Carotenoids in Algae: Distributions, Biosyntheses and Functions
Abstract
:1. Introduction
2. Distribution of Carotenoids
3. Carotenogenesis Pathways, Enzymes and Genes
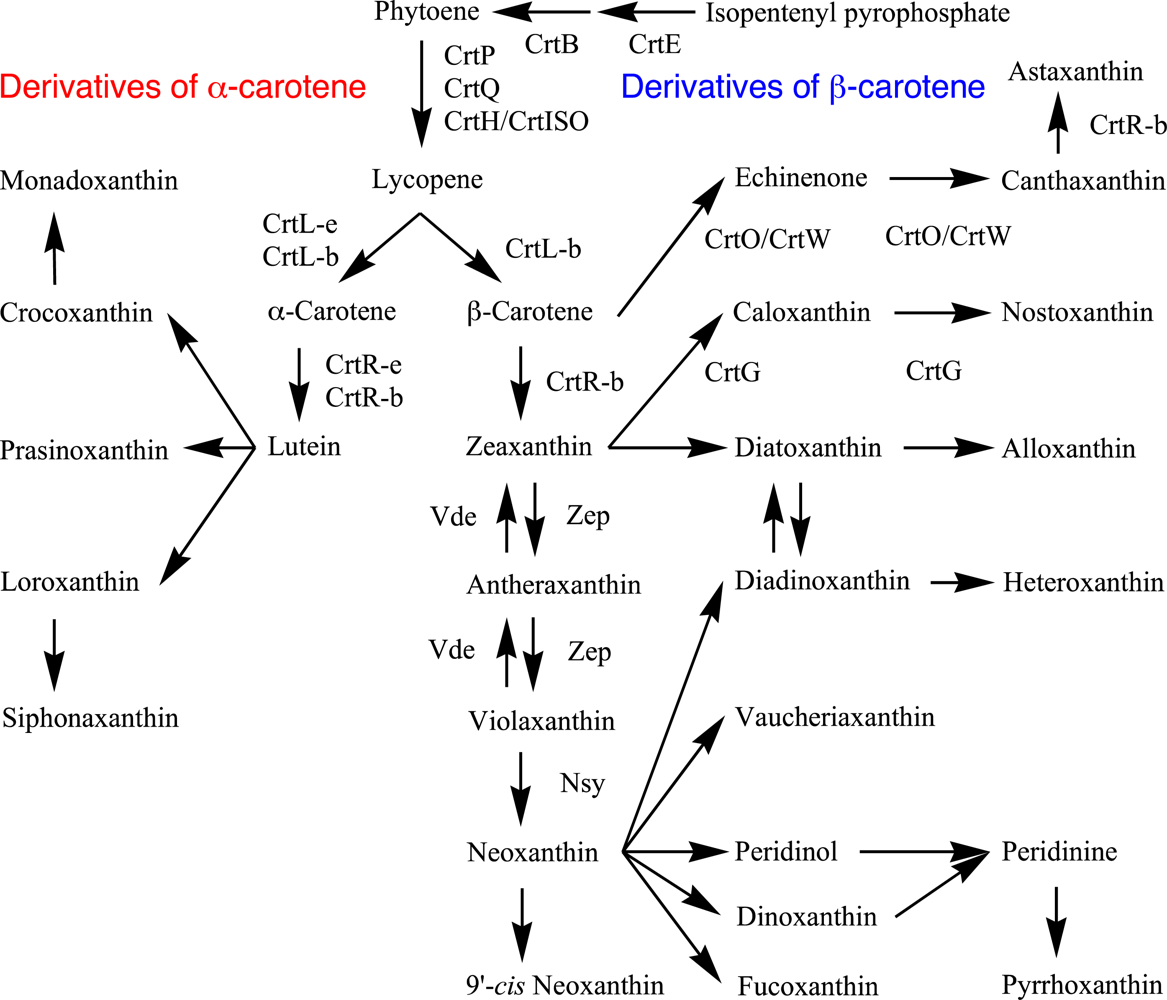
3.1. Lycopene Synthesis
3.1.1. Isopentenyl Pyrophosphate to Phytoene Synthesis
3.1.2. Phytoene to Lycopene Synthesis
3.2. β-Carotene and α-Carotene Synthesis by Lycopene Cyclases
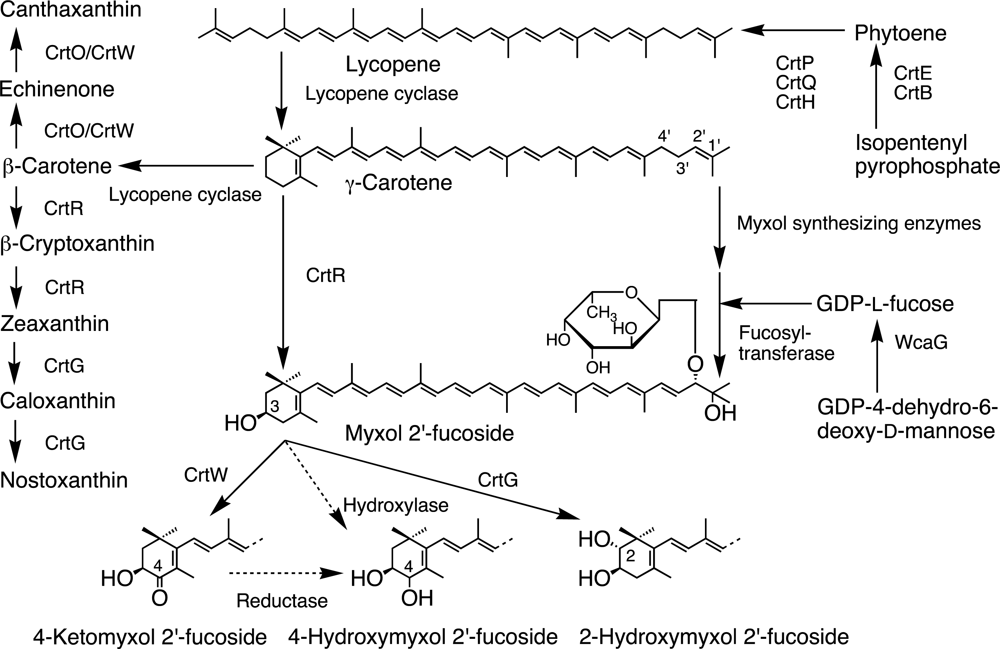
3.3. β-Carotene Derivatives and Their Synthesis
3.3.1. Cyanobacteria
3.3.2. Land Plants
3.3.3. Algae
3.4. α-Carotene Derivatives and Their Synthesis
4. Function of Carotenoids
References
- Britton, G; Liaaen-Jensen, S; Pfander, H. Carotenoids Handbook; Birkhäuser: Basel, Switzerland, 2004. [Google Scholar]
- Rowan, KS. Photosynthetic Pigments of Algae; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Bjørnland, T; Liaaen-Jensen, S. Distribution patterns of carotenoids in relation to chromophyte phylogeny and systematics. In The Chromophyte Algae: Problems and Perspectives; Green, JC, Leadbeater, BSC, Diver, WI, Eds.; Clarendon Press: Oxford, UK, 1989; pp. 37–60. [Google Scholar]
- Liaaen-Jensen, S. Marine carotenoids. New J Chem 1990, 14, 747–759. [Google Scholar]
- Mackey, MD; Mackey, DJ; Higgins, HW; Wright, SW. CHEMTAX-a program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Mar Ecol Prog Ser 1996, 144, 265–283. [Google Scholar]
- Jeffrey, SW; Vesk, M. Introduction to marine phytoplankton and their pigment signatures. In Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods; Jeffrey, SW, Mantoura, RFC, Wright, SW, Eds.; UNESCO Publishing: Paris, France, 1997; pp. 37–84. [Google Scholar]
- Liaaen-Jensen, S. Carotenoids in chemosystematics. In Carotenoids: Biosynthesis and Metabolism; Britton, G, Liaaen-Jensen, S, Pfander, H, Eds.; Birkhäuser: Basel, Switzerland, 1998; Volume 3, pp. 217–247. [Google Scholar]
- Frommolt, R; Werner, S; Paulsen, H; Goss, R; Wilhelm, C; Zauner, S; Maier, UG; Grossman, AR; Bhattacharya, D; Lohr, M. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol Biol Evol 2008, 25, 2653–2667. [Google Scholar]
- Bertrand, M. Carotenoid biosynthesis in diatoms. Photosynth Res 2010, 106, 89–102. [Google Scholar]
- Dembitsky, VM; Maoka, T. Allenic and cumulenic lipids. Prog Lipid Res 2007, 46, 328–375. [Google Scholar]
- Takaichi, S; Mimuro, M. Distribution and geometric isomerism of neoxanthin in oxygenic phototrophs: 9′-cis, a sole molecular form. Plant Cell Physiol 1998, 39, 968–977. [Google Scholar]
- Yoshii, Y; Takaichi, S; Maoka, T; Suda, S; Sekiguchi, H; Nakayama, T; Inouye, I. Variation of siphonaxanthin series among the genus Nephroselmis (Prasinophyceae, Chlorophyta), including a novel primary methoxy carotenoid. J Phycol 2005, 41, 827–834. [Google Scholar]
- Takaichi, S; Mochimaru, M. Carotenoids and carotenogenesis in cyanobacteria: Unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci 2007, 64, 2607–2619. [Google Scholar]
- Takaichi, S. Nippon Medical School, Kawasaki, Japan. Unpublished works, 2011.
- Takaichi, S; Maoka, T; Masamoto, K. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-α-l-fucoside), not rhamnoside. Plant Cell Physiol 2001, 42, 756–762. [Google Scholar]
- Schubert, N; García-Mendoza, E. Photoinhibition in red algal species with different carotenoid profiles. J Phycol 2008, 44, 1437–1446. [Google Scholar]
- Britton, G. Overview of carotenoid biosynthesis. In Carotenoids: Biosynthesis and Metabolism; Britton, G, Liaaen-Jensen, S, Pfander, H, Eds.; Birkhäuser: Basel, Switzerland, 1998; Volume 3, pp. 13–147. [Google Scholar]
- Lichtenthaler, HK. The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 1999, 50, 47–65. [Google Scholar]
- Eisenreich, W; Bacher, A; Arigoni, D; Rohdich, F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci 2004, 61, 1401–1426. [Google Scholar]
- Miziorko, HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys 2011, 505, 131–143. [Google Scholar]
- Ohto, C; Ishida, C; Nakane, H; Muramatsu, M; Nishino, T; Obata, S. A thermophilic cyanobacterium Synechococcus elongatus has three different Class I prenyltransferase genes. Plant Mol Biol 1999, 40, 307–321. [Google Scholar]
- Steiger, S; Jackisch, Y; Sandmann, G. Carotenoid biosynthesis in Gloeobacter violaceus PCC4721 involves a single crtI-type phytoene desaturase instead of typical cyanobacterial enzymes. Arch Microbiol 2005, 184, 207–214. [Google Scholar]
- Chamovitz, D; Misawa, N; Sandmann, G; Hirschberg, J. Molecular cloning and expression in Escherichia coli of a cyanobacterial gene coding for phytoene synthase, a carotenoid biosynthesis enzyme. FEBS Lett 1992, 296, 305–310. [Google Scholar]
- Martínez-Férez, I; Fernández-González, B; Sandmann, G; Vioque, A. Cloning and expression in Escherichia coli of the gene coding for phytoene synthase from the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys Acta 1994, 1218, 145–152. [Google Scholar]
- McCarthy, SS; Kobayashi, MC; Niyogi, KK. White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 2004, 168, 1249–1257. [Google Scholar]
- Steinbrenner, J; Linden, H. Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol 2001, 125, 810–817. [Google Scholar]
- Tsuchiya, T; Takaichi, S; Misawa, N; Maoka, T; Miyashita, H; Mimuro, M. The cyanobacterium Gloeobacter violaceus PCC 7421 uses bacterial-type phytoene desaturase in carotenoid biosynthesis. FEBS Lett 2005, 579, 2125–2129. [Google Scholar]
- Martínez-Férez, IM; Vioque, A. Nucleotide sequence of the phytoene desaturase gene from Synechocystis sp. PCC 6803 and characterization of a new mutation which confers resistance to the herbicide norflurazon. Plant Mol Biol 1992, 18, 981–983. [Google Scholar]
- Vila, M; Couso, I; León, R. Carotenoid content in mutants of the chlorophyte Chlamydomonas reinhardtii with low expression levels of phytoene desaturase. Process Biochem 2008, 43, 1147–1152. [Google Scholar]
- Huang, J; Liu, J; Li, Y; Chen, F. Isolation and characterization of the phytoene desaturase gene as a potential selective marker for genetic engineering of the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J Phycol 2008, 44, 684–690. [Google Scholar]
- Liu, J; Zhong, Y; Sun, Z; Huang, J; Sandmann, G; Chen, F. One amino acid substitution in phytoene desaturase makes Chlorella zofingiensis resistant to norflurazon and enhances the biosynthesis of astaxanthin. Planta 2010, 232, 61–67. [Google Scholar]
- Linden, H; Vioque, A; Sandmann, G. Isolation of a carotenoid biosynthesis gene coding for ζ-carotene desaturase from Anabaena PCC 7120 by heterologous complementation. FEMS Microbiol Lett 1993, 106, 99–104. [Google Scholar]
- Breitenbach, J; Fernández-González, B; Vioque, A; Sandmann, G. A higher-plant type ζ-carotene desaturase in the cyanobacterium Synechocystis PCC6803. Plant Mol Biol 1998, 36, 725–732. [Google Scholar]
- Masamoto, K; Wada, H; Kaneko, T; Takaichi, S. Identification of a gene required for cis-to-trans carotene isomerization in carotenogenesis of the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 2001, 42, 1398–1402. [Google Scholar]
- Breitenbach, J; Vioque, A; Sandmann, G. Gene sll0033 from Synechocystis 6803 encodes a carotene isomerase involved in the biosynthesis of all-E lycopene. Z Naturforsch 2001, 56c, 915–917. [Google Scholar]
- Cunningham, FX, Jr; Sun, Z; Chamovitz, D; Hirschberg, J; Gantt, E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp. strain PCC7942. Plant Cell 1994, 6, 1107–1121. [Google Scholar]
- Stickforth, P; Steiger, S; Hess, WR; Sandmann, G. A novel type of lycopene ɛ-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch Microbiol 2003, 179, 409–415. [Google Scholar]
- Cunningham, FX, Jr; Lee, H; Gantt, E. Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae. Eukaryot Cell 2007, 6, 533–545. [Google Scholar]
- Ramos, A; Coesel, S; Marques, A; Rodrigues, M; Baumgartner, A; Noronha, J; Rauter, A; Brenig, B; Varela, J. Isolation and characterization of a stress-inducible Dunaliella salina Lyc-β gene encoding a functional lycopene β-cyclase. Appl Microbiol Biotechnol 2008, 79, 819–828. [Google Scholar]
- Steinbrenner, J; Linden, H. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: Regulation by photosynthetic redox control. Plant Mol Biol 2003, 52, 343–356. [Google Scholar]
- Mochimaru, M; Msukawa, H; Maoka, T; Mohamed, HE; Vermaas, WFJ; Takaichi, S. Substrate specificities and availability of fucosyltransferase and β-carotene hydroxylase for myxol 2′-fucoside synthesis in Anabaena sp. strain PCC 7120 compared with Synechocystis sp. strain PCC 6803. J. Bacteriol 2008, 190, 6726–6733. [Google Scholar]
- Makino, T; Harada, H; Ikenaga, H; Matsuda, S; Takaichi, S; Shindo, K; Sandmann, G; Ogata, T; Misawa, N. Characterization of cyanobacterial carotenoid ketolase CrtW and hydroxylase CrtR by complementation analysis in Escherichia coli. Plant Cell Physiol 2008, 49, 1867–1878. [Google Scholar]
- Masamoto, K; Misawa, N; Kaneko, T; Kikuno, R; Toh, H. β-Carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 1998, 39, 560–564. [Google Scholar]
- Lagarde, D; Vermaas, W. The zeaxanthin biosynthesis enzyme β-carotene hydroxylase is involved in myxoxanthophyll synthesis in Synechocystis sp. PCC 6803. FEBS Lett 1999, 454, 247–251. [Google Scholar]
- Lagarde, D; Beuf, L; Vermaas, W. Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ Microbiol 2000, 66, 64–72. [Google Scholar]
- Linden, H. Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim Biophys Acta 1999, 1446, 203–212. [Google Scholar]
- Iwai, M; Maoka, T; Ikeuchi, M; Takaichi, S. 2,2′-β-Hydroxylase (CrtG) is involved in carotenogenesis of both nostoxanthin and 2-hydroxymyxol 2′-fucoside in Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol 2008, 49, 1678–1687. [Google Scholar]
- Baroli, I; Do, AD; Yamane, T; Niyogi, KK. Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlomydomonas reinhardtii from photooxidative stress. Plant Cell 2003, 15, 992–1008. [Google Scholar]
- Goss, R. Substrate specificity of the violaxanthin de-epoxidase of the primitive green alga Mantoniella squamata (Prasinophyceae). Planta 2003, 217, 801–812. [Google Scholar]
- Mochimaru, M; Msukawa, H; Takaichi, S. The cyanobacterium Anabaena sp. PCC 7120 has two distinct β-carotene ketolase: CrtO for echinenone and CrtW for ketomyxol synthesis. FEBS Lett 2005, 579, 6111–6114. [Google Scholar]
- Fernández-González, B; Sandmann, G; Vioque, A. A new type of asymmetrically acting β-carotene ketolase is required for the synthesis of echinenone in the cyanobacterium Synechocystis sp PCC 6803. J Biol Chem 1997, 272, 9728–9733. [Google Scholar]
- Steiger, S; Sandmann, G. Cloning of two carotenoid ketolase genes from Nostoc punctiforme for the heterologous production of canthaxanthin and astaxanthin. Biotechnol Lett 2004, 26, 813–817. [Google Scholar]
- Huang, J-C; Wang, Y; Sandmann, G; Chen, F. Isolation and characterization of a carotenoid oxygenase gene from Chlorella zofingiensis (Chlorophyta). Appl Microbiol Biotechnol 2006, 71, 473–479. [Google Scholar]
- Kajiwara, S; Kakizono, T; Saito, T; Kondo, K; Ohtani, T; Nishio, N; Nagai, S; Misawa, N. Isolation and functional identification of a novel cDNA from astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol Biol 1995, 29, 343–352. [Google Scholar]
- Huang, J-C; Chen, F; Sandmann, G. Stress-related differential expression of multiple β-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J Biotechnol 2006, 122, 176–185. [Google Scholar]
- Lotan, T; Hirschberg, J. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett 1995, 364, 125–128. [Google Scholar]
- Sandmann, G. Carotenoid biosynthesis in microorganisms and plants. Eur J Biochem 1994, 223, 7–24. [Google Scholar]
- Armstrong, GA. Genetics of eubacterial carotenoid biosynthesis: A colorful tale. Annu Rev Microbiol 1997, 51, 629–659. [Google Scholar]
- Misawa, N; Nakagawa, M; Kobayashi, K; Yamano, S; Izawa, Y; Nakamura, K; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 1990, 172, 6704–6712. [Google Scholar]
- Schneider, C; Böger, P; Sandmann, G. Phytoene desaturase: Heterologous expression in an active state, purification, and biochemical properties. Protein Expr Purif 1997, 10, 175–179. [Google Scholar]
- Takaichi, S. Distribution and biosynthesis of carotenoids. In The Purple Phototrophic Bacteria; Hunter, CN, Daldal, F, Thurnauer, MC, Beatty, JT, Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 97–117. [Google Scholar]
- Krubasik, P; Sandmann, G. Molecular evolution of lycopene cyclases involved in the formation of carotenoids with ionone end groups. Biochem Soc Trans 2000, 28, 806–810. [Google Scholar]
- Maresca, JA; Graham, JE; Wu, M; Eisen, JA; Bryant, DA. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc Natl Acad Sci USA 2007, 104, 11784–11789. [Google Scholar]
- Sandmann, G. Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiol Plant 2002, 116, 431–440. [Google Scholar]
- Harker, M; Hirschberg, J. Molecular biology of carotenoid biosynthesis in photosynthetic organisms. Methods Enzymol 1998, 297, 244–263. [Google Scholar]
- Cunningham, FX, Jr; Gantt, E. One ring or two? Determination of ring number in carotenoids by lycopene ɛ-cyclases. Proc Natl Acad Sci USA 2001, 98, 2905–2910. [Google Scholar]
- Hemmi, H; Ikejiri, S; Nakayama, T; Nishino, T. Fusion-type lycopene β-cyclase from a thermoacidophilic archaeon Sulfolobus solfataricus. Biochem Biophys Res Commun 2003, 305, 586–591. [Google Scholar]
- Maresca, JA; Frigaard, N-U; Bryant, DA. Identification of a novel class of lycopene cyclases in photosynthetic organisms. In Photosynthesis: Fundamental Aspects to Global Perspectives; van der Est, A, Bruce, D, Eds.; Allen Press: Lawrence, KS, USA, 2005; pp. 884–886. [Google Scholar]
- Swift, IE; Milborrow, BV; Jeffrey, SW. Formation of neoxanthin, diadinoxanthin and peridinin from [14C]zeaxanthin by a cell-free system from Amphidinium carterae. Phytochemistry 1982, 21, 2859–2864. [Google Scholar]
- Swift, IE; Milborrow, BV. Stereochemistry of allene biosynthesis and the formation of the acetylenic carotenoid diadinoxanthin and peridinin (C37) from neoxanthin. Biochem J 1981, 199, 69–74. [Google Scholar]
- Lemoine, Y; Schoefs, B. Secondary ketocarotenoid astaxanthin biosynthesis in algae: A multifunctional response to stress. Photosynth Res 2010, 106, 155–177. [Google Scholar]
- Kim, J; Smith, JJ; Tian, L; DellaPenna, D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol 2009, 50, 463–479. [Google Scholar]
- Durnford, DG. Structure and regulation of algal light-harvesting complex genes. In Photosynthesis in Algae; Larkum, AWD, Douglas, SE, Raven, JA, Eds.; Kluwer: Dordrecht, The Netherlands, 2003; pp. 63–82. [Google Scholar]
- Macpherson, AN; Hiller, RG. Light-harvesting systems in chlorophyll c-containing algae. In Light-Harvesting Antennas in Photosynthesis; Green, BR, Parson, WW, Eds.; Kluwer: Dordrecht, The Netherlands, 2003; pp. 323–352. [Google Scholar]
- Neilson, JAD; Durnford, DG. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth Res 2010, 106, 57–71. [Google Scholar]
- Kana, TM; Glibert, PM; Goericke, R; Welschmeyer, NA. Zeaxanthin and β-carotene in Synechococcus WH7803 respond differently to irradiance. Limnol Oceanogr 1998, 33, 1623–1627. [Google Scholar]
- Masamoto, K; Zsiros, O; Gombos, Z. Accumulation of zeaxanthin in cytoplasmic membranes of the cyanobacteirum Synechococcus sp. Strain PCC 7942 grown under high light condition. J Plant Physiol 1999, 155, 136–138. [Google Scholar]
- Kurisu, G; Zhang, H; Smith, JL; Cramer, WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 2003, 302, 1009–1014. [Google Scholar]
- Stroebel, D; Choquet, Y; Popot, J-L; Picot, D. An atypical haem in the cytochrome b6f complex. Nature 2003, 426, 413–418. [Google Scholar]
- Boronowsky, U; Wenk, S-O; Schneider, D; Jäger, C; Rögner, M. Isolation of membrane protein subunits in their native state: Evidence for selective binding of chlorophyll and carotenoid to the b6 subunit of the cytochrome b6f complex. Biochim Biophys Acta 2001, 1506, 55–66. [Google Scholar]
- Hofmann, E; Wrench, PM; Sharples, FP; Hiller, RG; Welte, W; Diederichs, K. Structural basis of light harvesting by carotenoids: Peridinin-chlorophyll-protein from Amphidinium carterae. Science 1996, 272, 1788–1791. [Google Scholar]
- Kerfeld, CA; Sawaya, MR; Brahmandam, V; Cascio, D; Ho, KK; Trevithick-Sutton, CC; Krogmann, DW; Yeates, TO. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure 2003, 11, 55–65. [Google Scholar]
- Wilson, A; Ajlani, G; Verbavatz, J-M; Vass, I; Kerfeld, CA; Kirilovsky, D. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 2006, 18, 992–1007. [Google Scholar]
- Englert, G; Bjørnland, T; Liaaen-Jensen, S. 1D and 2D NMR study of some allenic carotenoids of the fucoxanthin series. Magn Reson Chem 1990, 28, 519–528. [Google Scholar]
- Egeland, ES; Guillard, RRL; Liaaen-Jensen, S. Additional carotenoid prototype representatives and a general chemosystematic evaluation of carotenoids in Prasinophyceae (Chlorophyta). Phytochemistry 1997, 44, 1087–1097. [Google Scholar]
- Yoshii, Y; Takaichi, S; Maoka, T; Hanada, S; Inouye, I. Characterization of two unique carotenoid fatty acid esters from Pterosperma cristatum (Prasinophyceae, Chlorophyta). J Phycol 2002, 38, 297–303. [Google Scholar]
- Egeland, ES; Liaaen-Jensen, S. Ten minor carotenoids from Prasinophyceae (Chlorophyta). Phytochemistry 1995, 40, 515–520. [Google Scholar]
- Mimuro, M; Nagashima, U; Takaichi, S; Nishimura, Y; Yamazaki, I; Katoh, T. Molecular structure and optical properties of carotenoids for the in vivo energy transfer function in the algal photosynthetic pigment system. Biochim Biphys Acta 1992, 1098, 271–274. [Google Scholar]
- Akimoto, S; Yokono, M; Higuchi, M; Tomo, T; Takaichi, S; Murakami, A; Mimuro, M. Solvent effects on excitation relaxation dynamics of a keto-carotenoid, siphonaxanthin. Photochem Photobiol Sci 2008, 7, 1206–1209. [Google Scholar]
- Yamamoto, HY; Bugos, RC; Hieber, AD. Biochemistry and molecular biology of the xanthophyll cycle. In The Phytochemistry of Carotenoids; Frank, HA, Young, AJ, Britton, G, Cogdell, RJ, Eds.; Kluwer: Dordrecht, The Netherlands, 1999; pp. 293–303. [Google Scholar]
- Goss, R; Jakob, T. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth Res 2010, 106, 103–122. [Google Scholar]
- Grouneva, I; Jakob, T; Wilhelm, C; Goss, R. Influence of ascorbate and pH on the activity of the diatom xanthophyll cycle-enzyme diadinoxanthin de-epoxidase. Physiol Plant 2006, 126, 205–211. [Google Scholar]
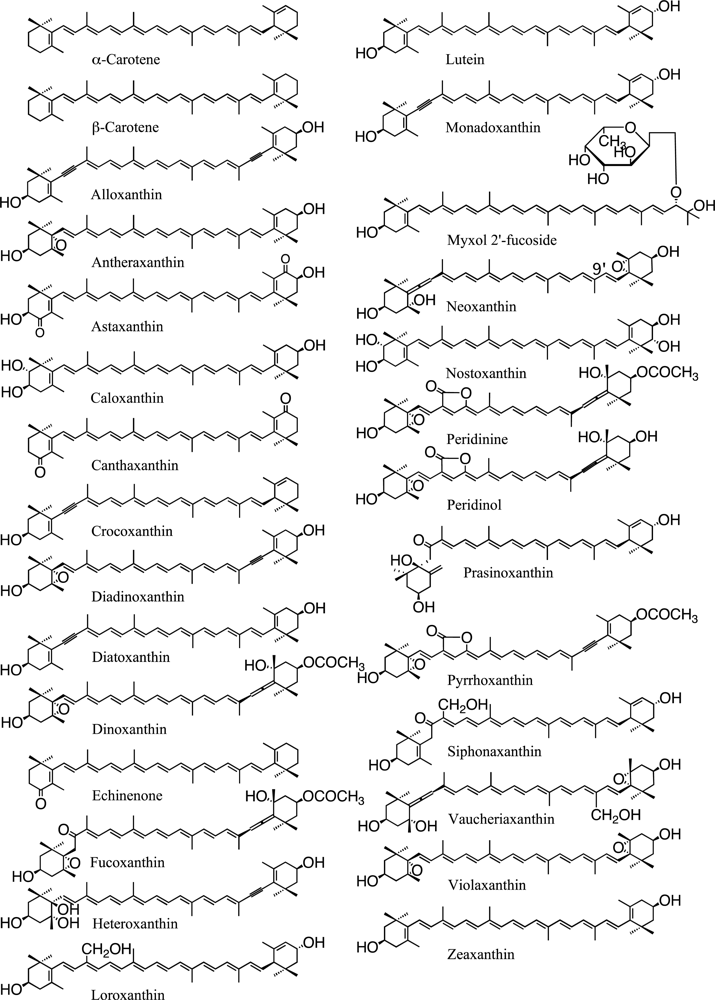
| Division Class | Carotene | Xanthophyll | Chlorophyll | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | α | Ze | Vi | Ne | Da | Dd | Fx | Va | Lu | Lo | Sx | Other xanthophyll(s) | a | b | c | |
| Cyanophyta | H | L | H | No, L; Ec, H; My, H | H | L | ||||||||||
| Glaucophyta | H | H | H | |||||||||||||
| Rhodophyta | ||||||||||||||||
| Unicellular type | H | H | H | |||||||||||||
| Macrophytic type | L | L | H | L | L | H | H | |||||||||
| Cryptophyta | H | L | Al, L; Cr, L; Mo, L | H | H | |||||||||||
| Heterokontophyta | ||||||||||||||||
| Chrysophyceae | H | L | L | L | H | L | H | H | ||||||||
| Raphidophyceae | H | H | L | L | L | L | H | H | ||||||||
| Bacillariophyceae | H | L | L | L | H | H | H | |||||||||
| Phaeophyceae | H | H | H | L | L | H | H | H | ||||||||
| Xanthophyceae | H | L | H | H | Va-FA, L | H | H | |||||||||
| Eustigmatophyceae | H | H | L | H | ||||||||||||
| Haptophyta | H | L | L | H | H | Fx-FA, L | H | H | ||||||||
| Dinophyta | L | L | L | H | L | Pe, H | H | H | ||||||||
| Euglenophyta | H | L | L | L | H | L | L | H | H | |||||||
| Chlorarachniophyta | H | L | L | L | L | L | Lo-FA, L | H | H | |||||||
| Chlorophyta | ||||||||||||||||
| Prasinophyceae | H | L | L | H | H | L | L | H | Pr, L; Lo-FA, L; Sx-FA, H | H | H | |||||
| Chlorophyceae | H | H | L | H | H | H | L | L | Sx-FA, L | H | H | |||||
| Ulvophyceae | H | L | L | H | H | L | L | L | Sx-FA, H | H | H | |||||
| Trebouxiophyceae | H | L | H | H | H | H | H | |||||||||
| Charophyceae | H | L | H | H | H | H | H | |||||||||
| Land Plants | H | L | L | H | H | H | H | H | ||||||||
| Gene | Enzyme | Species | References |
|---|---|---|---|
| crtE, ggps | Geranylgeranyl pyrophosphate synthase | Thermosynechococcus elongates BP-1 | [21] |
| crtB, pys, psy | Phytoene synthase | Gloeobacter violaceus PCC 7421 | [22] |
| Synechococcus elongatus PCC 7942 | [23] | ||
| Synechocystis sp. PCC 6803 | [24] | ||
| Chlamydomonas reinhardtii | [25] | ||
| Haematococcus pluvialis NIES-144 | [26] | ||
| crtI | Phytoene desaturase (bacterial type) | Gloeobacter violaceus PCC 7421 | [22,27] |
| crtP, pds | Phytoene desaturase (plant type) | Synechococcus elongatus PCC 7942 | [23] |
| Synechocystis sp. PCC 6803 | [28] | ||
| Chlamydomonas reinhardtii | [29] | ||
| Chlorella zofingiensis ATCC 30412 | [30,31] | ||
| crtQ, zds | ζ-Carotene desaturase | Anabaena sp. PCC 7120 | [32] |
| Synechocystis sp. PCC 6803 | [33] | ||
| crtH, crtISO | Carotene isomerase | Synechocystis sp. PCC 6803 | [34,35] |
| crtL, crtL-b, lcy-b | Lycopene β-cyclase | Synechococcus elongatus PCC 7942 | [36] |
| Prochlorococcus marinus MED4 | [37] | ||
| Cyanidioschyzon merolae NIES-1332 | [38] | ||
| Dunaliella salina CCAP 19/30 | [39] | ||
| Haematococcus pluvialis NIES-144 | [40] | ||
| crtL-e, lcy-e | Lycopene ɛ-cyclase | Prochlorococcus marinus MED4 | [37] |
| crtR | β-Carotene hydroxylase | Anabaena sp. PCC 7120 | [41,42] |
| Anabaena variabilis ATCC 29413 | [42] | ||
| Synechocystis sp. PCC 6803 | [42–45] | ||
| Haematococcus pluvialis NIES-144 | [46] | ||
| crtG | β-Carotene 2-hydroxylase | Thermosynechococcus elongates BP-1 | [47] |
| zep, npq | Zeaxanthin epoxidase | Chlamydomonas reinhardtii CC-125 | [48] |
| vde | Violaxanthin de-epoxidase | Mantonilla squamata | [49] |
| crtO | β-Carotene ketolase | Anabaena sp. PCC 7120 | [50] |
| Gloeobacter violaceus PCC 7421 | [22] | ||
| Synechocystis sp. PCC 6803 | [42,45,51] | ||
| crtW, bkt | β-Carotene ketolase | Anabaena sp. PCC 7120 | [42,50] |
| Gloeobacter violaceus PCC 7421 | [22,27,42] | ||
| Nostoc punctiforme PCC 73102 | [42,52] | ||
| Chlorella zofingiensis ATCC 30412 | [53] | ||
| Haematococcus pluvialis NIES-144 | [54,55] | ||
| Haematococcus pluvialis strain 34/7 | [56] | ||
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Takaichi, S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Mar. Drugs 2011, 9, 1101-1118. https://doi.org/10.3390/md9061101
Takaichi S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Marine Drugs. 2011; 9(6):1101-1118. https://doi.org/10.3390/md9061101
Chicago/Turabian StyleTakaichi, Shinichi. 2011. "Carotenoids in Algae: Distributions, Biosyntheses and Functions" Marine Drugs 9, no. 6: 1101-1118. https://doi.org/10.3390/md9061101
APA StyleTakaichi, S. (2011). Carotenoids in Algae: Distributions, Biosyntheses and Functions. Marine Drugs, 9(6), 1101-1118. https://doi.org/10.3390/md9061101




