Morin Protects Human Respiratory Cells from PM2.5 Induced Genotoxicity by Mitigating ROS and Reverting Altered miRNA Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Extraction of PM2.5
2.2. Characterization of PM2.5
2.3. Estimation of Endotoxin
2.4. Estimation of Oxidative Potential of PM2.5
2.5. Cell Line and Culture Conditions
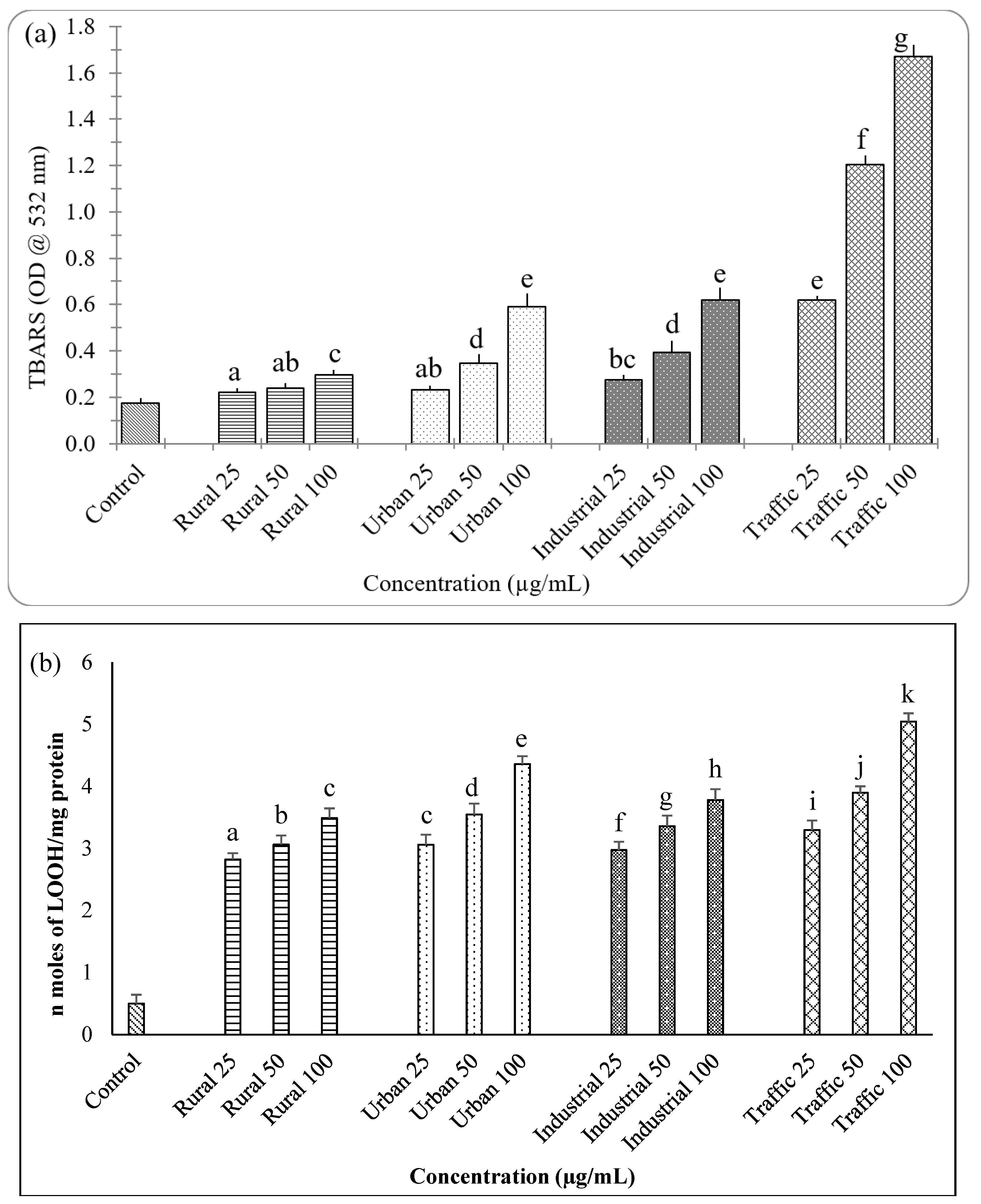
2.6. Estimation of Lipid Peroxidation
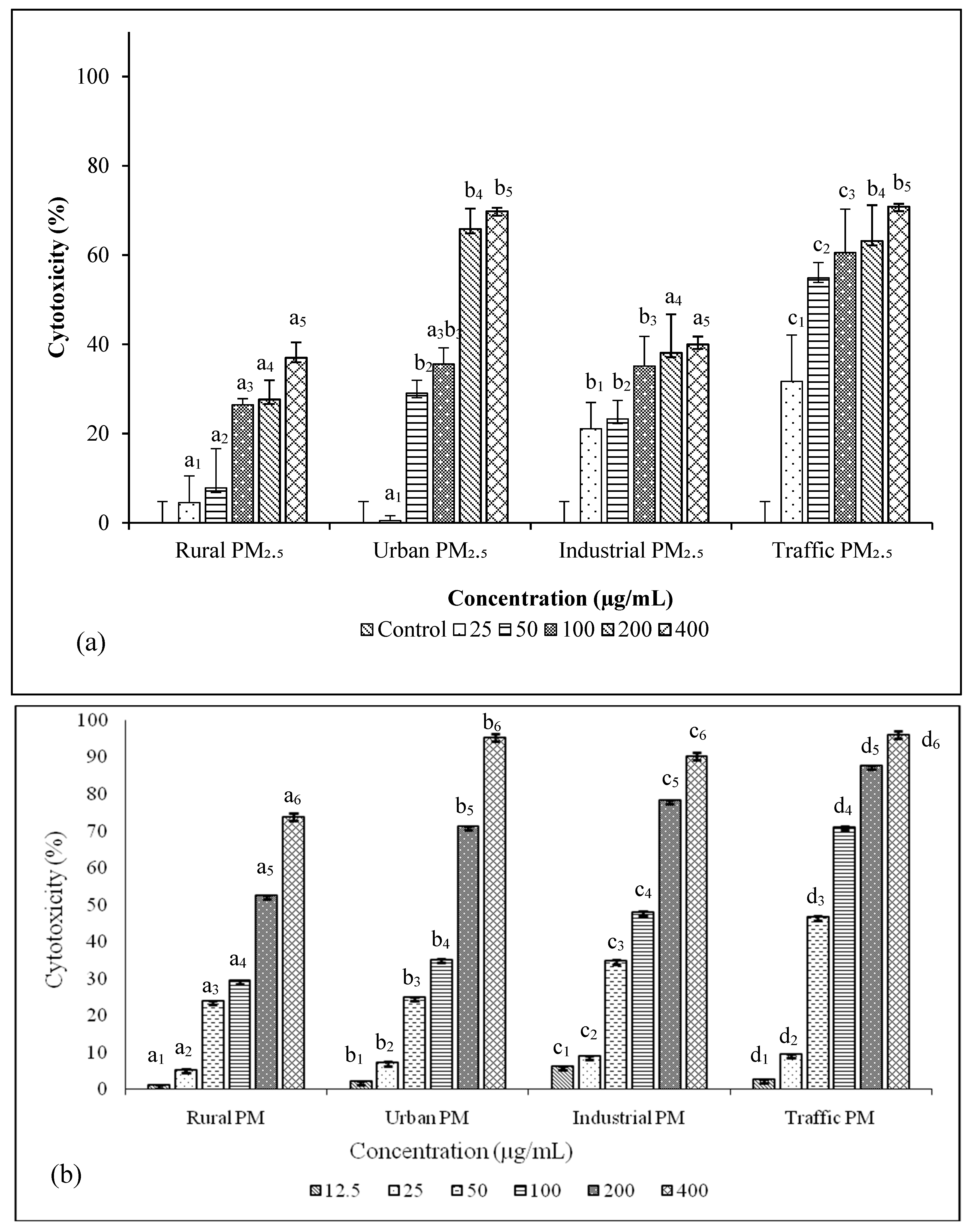
2.7. Estimation of Cytotoxicity of PM2.5
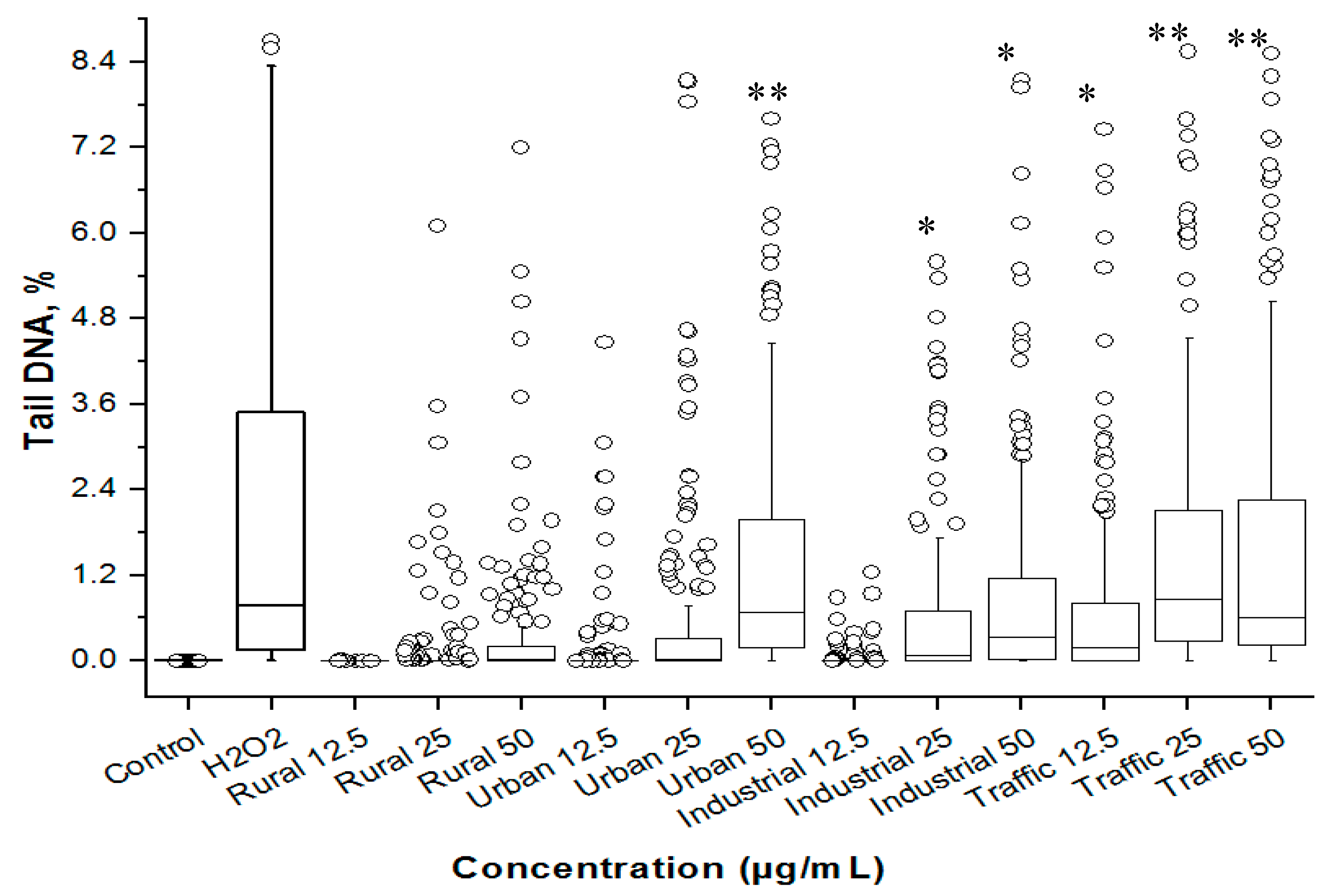
2.8. Estimation of Genotoxicity of PM2.5
2.9. Measurement of Intracellular Reactive Oxygen Species
2.10. Effect of PM2.5 and Morin on miRNA and Gene Expression
2.11. Statistical Analysis
3. Results and Discussion
3.1. Characterization of PM2.5
3.2. Metals and PAHs
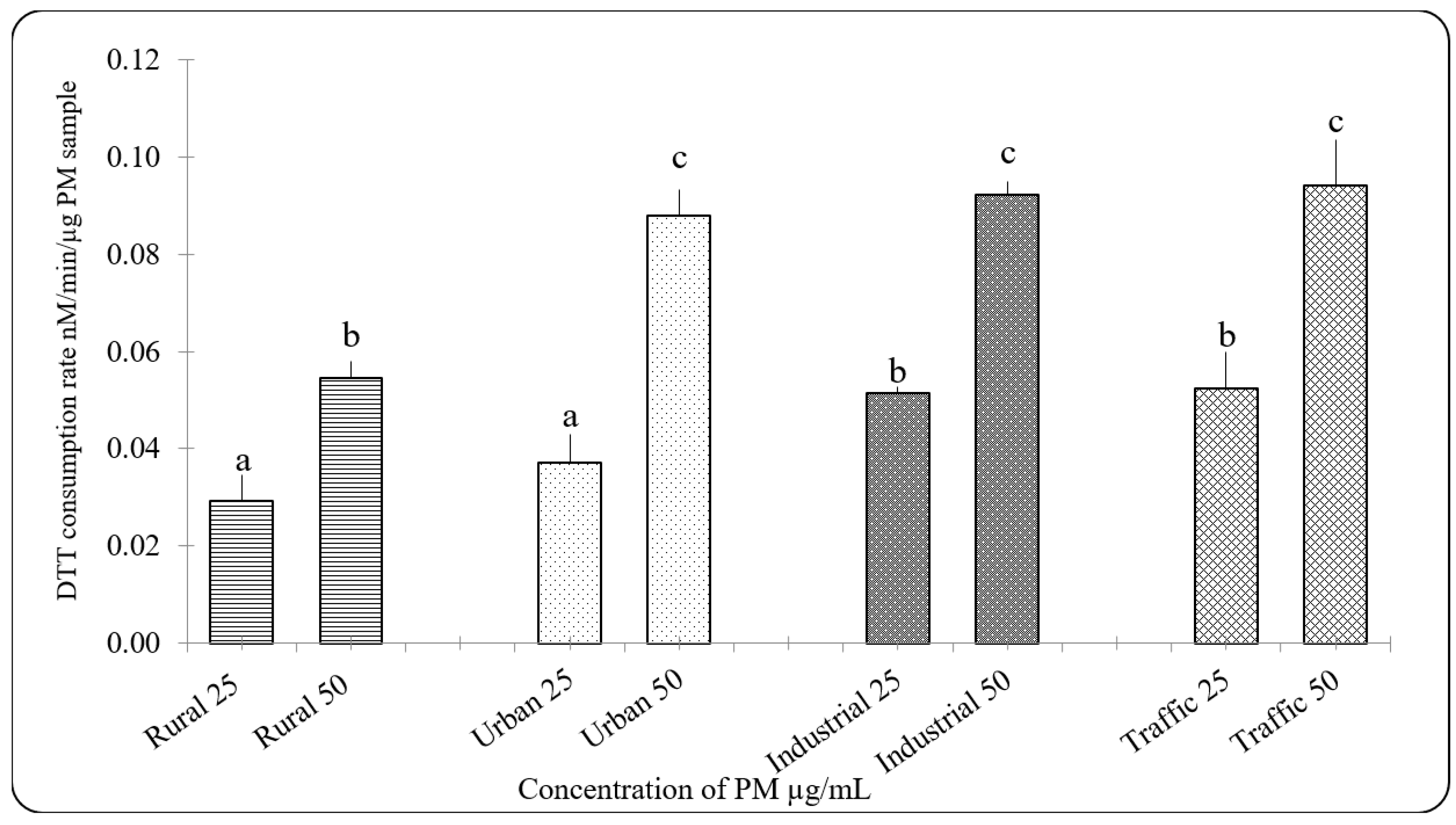
3.3. Oxidative Potential of PM2.5
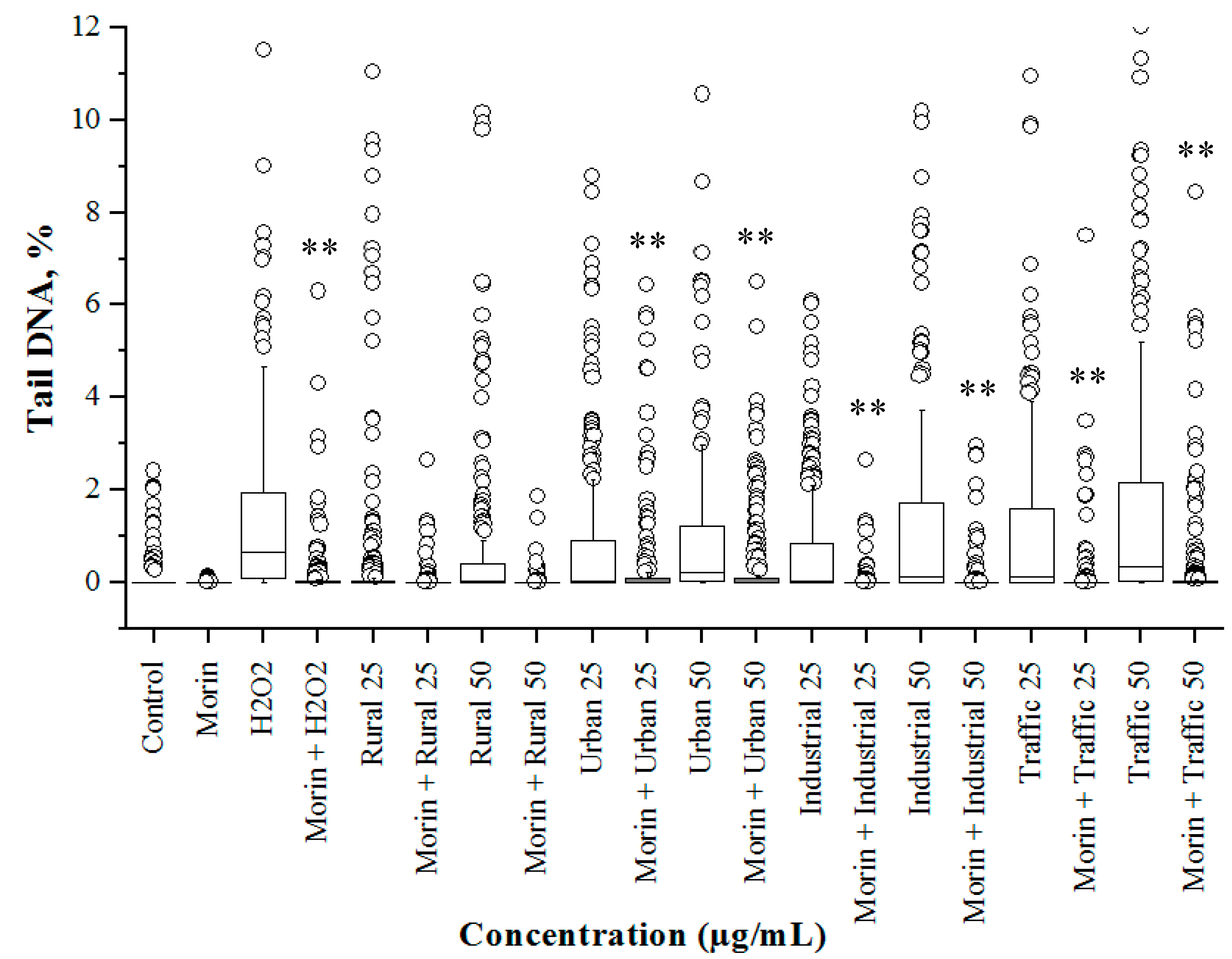
3.4. Cytotoxicity and Genetoxicity in A549 Cells
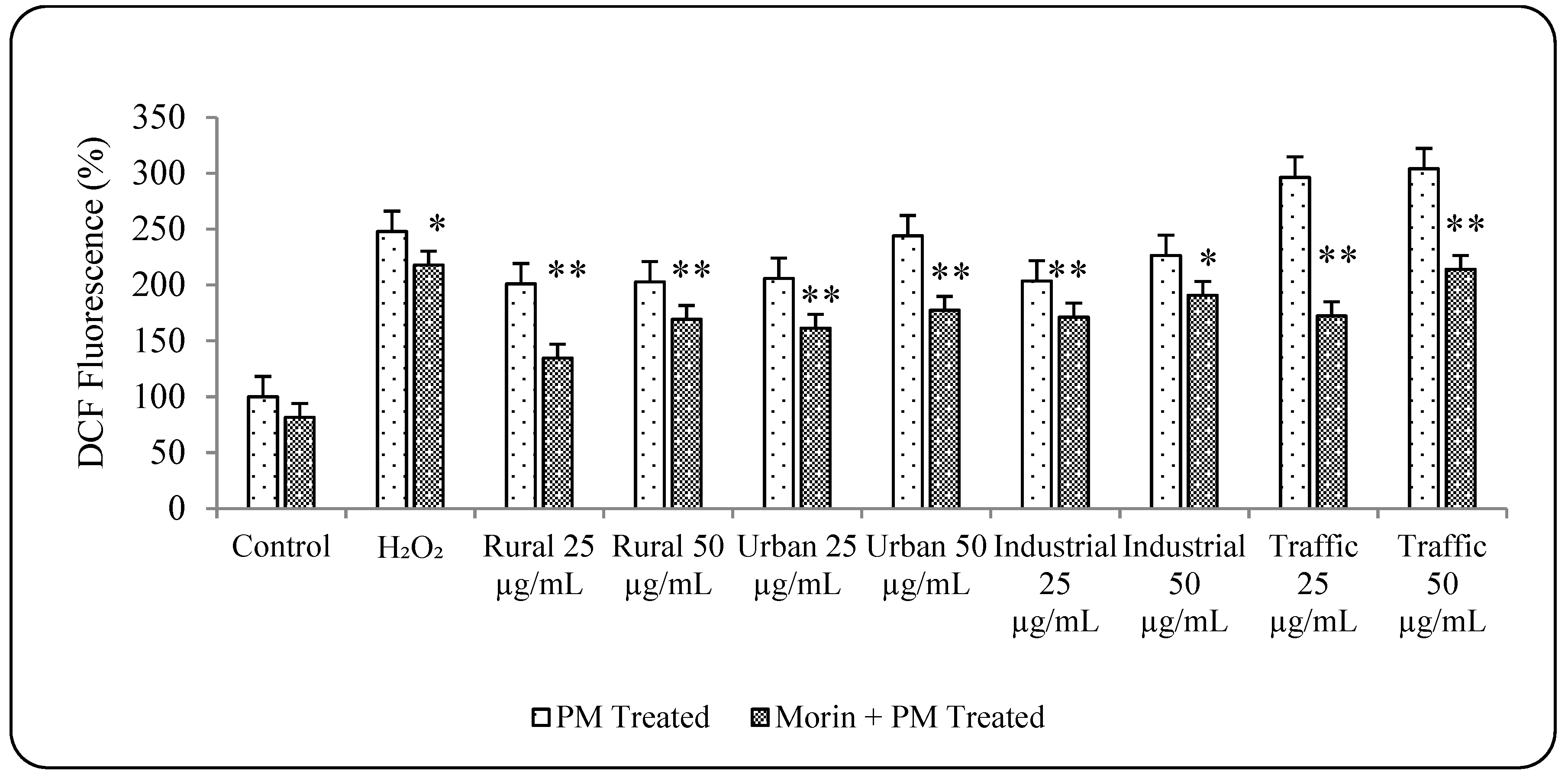
3.5. Morin Protects Cells from PM2.5 Induced Genotoxicity by Reducing ROS Production
3.6. PM2.5 from Traffic Region Confers More Toxicity
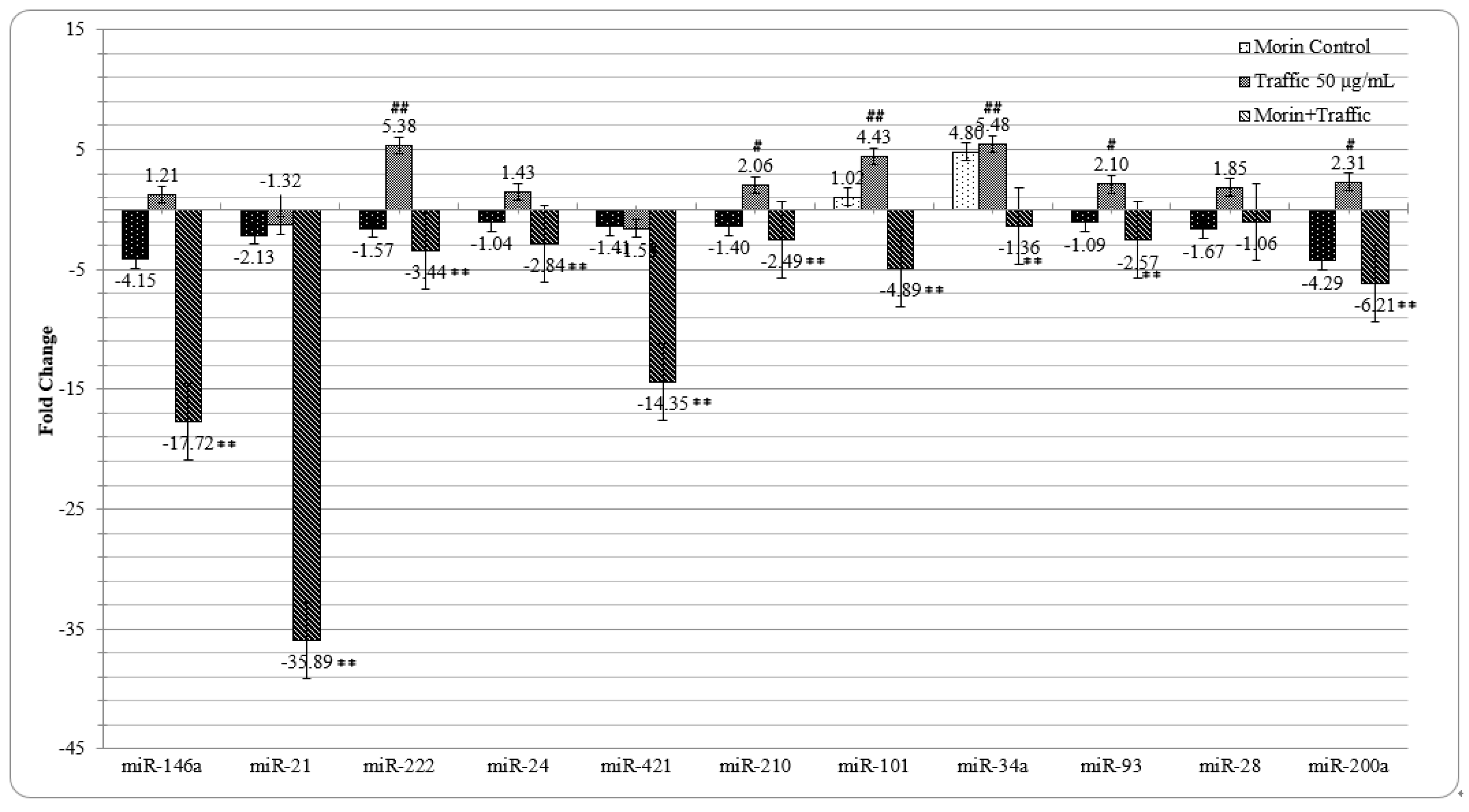
3.7. Morin Reverted the Traffic PM2.5 Induced Altered miRNA Expression in A549 cells
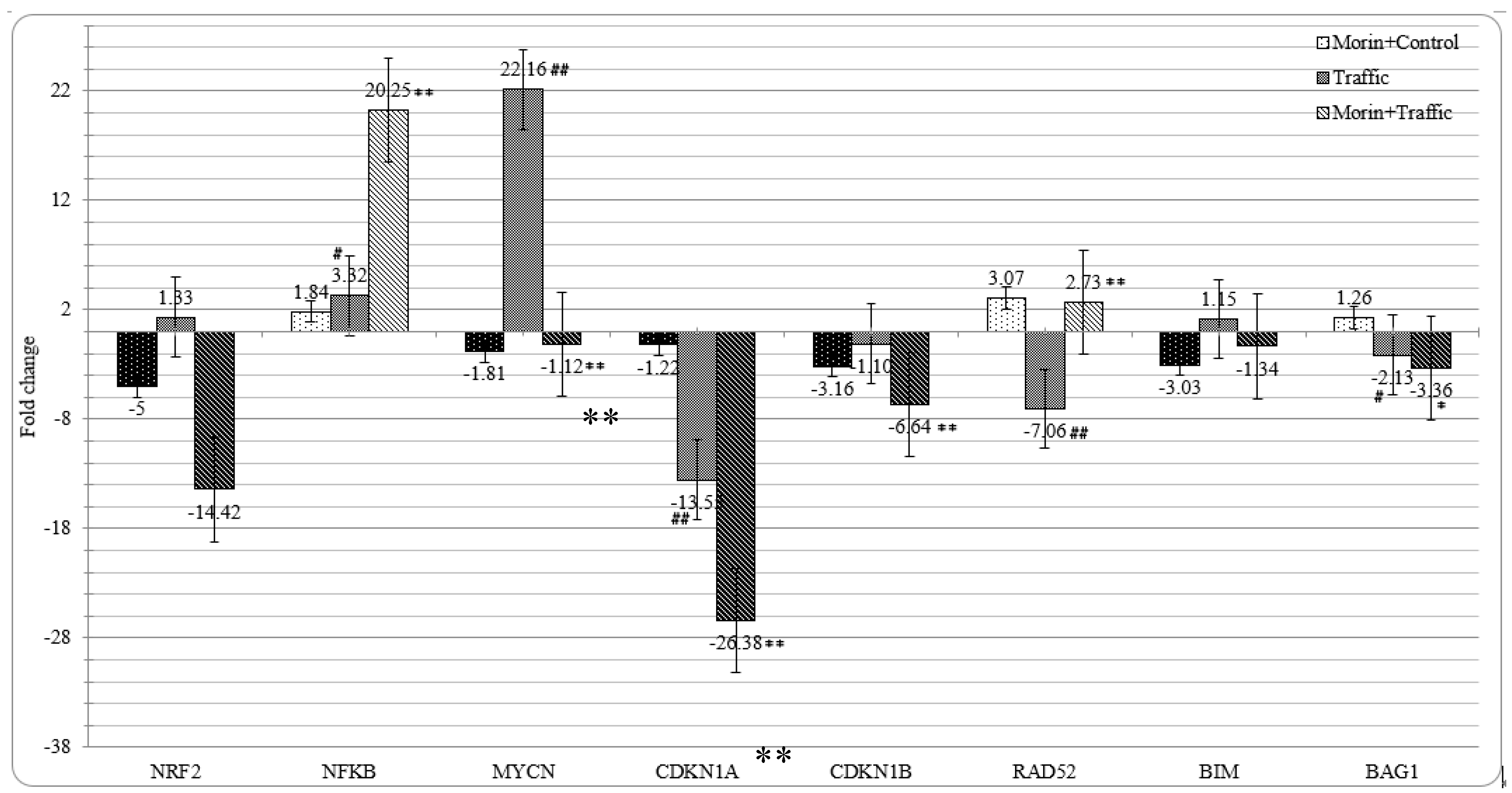
3.8. Morin Revised the Traffic PM2.5 Altered Gene Expression in A549 cells
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Strosnider, H.; Kennedy, C.; Monti, M.; Yip, F. Rural and Urban differences in Air Quality 2008–2012, and Community Drinking Water Quality, 2010–2015—United States. MMWR Surveill. Summ. 2017, 66, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ambient (Outdoor) Air Quality and Health. Available online: www.who.int/news-room/fact-sheets/detail/ambient-(outdoor)-air-quality-and-health (accessed on 18 May 2019).
- Schwartz, J. Air Pollution and Blood markers of Cardiovascular Risk. Environ. Health Perspect. 2001, 109, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Ruckerl, R.; Schneider, A.; Breitner, S.; Cyrys, J.; Peters, A. Health effects of Particulate Air Pollution: A review of epidemiological evidence. Inhal. Toxicol. 2011, 23, 555–592. [Google Scholar] [CrossRef] [PubMed]
- Brook, R.D.; Franklin, B.; Cascio, W.; Hong, Y.; Howard, G.; Lipsett, M.; Luepkar, R.; Mittleman, M.; Samet, J.; Smith, S.C.; et al. Air Pollution and Cardiovascular Disease. A Statement for Healthcare Professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 2004, 109, 2655–2671. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, S.; Manju, A.; Muthuselvam, P.; Shalini, D.; Indhumathi, V.; Kalaiselvi, K.; Palanivel, M.; Chandrasekar, P.P.; Rajaguru, P. Characterization and genotoxicity evaluation of particulate matter collected from industrial atmosphere in Tamil Nadu State, India. J. Hazard. Mater. 2014, 274, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Staniswalis, J.G.; Parks, N.J.; Bader, J.O.; Maldonado, Y.M. Temporal analysis of Air-borne Particulate Matterreveals a dose-rate effect on mortality in El Paso: Indications of Differential toxicity for different particle mixtures. J. Air Waste Manag. Assoc. 2005, 55, 893–902. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davy, P.K.; Ancelet, T.; Trompetter, W.J.; Markwitz, A.; Weatherburn, D.C. Composition and source contributions of air particulate matter pollution in a New Zealand suburban town. Atmos. Pollut. Res. 2012, 3, 143–147. [Google Scholar] [CrossRef]
- Gonzalez, L.T.; Rodriguez, F.E.L.; Dominguez, M.S.; Cavazos, A.; Porras, C.L.; Vidaurri, L.G.S.; Askar, K.A.; Kharissov, B.I.; Chiu, J.F.V.; Barbosa, J.M.A. Determination of trace metals in TSP and PM2.5 materials collected in the metropolitan area of Monterrey, Mexico: A Characterization study by XPS, ICP-AES and SEM-EDS. Atmos. Res. 2017, 196, 8–22. [Google Scholar] [CrossRef]
- Hanningan, M.P.; Busby, W.F., Jr.; Cass, G.R. Source contributions to the mutagenicity of Urban Particulate Air pollution. J. Air Waste Manag. Assoc. 2005, 55, 399–410. [Google Scholar] [CrossRef]
- Hong, Z.; Guo, Z.; Zhang, R.; Xu, J.; Dong, W.; Zhuang, G.; Deng, C. Air borne fine Particulate Matter induces oxidative stress and inflammation in human nasal epithelial cells. Tohoku J. Exp. Med. 2016, 239, 117–125. [Google Scholar] [CrossRef]
- Mahapatra, P.S.; Jain, S.; Shrestha, S.; Senapati, S.; Puppala, S.P. Ambient endotoxin in PM10 and association with inflammatory activity, air pollutants and meteorology in Chitwan, Nepal. Sci. Total Environ. 2018, 618, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Harmon, A.C.; Hebert, V.Y.; Cormier, S.A.; Subramanian, B.; Reed, J.R.; Backes, W.L.; Dugas, T.R. Particulate Matter containing environmentally persistent free radicals induces AhR-dependent cytokine and reactive oxygen species production in human bronchial epithelial cells. PLoS ONE 2018, 13, e0205412. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, P.; Lu, Y.; Huo, L.; Bai, M.; Yu, F.; Tie, Y. Potential injurious effects of the fine particulate PM2.5 on the progression of atherosclerosis in apoE-deficient mice by activating platelets and leukocytes. Arch. Med. Sci. 2019, 15, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.S.; Kang, K.A.; Piao, M.J.; Ahn, M.J.; Yi, J.M.; Hyun, Y.M.; Kim, S.H.; Ko, M.K.; Park, C.O.; Hyun, J.W. Particulate Matter induces inflammatory cytokine production via activation of NFκB by TLR5-NOX4-ROS signaling in human skin keratinocyte and mouse skin. Redox. Biol. 2019, 21, 101080. [Google Scholar] [CrossRef] [PubMed]
- Janssen, N.A.H.; Yang, A.; Strak, M.; Steenhof, M.; Hellack, B.; Gerlofs-Nijland, M.E.; Kuhlbusch, T.; Kelly, F.; Harrison, R.; Brunekreef, B.; et al. Oxidative potential of particulate matter collected at sites with different source characteristics. Sci. Total Environ. 2014, 472, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Moreno, T.; Kelly, F.J.; Dunster, C.; Oliete, A.; Martins, V.; Reche, C.; Minguillon, M.C.; Amato, F.; Capdevila, M.; Miguel, E.; et al. Oxidative potential of subway PM2.5. Atmos. Environ. 2017, 148, 230–238. [Google Scholar] [CrossRef]
- Ambroz, A.; Vlkova, V.; Rossner, P., Jr.; Rossnerova, A.; Svecova, V.; Milcova, A.; Pulkrabova, J.; Hajslov, J.; Veleminsky, M., Jr.; Solansky, I.; et al. Impact of air pollution on oxidative DNA damage and lipid peroxidation in mothers and their newborns. Int. J. Hyg. Environ. Health 2016, 219, 545–556. [Google Scholar] [CrossRef] [PubMed]
- De Prins, S.; Koppen, G.; Jacobs, G.; Dons, E.; Van de Mieroop, E.; Nelen, V.; Fierens, F.; Int Panis, L.; De Boever, P.; Cox, B.; et al. Influence of ambient air pollution on global DNA methylation in healthy adults: A seasonal follow-up. Environ. Int. 2013, 59, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Plusquin, M.; Guida, F.; Polidoro, S.; Vermeulen, R.; Raaschou Nielsen, O.; Campanella, G.; Hoek, G.; Kyrtopoulos, S.A.; Georgiadis, P.; Naccarati, A.; et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ. Int. 2017, 108, 127–136. [Google Scholar] [CrossRef]
- Bollati, V.; Marinelli, B.; Apostoli, P.; Bonzini, M.; Nordio, F.; Hoxha, M.; Pegoraro, V.; Motta, V.; Tarantini, L.; Cantone, L.; et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health Perspect. 2010, 118, 763–768. [Google Scholar] [CrossRef]
- Jardim, M.J.; Fry, R.C.; Jaspers, I.; Dailey, L.; Diaz-Sanchez, D. Disruption of MicroRNA Expression in Human Airway Cells by Diesel Exhaust Particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 2009, 117, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Tsamou, M.; Vrijens, K.; Madhloum, N.; Lefebvre, W.; Vanpoucke, C.; Nawrot, T.S. Air pollution–induced placental epigenetic alterations in early life: A candidate miRNA approach. Epigenetics 2018, 13, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shi, X.; Zhang, S.; Liu, M. miR-210 inhibits oxidative stress damage in myocardial cell injury model. Int. J. Clin. Exp. Pathol. 2017, 10, 3077–3084. [Google Scholar]
- Dando, I.; Cordani, M.; Pozza, E.D.; Biondani, G.; Donadelli, M.; Palmieri, M. Antioxidant mechanisms and ROS related MiRNAs in Cancer Stem cells. Oxidative Med. Cell. Longev. 2015, 2015, 425708. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011, 132, 75–85. [Google Scholar] [CrossRef]
- Eades, G.; Yang, M.; Yao, Y.; Zhang, Y.; Zhou, Q. miR-200a Regulates Nrf2 Activation by Targeting Keap1mRNA in Breast Cancer Cells. J. Biol. Chem. 2011, 286, 40725–40733. [Google Scholar] [CrossRef]
- Marsit, C.J.; Eddy, K.; Kelsey, K.T. MicroRNA responses to cellular stress. Cancer Res. 2006, 66, 10843–10848. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Pogue, A.I. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J. Inorg. Biochem. 2007, 101, 1265–1269. [Google Scholar] [CrossRef]
- Dong, R.; Wang, D.; Wang, X.; Zhang, K.; Chen, P.; Yang, C.S.; Zhang, J. Epigallocatechin-3-gallate enhances key enzymatic activities of hepatic thioredoxin and glutathione systems in selenium-optimal mice but activates hepatic Nrf2 responses in selenium-deficient mice. Redox. Biol. 2016, 10, 221–232. [Google Scholar] [CrossRef]
- Li, S.G.; Ding, Y.S.; Niu, Q.; Xu, S.Z.; Pang, L.J.; Ma, R.L.; Jing, M.X.; Feng, G.L.; Liu, J.M.; Guo, S.X. Grape Seed Proanthocyanidin Extract Alleviates Arsenic-induced Oxidative Reproductive Toxicity in Male Mice. Biomed. Environ. Sci. 2015, 28, 272–280. [Google Scholar] [CrossRef]
- Wang, H.; Bian, S.; Yang, C.S. Green Tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF1α. Carcinogenesis 2011, 32, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Calin, G.A.; Steele, V.E.; Cartiglia, C.; Longobardi, M.; Croce, C.M.; De Flora, S. Chemoprevention of cigarette smoke–induced alterations of MicroRNA expression in rat lungs. Cancer Prev. Res. 2010, 3, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, E.; Sanchez-Gomez, M.V.; Ruiz, A.; Cavaliere, F.; Ortiz-Sanz, C.; Quintela-Lopez, T.; Capetillo-Zarate, E.; Sole-Domenech, S.; Matute, C. Mangiferin and Morin Attenuate Oxidative Stress, Mitochondrial Dysfunction, and Neurocytotoxicity Induced by Amyloid Beta Oligomers. Oxidative Med. Cell. Longev. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ge, A.; Zhu, W.; Liu, Y.N.; Ji, N.F.; Zha, W.J.; Zhang, J.X.; Zeng, X.N.; Huang, M. Morin attenuates Ovalbumin–induced Airway Inflammation by Modulating Oxidative Stress–Responsive MAPK signaling. Oxidative Med. Cell. Longev. 2016, 2016, 5843672. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kang, K.A.; Kim, K.C.; Cha, J.W.; Kim, E.H.; Hyun, J.W. Morin Induces Heme Oxygenase-1 via ERK-Nrf2 Signaling Pathway. J. Cancer Prev. 2013, 18, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Vanitha, P.; Senthilkumar, S.; Dornadula, S.; Anandhakumar, S.; Rajaguru, P.; Ramkumar, K.M. Morin activates the Nrf2-ARE pathway and reduces oxidative stress-induced DNA damage in pancreatic beta cells. Eur. J. Pharm. 2017, 801, 9–18. [Google Scholar] [CrossRef]
- Yao, D.; Cui, H.; Zhou, S.; Guo, L. Morin inhibited lung cancer cells viability, growth, and migration by suppressing miR-135b and inducing its target CCNG2. Tumor. Biol. 2017, 39, 1010428317712443. [Google Scholar] [CrossRef]
- Li, B.; Jin, X.; Meng, H.; Hu, B.; Zhang, T.; Yu, J.; Chen, S.; Guo, X.; Wang, W.; Jiang, W.; et al. Morin promotes prostate cancer cells chemosensitivity to paclitaxel through miR-155/GATA3 axis. Oncotarget 2017, 8, 47849–47860. [Google Scholar] [CrossRef]
- Brief Industrial Profile of Coimbatore District 2015-16, Ministry of MSME, Govt of India. 2016; 15. Available online: http://dcmsme.gov.in/dips/IPS%20coimbatore%202012_t.pdfs (accessed on 1 July 2019).
- Palaniappan, V.S. Work Begins on Green Corridor Concept for Mettupalayam Road. The Hindu. 26 October 2012. Available online: https://www.thehindu.com/news/cities/Coimbatore/work-begins-on-green-corridor-concept-for-mettupalayam-road/article4034204.ece (accessed on 28 April 2019).
- Shalini, D.; Senthilkumar, S.; Rajaguru, P. Effect of Size and Shape on toxicity of zinc oxide (ZnO) nanomaterials in human peripheral blood lymphocytes. Toxicol. Mech. Methods 2018, 28, 87–94. [Google Scholar] [CrossRef]
- De Vizcaya-Ruiz, A.; Gutierrez-Castillo, M.E.; Uribe-Ramirez, M.; Cebrian, M.E.; Mugica-Alvarez, V.; Sepulveda, J.; Rosas, I.; Salinas, E.; Garcia-Cuellar, C.; Martinez, F.; et al. Characterization and in vitro biological effects of concentrated particulate matter from Mexico City. Atmos. Environ. 2016, 40, 583–592. [Google Scholar] [CrossRef]
- Nichens Niehaus, W.G.; Samuelson, B. Formation of malondialdehyde from phospholipids arachidonate during microsomal lipid peroxidation. Eur. J. Biochem. 1996, 6, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Y.; Hunt, J.V.; Wolf, S.P. Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem. 1992, 202, 384–389. [Google Scholar] [CrossRef]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Chahine, T.; Baccarelli, A.; Litonjua, A.; Wright, R.O.; Suh, H.; Gold, D.R.; Sparrow, D.; Vokonas, P.; Schwartz, J. Particulate Air Pollution, Oxidative Stress Genes, and Heart Rate Variability in an Elderly Cohort. Environ. Health Perspect. 2007, 115, 1617–1622. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, M.I.; Jaakkola, M.S.; London, S.J.; Nel, A.E.; Rogers, C.A. How Exposure to Environmental Tobacco Smoke, Outdoor Air Pollutants, and Increased Pollen Burdens Influences the Incidence of Asthma. Environ. Health Perspect. 2006, 114, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Kelly, F.J.; Fussell, J.C. Role of oxidative stress in cardiovascular disease outcomes following exposure to ambient air pollution. Free Radic. Biol. Med. 2017, 110, 345–367. [Google Scholar] [CrossRef] [PubMed]
- WHO. Air Quality Guidelines for Particulate Matter, Ozone, Nitrogen Dioxide and Sulfur Dioxide, Global Update 2005; World Health Organization: Geneva, Switzerland, 2006; p. 9. [Google Scholar]
- Kato, N.; Mastui, Y.; Takaoka, M.; Yoneda, M. Measurement of nanoparticle exposure in crematoriums and estimation of respiratory deposition of the nanoparticles by number and size distribution. J. Occup. Health 2017, 59, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Fishler, R.; Hofemeier, P.; Etzion, Y.; Dubowski, Y.; Sznitman, J. Particle dynamics and deposition in true-scale pulmonary acinar models. Sci. Rep. 2015, 5, 14071. [Google Scholar] [CrossRef] [PubMed]
- Leeuwen, F.X.R.V.; Krzyzanowski, M. Book Chapter: 7.3. In Air Quality Guidelines–Second Edition; WHO Regional Office for Europe: Copenhagen, Denmark, 2000; pp. 1–2. [Google Scholar]
- Gangwar, C.; Choudhari, R.; Chauhan, A.; Kumar, A.; Singh, A.; Tripathi, A. Assessment of air pollution caused by illegal e-waste burning to evaluate the human health risk. Environ. Int. 2019, 125, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Tinaroth. Iron: Health and Environment Effects. Iron Map 2012. Available online: https://www.ironmap.com/iron-health-and-environment-effects/ (accessed on 11 May 2019).
- Aitio, A.; Boudet, C.; Chikviladze, K.; Clasrkson, S.; Temmerman, L.D.; Englert, N.; Eriksson, J.; Gregor, H.; Gupta, S.; Hannien, O.; et al. Health Risks of Heavy Metals from Long-Range Transboundary Air Pollution; Chapter 3; World Health Organization: Copenhagen, Denmark, 2007; p. 43. [Google Scholar]
- Alka, B.; Madhurima, P.; Piyush kant, P. A review of hexavalent chromium contamination in India. Res. J. Chem. Sci. 2017, 7, 39–44. [Google Scholar]
- Bucchieri, F.; Puddicombe, S.M.; Lordan, J.L.; Richter, A.; Buchanan, D.; Wilson, S.J.; Ward, J.; Zummo, G.; Howarth, P.H.; Djukanovi, R.; et al. Asthmatic Bronchial Epithelium is More Susceptible to Oxidant-Induced Apoptosis. Am. J. Respir. Cell. Mol. Biol. 2002, 27, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Yanosky, J.D.; Tonne, C.C.; Beevers, S.D.; Wilkinson, P.; Kelly, F.J. Modeling Exposures to the Oxidative Potential of PM10. Environ. Sci. Technol. 2012, 46, 7612–7620. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, U.S.; Rastogi, N.; McWhinney, R.D.; Urch, B.; Chow, C.W.; Evans, G.J.; Scott, J.A. The combined effects of physicochemical properties of size-fractionated ambient PM on in vitro toxicity in human A549 lung epithelial cells. Toxicol. Rep. 2014, 1, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.M.; Breznan, D.; Karthikeyan, S.; MacKinnon-Roy, C.; Charland, J.P.; Dabek-Zlotorzynska, E.; Celo, V.; Kumarathasan, P.; Brook, J.R.; Vincent, R. Cytotoxic and inflammatory potential of size-fractionated particulate matter collected repeatedly within a small urban area. Part. Fibre Toxicol. 2015, 12, 1–19. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, C.; Liu, N.; Gao, C.; Liu, F. Morin Exhibits Anti-Inflammatory Effectson IL-1β-Stimulated Human Osteoarthritis Chondrocytes by Activating the Nrf2 Signaling Pathway. Cell. Physiol. Biochem. 2018, 51, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Subash, S.; Subramanian, P. Effect of morin on the levels of circulatory liver markers and redox status in experimental chronic hyperammonaemic rats. Singap. Med. J. 2008, 49, 650–655. [Google Scholar]
- Zhang, R.; Kang, K.A.; Kang, S.S.; Park, J.W.; Hyun, J.W. Morin (2,3,4,5,7-Pentahydroxyflavone) protected cells against γ-radiation-induced Oxidative stress. Basic Clin. Pharm. Toxicol. 2011, 108, 63–72. [Google Scholar] [CrossRef]
- Sinharay, R.; Gong, J.; Barratt, B.; Ohman-Strickland, P.; Ernst, S.; Kelly, F.J.; Zhang, J.J.; Collins, P.; Cullinan, P.; Chung, K.F. Respiratory and cardiovascular responses to walking down a traffic-polluted road compared with walking in a traffic-free area in participants aged 60 years and older with chronic lung or heart disease and age-matched healthy controls: A randomized, crossover study. Lancet 2018, 391, 339–349. [Google Scholar] [CrossRef]
- Zheng, Y.; Sanchez-Guerra, M.; Zhang, Z.; Joyce, B.T.; Zhong, J.; Kresovich, J.K.; Liu, L.; Zhang, W.; Gao, T.; Chang, D.; et al. Traffic-derived particulate matter exposure and histone H3 modification: A repeated measures study. Environ. Res. 2017, 153, 112–119. [Google Scholar] [CrossRef]
- Pindus, M.; Orru, H.; Maasikmets, M.; Kaasik, M.; Jogi, R. Association between Health symptoms and Particulate matter from Traffic and Residential Heating-Results from RHINE III in Tartu. Open Respir. Med. J. 2016, 10, 58–69. [Google Scholar] [CrossRef]
- Guo, S.; Xu, X.; Ouyang, Y.; Wang, Y.; Yang, J.; Yin, L.; Ge, J.; Wang, H. Microarray expression profile analysis ofcircular RNAs in pancreatic cancer. Mol. Med. Rep. 2018, 17, 7661–7671. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, Y.; Zhang, C.; Gong, W.; Zhang, X.; Nie, S. Polycyclic Aromatic Hydrocarbons from Particulate Matter 2.5 (PM2.5) in Polluted Air Changes miRNA Profile Related to Cardiovascular Disease. Med. Sci. Monit. 2018, 24, 5925–5934. [Google Scholar] [CrossRef] [PubMed]
- Mateu-Jimenez, M.; Curull, V.; Rodriguez-Fuster, A.; Aguiló, R.; Sanchez-Font, A.; Pijuan, L.; Gea, J.; Barreiro, E. Profile of epigenetic mechanisms in lung tumors of patients with underlying chronic respiratory conditions. Clin. Epigenetics 2018, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Hou, J.; Zhang, Q.; Zhong, W.; Li, B.; Li, C.; Liu, Z.; Yang, M.; Zhao, P. Circulating microRNA expression profiling and bioinformatics analysis of dysregulated microRNAs of patients with coronary artery disease. Medicine 2018, 97, e11428. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Pivarcsi, A. MicroRNAs in inflammation and response to injuries induced by environmental pollution. Mutat. Res. 2011, 717, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Xie, X.Y.; Xu, S.K.; Wang, Y.N.; Jiang, M.; Wen, L.R.; Lai, W.; Guan, L. PM2.5 Exposure Elicits Oxidative Stress Responses and Mitochondrial Apoptosis Pathway Activation in HaCaT Keratinocytes. Chin. Med. J. 2017, 130, 2205–2214. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Tong, L.; Liu, X.; Weng, X.; Chen, X.; Wang, D.; Dudley, S.C.; Weir, E.K.; Ding, W.; Lu, Z.; et al. Short term PM2.5 exposure caused a robust lung inflammation, vascular remodeling, and exacerbated transition from left ventricular failure to right ventricular hypertrophy. Redox. Biol. 2019, 22, 101161. [Google Scholar] [CrossRef]
- Gurgueira, S.A.; Lawrence, J.; Coull, B.; Murthy, G.G.; Gonzalez-Flecha, B. Rapid Increases in the Steady-State Concentration of Reactive Oxygen Species in the Lungs and Heart after Particulate Air Pollution Inhalation. Environ. Health Perspect. 2002, 110, 749–755. [Google Scholar] [CrossRef]
- Zhao, L.; Quan, J.; Li, Z.; Pan, X.; Wang, J.; Xu, J.; Xu, W.; Guan, X.; Li, H.; Yang, S.; et al. MicroRNA-222-3p promotes tumor cell migration and invasion and inhibits apoptosis, and is correlated with an unfavorable prognosis of patients with renal cell carcinoma. Int. J. Mol. Med. 2019, 43, 525–534. [Google Scholar] [CrossRef]
- Devlin, C.; Greco, S.; Martelli, F.; Ivan, M. miR-210: More than a silent player in Hypoxia. IUBMB Life 2011, 63, 94–100. [Google Scholar] [CrossRef]
- miRNAs Involved in DNA Damage Response. 2019. Available online: https://pathcards.genecards.org/card/mirnas_involved_in_dna_damage_response (accessed on 18 May 2019).
- Singh, B.; Ronghe, A.M.; Chatterjee, A.; Bhat, N.K.; Bhat, H.K. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis 2013, 34, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Lin, C.; Shi, Y.H.; Kuerban, G. MicroRNA-101 inhibits cell proliferation, invasion and promotes apoptosis by regulating Cycloxygenase-2 in Hela cervical carcinoma cells. Asian Pac. J. Cancer Prev. 2013, 14, 5915–5920. [Google Scholar] [CrossRef] [PubMed]
- Slabakova, E.; Culig, Z.; Remsik, J.; Soucek, K. Alternative mechanisms of miR34-a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.H.; Jang, J.Y.; Jeon, Y.K.; Kim, W.Y.; Kim, T.M.; Heo, D.S.; Kim, C.W. MicroRNA-146a down regulates NFκB activity via targeting TRAF6 and functions as tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin. Cancer Res. 2011, 17, 4761–4771. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Mungur, R.; Lan, P.; Wang, P.; Wan, S. MicroRNA-21 and microRNA-146a negatively regulate the secondary inflammatory response of microglia after intracerebral hemorrhage. Int. J. Clin. Exp. Pathol. 2018, 11, 3348–3356. [Google Scholar]
- Li, M.; Xu, T.; Zhou, F.; Wang, M.; Song, H.; Xiao, X.; Lu, B. Neuroprotective effects of four Phenylethanoid glycosides on H2O2–induced apoptosis on PC12 cells via the Nrf2/ARE pathway. Int. J. Mol. Sci. 2018, 19, 1135. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, S.; Muthuselvam, P.; Pugalenthi, V.; Subramanian, N.; Ramkumar, K.M.; Suresh, T.; Suzuki, T.; Rajaguru, P. Toxicoproteomic analysis of human lung epithelial cells exposed to Steel industry ambient particulate matter (PM) reveals possible mechanism of PM related carcinogenesis. Environ. Pollut. 2018, 239, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, M.V.; Henley, A.B.; Arsenian-Henrikson, M. The MYCN Protein in Health and Disease. Genes 2017, 8, 113. [Google Scholar] [CrossRef]
- Crosby, M.E.; Kulshreshtha, R.; Ivan, M.; Glazer, P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009, 69, 1221–1229. [Google Scholar] [CrossRef]
- Huang, J.W.; Wang, Y.; Dhillon, K.K.; Calses, P.; Villegas, E.; Mitchell, P.S.; Tewari, M.; Kemp, C.J.; Taniguchi, T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol. Cancer Res. 2013, 11, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Ng, W.L.; Zhang, X.; Wang, P.; Zhang, Z.; Mo, Y.Y.; Mao, H.; Hao, C.; Olson, J.J.; Curran, W.J.; et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE 2010, 5, e11397. [Google Scholar] [CrossRef] [PubMed]
- Feurstein, S.; Rucker, F.G.; Bullinger, L.; Hofmann, W.; Manukjan, G.; Gohring, G.; Lehmann, U.; Heuser, M.; Ganser, A.; Dohner, K.; et al. Haploinsufficiency of ETV6 and CDKN1B in patients with acute myeloid leukemia and complex karyotype. BMC Genom. 2014, 15, 784. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ie Sage, C.; Nagel, R.; Agami, R. Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle 2007, 6, 2742–2749. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Forward Primer (5′-3′) |
|---|---|
| miR 146a (MI0000477) | TGAGAACTGAATTCCATGGGT |
| miR 21 (MI0000077) | TAGCTTATCAGACTGATGTTGA |
| miR 222 (MI0000299) | CTCAGTAGCCAGTGTAGATCCT |
| miR 24 (MI0000080) | TGCCTACTGAGCTGATATC |
| miR 421 (MI0003685) | ATCAACAGACATTAATTGGGCGC |
| miR 210 (MI0000286) | AGCCCCTGCCCACCGCACACTG |
| miR 101 (MI0000103) | CAGTTATCACAGTGCTGATGCT |
| miR 34a (MI0000268) | TGGCAGTGTCTTAGCTGGTTGT |
| miR 93 (MI0000095) | CAAAGTGCTGTTCGTGCAGGTAG |
| miR 28 (MI0000086) | AAGGAGCTCACAGTCTATTGAG |
| miR 200a (MI0000737) | CATCTTACCGGACAGTGCTG |
| RNU48 | AGTGATGATGACCCCAGGTAA |
| Universal Reverse Primer for 11 miRNAs (5′-3′) | ATTCTAGAGGCCGAGGCGGCCGACATGT |
| Target Gene | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| NrF2 | ATGCCCTCACCTGCTACTTT | GCCAAGTAGTGTGTCTCC |
| NFκB | ATCCTTCTTTGACTCATACA | CCTTTGCTGGTCCCACATAG |
| MycN | AAGCCCTGGACGGGATTGCG | CGCAGAAGCCATTACTCCC |
| CDKN1A | TGGGGATGTCCGTCAGAACC | CCTCCTCCCAACTCATCCCG |
| CDKN1B | CCGCCCTCCCGCTCGCCAG | GTCCATCCGCTCCAGGCTA |
| RAD52 | CTCCCACCTCTGCCTTACAA | CCCATCCCCAAGGTCTCATT |
| BIM | TGACTCTGACTCTCGGACTG | TCCAATACGCCGCAACTCT |
| BAG1 | TGGGAAGTAGTCGGGCGGGG | CGAGAGGGAGGCGGACCACG |
| β-actin | GGCGGACTATGACTTAGTTG | AAACAACAATGTGCAATCAA |
| PM2.5 Sampling Area | Concentration of PM2.5 (µg/m3 of air) | PM2.5 Size Range (nm) | Peak Intensity Size (nm) | Polydispersity Index (PDI) | Zeta Potential (mV) | Conductivity (mS/cm) |
|---|---|---|---|---|---|---|
| Rural | 30.34 | 750–2585 | 759.4 | 1.000 | −34.7 | 0.021 |
| Urban | 36.53 | 550–2330 | 552.9 | 1.000 | −36.6 | 0.043 |
| Industrial | 41.93 | 440–1060 | 444.0 | 0.962 | −29.5 | 0.113 |
| Traffic | 51.47 | 605–1980 | 605.1 | 1.000 | −31.0 | 0.023 |
| Rural | Urban | Industrial | Traffic | |
|---|---|---|---|---|
| As | 0.003 | 0.006 | 0.009 | 0.002 |
| Cd | - | 0.001 | - | - |
| Cr | 0.057 | 0.065 | 0.628 | 0.056 |
| Cu | 0.001 | 0.001 | 0.001 | 0.001 |
| Fe | 0.985 | 1.116 | 1.699 | 1.357 |
| Mn | 0.039 | 0.048 | 0.006 | 0.029 |
| Ni | 0.001 | 0.002 | 0.001 | 0.002 |
| Se | 0.001 | 0.001 | 0.002 | 0.001 |
| Zn | 0.001 | 0.002 | 0.004 | 0.003 |
| Al | 0.004 | 0.002 | 0.019 | 0.002 |
| V | 0.003 | 0.008 | 0.012 | 0.005 |
| Pb | 0.228 | 0.322 | 0.318 | 0.710 |
| Total metal | 1.323 | 1.574 | 2.696 | 2.167 |
| Name of PAH | Rural | Urban | Industrial | Traffic |
|---|---|---|---|---|
| Total PAH | 5.6 | 20.1 | 25.0 | 24.8 |
| NaP | - | - | - | 0.2 |
| Acy | - | - | - | 0.7 |
| Ace | - | - | 0.2 | 1 |
| Fl | - | - | 0.1 | - |
| Phe | 1.6 | - | 3.6 | - |
| Ant | 0.2 | - | 3 | 1.4 |
| Flu | 0.8 | 0.4 | 11.5 | 0.5 |
| Pyr | - | 1.4 | 4.1 | 1.8 |
| BaA | - | 3.6 | - | 2.8 |
| Chr | - | 7.6 | - | 8.2 |
| BbF | - | 3.2 | - | 3.9 |
| BkF | - | - | - | 0.1 |
| BaP | 1.5 | 2.6 | 1.8 | 2.5 |
| DBA | 1.5 | 1.3 | 0.7 | 1.7 |
| BghiP | - | - | - | - |
| IND | - | - | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veerappan, I.; Sankareswaran, S.K.; Palanisamy, R. Morin Protects Human Respiratory Cells from PM2.5 Induced Genotoxicity by Mitigating ROS and Reverting Altered miRNA Expression. Int. J. Environ. Res. Public Health 2019, 16, 2389. https://doi.org/10.3390/ijerph16132389
Veerappan I, Sankareswaran SK, Palanisamy R. Morin Protects Human Respiratory Cells from PM2.5 Induced Genotoxicity by Mitigating ROS and Reverting Altered miRNA Expression. International Journal of Environmental Research and Public Health. 2019; 16(13):2389. https://doi.org/10.3390/ijerph16132389
Chicago/Turabian StyleVeerappan, Indhumathi, Senthil Kumar Sankareswaran, and Rajaguru Palanisamy. 2019. "Morin Protects Human Respiratory Cells from PM2.5 Induced Genotoxicity by Mitigating ROS and Reverting Altered miRNA Expression" International Journal of Environmental Research and Public Health 16, no. 13: 2389. https://doi.org/10.3390/ijerph16132389
APA StyleVeerappan, I., Sankareswaran, S. K., & Palanisamy, R. (2019). Morin Protects Human Respiratory Cells from PM2.5 Induced Genotoxicity by Mitigating ROS and Reverting Altered miRNA Expression. International Journal of Environmental Research and Public Health, 16(13), 2389. https://doi.org/10.3390/ijerph16132389





