity I, II, and III (%) | 21.21 | ± | 21.43 | 18.18 | 23.68 | |||||||||
| Glycated hemoglobin (%) | - | 8.17 | ± | 0.52 | 8.31 | ± | 0.34 | |||||||
| Number of missing teeth ‡ | 1.29 | ± | 0.21 | 2.00 | ± | 0.59 | 3.37 | ± | 0.29 | 3.82 | ± | 0.41 | ||
| Mean pocket-depth (mm) ‡ € | 2.16 | ± | 0.04 | 2.32 | ± | 0.09 | 3.90 | ± | 0.12 | 3.19 | ± | 0.13 | ‡ | |
| Mean attachment level (AL, mm) ‡ € | 1.89 | ± | 0.04 | 1.40 | ± | 0.08 | ‡ | 4.32 | ± | 0.15 | 3.45 | ± | 0.17 | † |
| Mean number sites with AL ≥ 5 mm ‡ € | 0 | ± | 0 | 0 | ± | 0 | 50.52 | ± | 3.64 | 35.97 | ± | 4.37 | ||
| % sites with: | ||||||||||||||
| Plaque accumulation | 45.88 | 4.92 | 86.28 | 4.11 | † | 51.71 | 4.30 | 92.86 | 1.53 | ‡ | ||||
| Gingival erythema | 19.87 | ± | 3.46 | 22.81 | ± | 8.04 | 25.00 | ± | 3.70 | 34.81 | ± | 3.49 | ||
| Bleeding on probing ‡ † | 16.35 | ± | 2.54 | 30.34 | ± | 3.75 | * | 45.95 | ± | 2.76 | 58.13 | ± | 3.79 | |
| Suppuration ‡ € | 0 | ± | 0 | 0 | ± | 0 | 4.91 | ± | 0.91 | 18.34 | ± | 2.51 | ‡ | |
| Species | ATCC | Complex | Species | ATCC | Complex |
|---|---|---|---|---|---|
| Actinomyces georgiae | 49285 | Actinomyces | Neisseria mucosa | 19696 | Other |
| Actinomyces israelii | 12102 | Actinomyces | Parvimonas micra | 33270 | Orange |
| Actinomyces naeslundii | 12104 | Actinomyces | Porphyromonas endodontalis | 35406 | Other |
| Actinomyces odontolyticus | 17929 | Purple | Porphyromonas gingivalis | 33277 | Red |
| Actinomyces viscosus | 43146 | Actinomyces | Prevotella intermedia | 25611 | Orange |
| Aggregatibacter actinomycetemcomitans | * | Ungrouped | Prevotella melaninogenica | 25845 | Other |
| Campylobacter gracilis | 33236 | Orange | Prevotella nigrescens | 33563 | Orange |
| Campylobacter rectus | 33238 | Orange | Propionibacterium acnes | 6919 | Other |
| Campylobacter showae | 51146 | Orange | Selenomonas artemidis | 43528 | Other |
| Capnocytophaga gingivalis | 33624 | Green | Selenomonas noxia | 43541 | Ungrouped |
| Capnocytophaga ochracea | 27872 | Green | Streptococcus anginosus | 33397 | Yellow |
| Capnocytophaga sputigena | 33612 | Green | Streptococcus constellatus | 27823 | Orange |
| Corynebacterium matruchotii | 14266 | Other | Streptococcus gordonii | 10558 | Yellow |
| Eikenella corrodens | 23834 | Green | Streptococcus intermedius | 27335 | Yellow |
| Eubacterium saburreum | 33271 | Other | Streptococcus mitis | 49456 | Yellow |
| Eubacterium sulci | 35585 | Other | Streptococcus oralis | 35037 | Yellow |
| Fusobacterium nucleatum | ** | Orange | Streptococcus sanguinis | 10556 | Yellow |
| Fusobacterium periodonticum | 33693 | Orange | Tannerella forsythia | 43037 | Red |
| Gemella morbillorum | 27824 | Other | Treponema denticola | 35405 | Red |
| Leptotrichia buccalis | 14201 | Other | Veillonella parvula | 10790 | Purple |
| Parameter | Obesity (−) (n = 40) | Obesity (+) (n = 12) | MW | HbA1c ≤ 8% (n = 31) | HbA1c >8% (n = 17) | MW | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Media | SEM | Media | SEM | Media | SEM | Media | SEM | |||||||
| Mean pocket-depth (mm) | 2.9 | ± | 0.1 | 3.1 | ± | 0.2 | 2.7 | ± | 0.1 | 3.3 | ± | 0.2 | † | |
| Mean attachment level (AL, mm) | 2.9 | ± | 0.2 | 2.9 | ± | 0.3 | 2.6 | ± | 0.2 | 3.2 | ± | 0.3 | ||
| Mean number sites with AL ≥ 5 mm | 25.6 | ± | 4.3 | 28.7 | ± | 9.2 | 19.5 | ± | 3.7 | 36.5 | ± | 7.9 | ||
| Plaque accumulation | 90.4 | ± | 2.0 | 93.3 | ± | 1.9 | 89.0 | ± | 2.5 | 94.2 | ± | 1.6 | ||
| Gingival erythema | 31.4 | ± | 3.9 | 32.3 | ± | 7.0 | 27.3 | ± | 3.9 | 32.1 | ± | 5.1 | ||
| Bleeding on probing | 50.4 | ± | 3.9 | 51.5 | ± | 7.4 | 44.7 | ± | 3.7 | 58.1 | ± | 6.5 | ||
| Suppuration | 13.5 | ± | 2.4 | 14.6 | ± | 4.8 | 10.8 | ± | 2.5 | 18.8 | ± | 4.0 | ||
| Total lipids (mg/L) | 663.7 | ± | 24.6 | 677.3 | ± | 63.8 | 643.2 | ± | 24.6 | 710.0 | ± | 48.6 | ||
| Triglycerides (mg/100 mL) | 203.8 | ± | 16.2 | 303.6 | ± | 131.1 | 190.2 | ± | 16.4 | 293.2 | ± | 84.8 | ||
| Total cholesterol (mg/100 mL) | 196.1 | ± | 9.0 | 196.9 | ± | 16.0 | 191.5 | ± | 8.7 | 205.1 | ± | 15.3 | ||
| High Density Lipids (HDL) | 38.9 | ± | 1.5 | 40.1 | ± | 2.7 | 40.3 | ± | 1.6 | 37.0 | ± | 2.3 | ||
| Low Density Lipids (LDL) | 142.4 | ± | 7.8 | 145.9 | ± | 15.2 | 139.1 | ± | 7.7 | 150.7 | ± | 13.7 | ||
| Atherogenic index (LDL/HDL) | 3.8 | ± | 0.2 | 3.9 | ± | 0.7 | 3.5 | ± | 0.2 | 4.3 | ± | 0.5 | ||
| Species | ||||||||||||||
| S. intermedius_levels | 4.4 | ± | 0.4 | 9.9 | ± | 1.8 | † | 5.0 | ± | 0.6 | 7.0 | ± | 1.4 | |
| S. intermedius_proportion | 1.5 | ± | 0.2 | 2.2 | ± | 0.4 | * | 1.7 | ± | 0.3 | 1.7 | ± | 0.3 | |
| T. denticola_levels | 5.0 | ± | 0.8 | 7.4 | ± | 1.4 | * | 4.3 | ± | 0.7 | 6.3 | ± | 1.1 | |
| N. mucosa_levels | 6.4 | ± | 0.8 | 14.3 | ± | 4.2 | * | 8.0 | ± | 1.2 | 9.8 | ± | 3.0 | |
| G. morbillorum_levels | 7.7 | ± | 0.9 | 10.3 | ± | 2.4 | 7.2 | ± | 1.1 | 10.8 | ± | 1.7 | * | |
| Species | TL ≤ 800 mg/100 mL (n = 38) | TL > 800 mg/100 mL (n = 10) | MW | TC < 185 mg/100 mL (n = 25) | TC ≥ 185 mg/100 mL (n = 23) | MW | LDL < 100 mg/100 mL (n = 9) | LDL ≥ 100 mg/100 mL (n = 39) | MW | LDL/HDL < 3 mg/100 mL (n = 15) | LDL/HDL ≥ 3 mg/100 mL (n = 33) | MW | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Media | SEM | Media | SEM | Media | SEM | Media | SEM | Media | SEM | Media | SEM | Media | SEM | Media | SEM | |||||||||||||
| A. israelii_levels | 10.17 | ± | 2.36 | 15.60 | ± | 4.15 | * | 8.08 | ± | 1.49 | 14.81 | ± | 3.92 | 7.57 | ± | 2.59 | 12.17 | ± | 2.46 | 8.68 | ± | 2.11 | 12.50 | ± | 2.85 | |||
| A. naeslundii_prevalence | 79.47 | ± | 3.27 | 95.82 | ± | 2.04 | † | 79.66 | ± | 4.35 | 86.37 | ± | 3.33 | 65.86 | ± | 10.00 | 86.80 | ± | 2.19 | 77.40 | ± | 6.89 | 85.36 | ± | 2.56 | |||
| A. naeslundii_proportion | 5.03 | ± | 0.80 | 9.84 | ± | 3.31 | 3.79 | ± | 0.45 | 8.46 | ± | 1.83 | * | 3.81 | ± | 0.78 | 6.54 | ± | 1.16 | 3.66 | ± | 0.52 | 7.11 | ± | 1.34 | |||
| A. viscosus_proportion | 4.22 | ± | 0.86 | 5.80 | ± | 1.43 | 4.15 | ± | 1.26 | 4.98 | ± | 0.75 | 3.54 | ± | 0.71 | 4.79 | ± | 0.90 | 2.74 | ± | 0.44 | 5.38 | ± | 1.03 | * | |||
| A. actinomycetem._levels | 2.20 | ± | 0.33 | 1.81 | ± | 0.66 | 2.33 | ± | 0.50 | 1.89 | ± | 0.30 | 1.11 | ± | 0.25 | 2.35 | ± | 0.35 | * | 1.79 | ± | 0.32 | 2.27 | ± | 0.41 | |||
| C. rectus_proportion | 1.12 | ± | 0.15 | 0.66 | ± | 0.20 | 1.31 | ± | 0.21 | 0.73 | ± | 0.13 | 1.22 | ± | 0.42 | 0.98 | ± | 0.13 | 1.20 | ± | 0.32 | 0.95 | ± | 0.13 | ||||
| C. gingivalis_prevalence | 47.88 | ± | 4.87 | 74.36 | ± | 10.63 | * | 39.88 | ± | 4.48 | 67.73 | ± | 7.32 | † | 45.56 | ± | 9.76 | 55.39 | ± | 5.32 | 42.98 | ± | 6.87 | 58.45 | ± | 5.93 | ||
| C. gingivalis_proportion | 1.01 | ± | 0.22 | 2.05 | ± | 0.50 | † | 0.77 | ± | 0.09 | 1.72 | ± | 0.40 | 0.77 | ± | 0.19 | 1.34 | ± | 0.25 | 0.74 | ± | 0.12 | 1.46 | ± | 0.30 | |||
| C. matruchotii_proportion | 5.76 | ± | 0.86 | 7.41 | ± | 1.05 | 4.40 | ± | 0.48 | 7.95 | ± | 1.32 | † | 5.50 | ± | 0.86 | 6.24 | ± | 0.86 | 4.66 | ± | 0.59 | 6.75 | ± | 1.00 | |||
| E. saburreum_prevalence | 63.95 | ± | 4.60 | 80.37 | ± | 8.79 | * | 62.46 | ± | 5.53 | 72.70 | ± | 6.18 | 53.41 | ± | 11.86 | 70.59 | ± | 4.24 | 63.84 | ± | 8.16 | 68.97 | ± | 4.83 | |||
| E. sulci_prevalence | 50.87 | ± | 4.69 | 79.74 | ± | 7.27 | † | 48.50 | ± | 5.17 | 66.00 | ± | 6.69 | * | 51.93 | ± | 10.10 | 58.03 | ± | 4.84 | 54.17 | ± | 6.86 | 58.12 | ± | 5.53 | ||
| F. periodonticum_prevalence | 64.25 | ± | 5.30 | 79.69 | ± | 7.30 | 60.79 | ± | 5.87 | 75.72 | ± | 6.70 | * | 59.03 | ± | 11.37 | 69.69 | ± | 4.89 | 65.90 | ± | 7.59 | 68.43 | ± | 5.67 | |||
| S. gordonii_prevalence | 65.36 | ± | 5.05 | 85.47 | ± | 8.53 | * | 63.30 | ± | 5.74 | 76.08 | ± | 6.74 | * | 62.91 | ± | 10.11 | 71.56 | ± | 5.05 | 73.85 | ± | 5.79 | 68.02 | ± | 6.01 | ||
| T. forsythia_proportion | 2.88 | ± | 0.43 | 3.54 | ± | 1.06 | 2.20 | ± | 0.46 | 3.90 | ± | 0.64 | * | 2.91 | ± | 1.16 | 3.04 | ± | 0.43 | 2.48 | ± | 0.71 | 3.26 | ± | 0.49 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Hernández, A.-P.; Márquez-Corona, M.d.L.; Pontigo-Loyola, A.P.; Medina-Solís, C.E.; Ximenez-Fyvie, L.-A. Subgingival Microbiota of Mexicans with Type 2 Diabetes with Different Periodontal and Metabolic Conditions. Int. J. Environ. Res. Public Health 2019, 16, 3184. https://doi.org/10.3390/ijerph16173184
Rodríguez-Hernández A-P, Márquez-Corona MdL, Pontigo-Loyola AP, Medina-Solís CE, Ximenez-Fyvie L-A. Subgingival Microbiota of Mexicans with Type 2 Diabetes with Different Periodontal and Metabolic Conditions. International Journal of Environmental Research and Public Health. 2019; 16(17):3184. https://doi.org/10.3390/ijerph16173184
Chicago/Turabian StyleRodríguez-Hernández, Adriana-Patricia, María de Lourdes Márquez-Corona, América Patricia Pontigo-Loyola, Carlo Eduardo Medina-Solís, and Laurie-Ann Ximenez-Fyvie. 2019. "Subgingival Microbiota of Mexicans with Type 2 Diabetes with Different Periodontal and Metabolic Conditions" International Journal of Environmental Research and Public Health 16, no. 17: 3184. https://doi.org/10.3390/ijerph16173184
APA StyleRodríguez-Hernández, A.-P., Márquez-Corona, M. d. L., Pontigo-Loyola, A. P., Medina-Solís, C. E., & Ximenez-Fyvie, L.-A. (2019). Subgingival Microbiota of Mexicans with Type 2 Diabetes with Different Periodontal and Metabolic Conditions. International Journal of Environmental Research and Public Health, 16(17), 3184. https://doi.org/10.3390/ijerph16173184






