Carotenoid β-Ring Hydroxylase and Ketolase from Marine Bacteria—Promiscuous Enzymes for Synthesizing Functional Xanthophylls
Abstract
:1. Introduction
2. Bacterial Strains Producing Ketocarotenoids
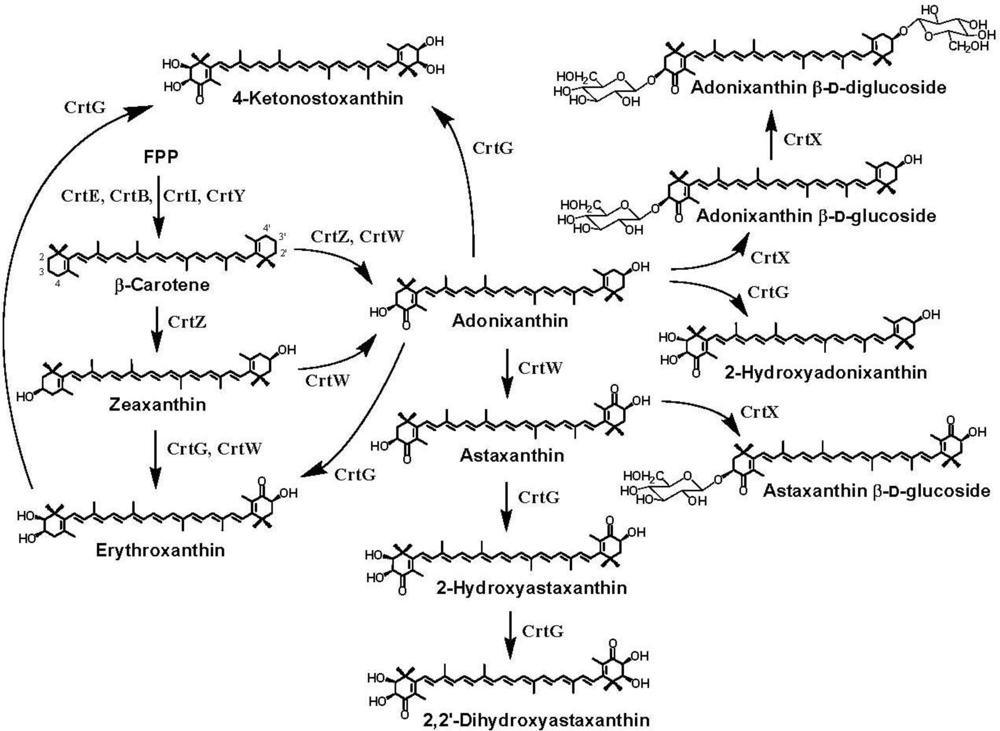
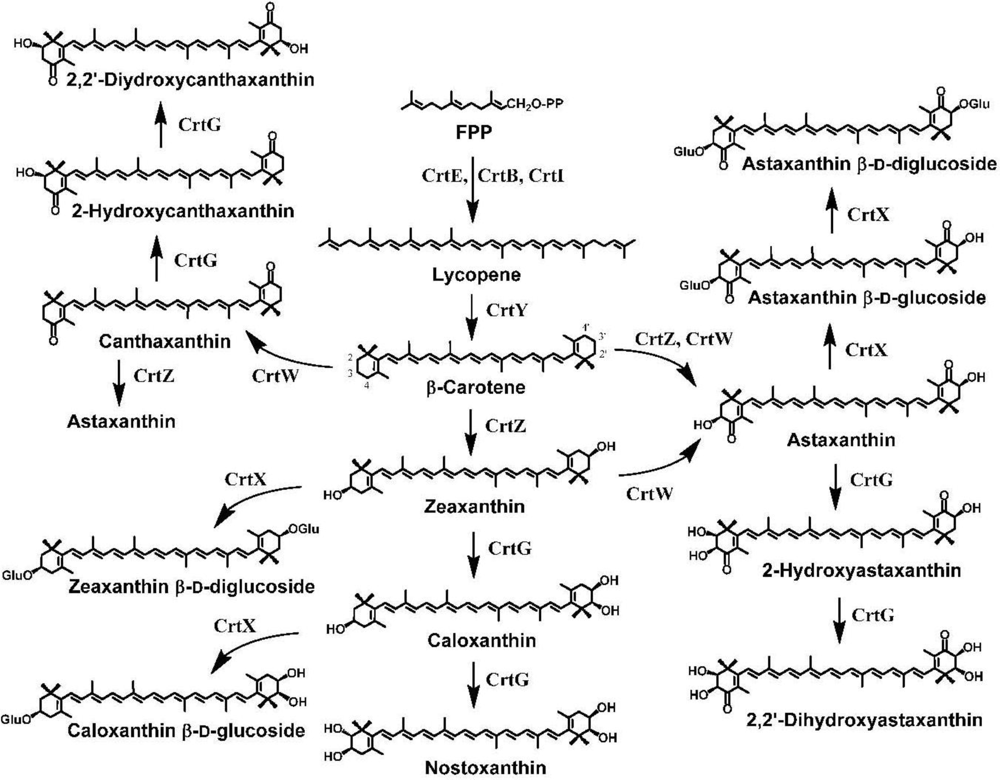
3. Ketocarotenoid Biosynthesis Genes
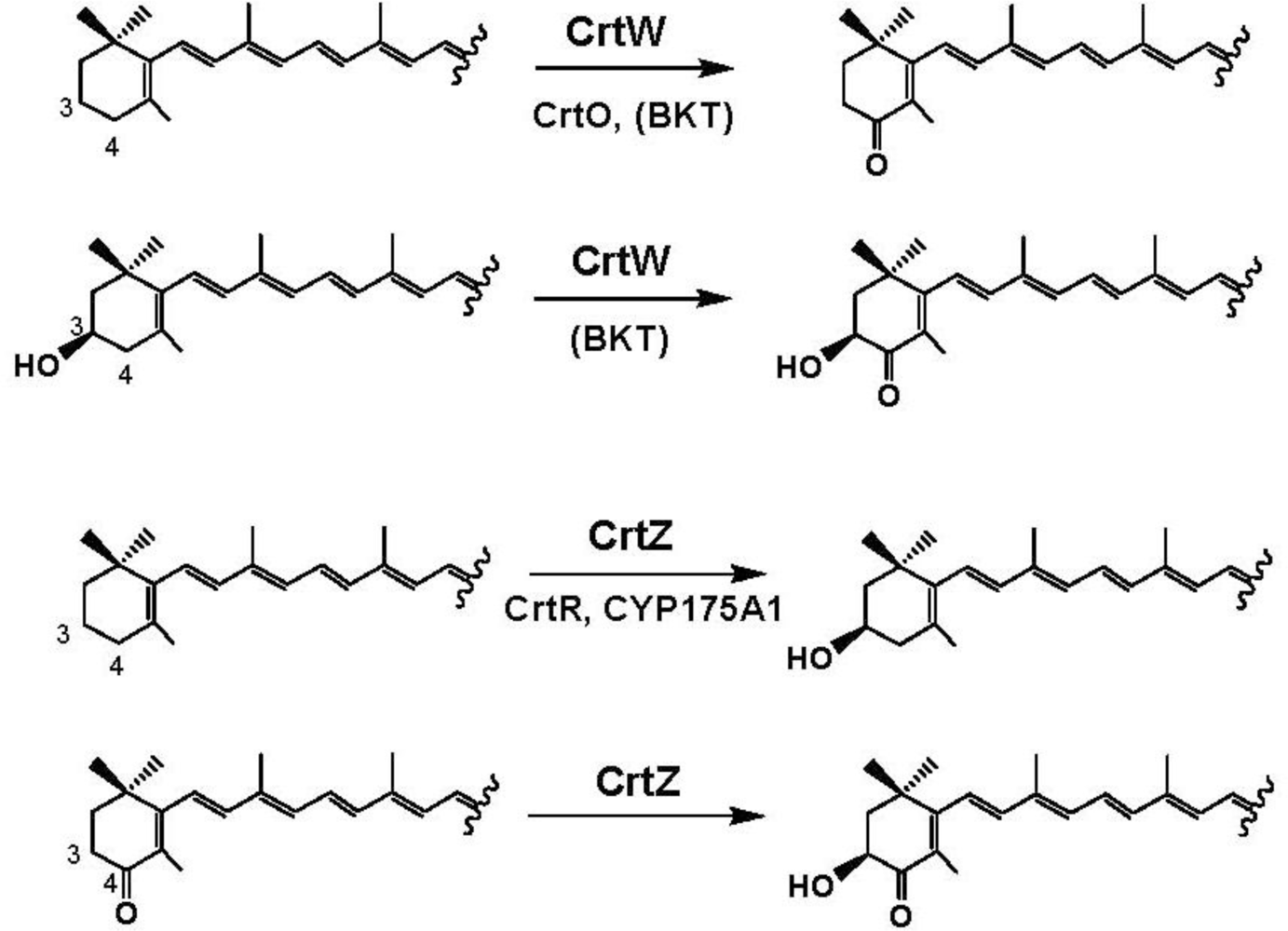
4. Carotenoid 4,4′-Ketolase
5. Carotenoid 3,3′-Hydroxylase
6. Carotenoid 2,2′-Hydroxylase
7. Pathway Engineering for the Synthesis of Functional Xanthophylls via the Incorporation of crtW, crtZ, and/or crtG Genes
8. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References
- Britton, G; Liaaen-Jensen, S; Pfander, H. Carotenoids Handbook; Birkhauser Verlag: Basel, Switzerland, 2004. [Google Scholar]
- Shindo, K; Endo, M; Miyake, Y; Wakasugi, K; Morritt, D; Bramley, PM; Fraser, PD; Kasai, H; Misawa, N. Methyl glucosyl-3,4-dehydro-apo-8′-lycopenoate, a novel antioxidative glyco-C30-carotenoic acid produced by a marine bacterium Planococcus maritimus. J Antibiot 2008, 61, 729–735. [Google Scholar]
- Shindo, K; Mikami, K; Tamesada, E; Takaichi, S; Adachi, K; Misawa, N; Maoka, T. Diapolycopenedioc acid xylosyl ester, a novel glyco-C30-carotenoic acid produced by a new marine bacterium Rubritalea squalenifaciens. Tetrahedron Lett 2007, 48, 2725–2727. [Google Scholar]
- Tao, L; Yao, H; Kasai, H; Misawa, N; Cheng, Q. A carotenoid synthesis gene cluster from Algoriphagus sp. KK10202C with a novel fusion-type lycopene-β-cyclase gene. Mol Genet Genomics 2006, 276, 79–86. [Google Scholar]
- Shindo, K; Kikuta, K; Suzuki, A; Katsuta, A; Kasai, H; Yasumoto-Hirose, M; Matuo, Y; Misawa, N; Takaichi, S. Rare carotenoids, (3R)-Saproxanthin and (3R,2′S)-myxol, isolated from novel marine bacteria (Flavobacteriaceae) and their antioxidant activities. Appl Microbiol Biotechnol 2007, 74, 1350–1357. [Google Scholar]
- Teramoto, M; Takaichi, S; Inomata, Y; Ikenaga, H; Misawa, N. Structural and functional analysis of a lycopene β-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett 2003, 545, 120–126. [Google Scholar]
- Yokoyama, A; Izumida, H; Miki, W. Production of astaxanthin and 4-ketozeaxanthin by the marine bacterium, Agrobacterium aurantiacum. Biosci Biotechnol Biochem 1994, 58, 1842–1844. [Google Scholar]
- Yokoyama, A; Izumida, H; Shizuri, Y. New carotenoid sulfates isolated from a marine bacterium. Biosci Biotechnol Biochem 1996, 60, 1877–1878. [Google Scholar]
- Yokoyama, A; Miki, W; Izumida, H; Shizuri, Y. New trihydroxy-keto-carotenoids isolated from an astaxanthin-producing marine bacterium. Biosci Biotechnol Biochem 1996, 60, 200–203. [Google Scholar]
- Maruyama, T; Kasai, H; Choi, SK; Ramasamy, AK; Inomata, Y; Misawa, N. Structure of a complete carotenoid biosynthesis gene cluster of marine bacterium Paracoccus sp. strain N81106. Carotenoid Sci 2007, 11, 50–55. [Google Scholar]
- Nishida, Y; Adachi, K; Kasai, H; Shizuri, Y; Shindo, K; Sawabe, A; Komemushi, S; Miki, W; Misawa, N. Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol 2005, 71, 4286–4296. [Google Scholar]
- Nishino, H; Murakoshi, M; Ii, T; Takemura, M; Kuchide, M; Kanazawa, M; Mou, XY; Wada, S; Masuda, M; Ohsaka, Y; et al. Carotenoids in cancer chemoprevention. Cancer Metastasis Rev 2002, 21, 257–264. [Google Scholar]
- Pashkow, FJ; Watumull, DG; Campbell, CL. Astaxanthin: A novel potential treatment for oxidative stress and inflammation in cardiovascular disease. Am J Cardiol 2008, 101, 58D–68D. [Google Scholar]
- Camera, E; Matrofrancesco, A; Fabbri, C; Daubrawa, F; Picardo, M; Sies, H; Stahl, W. Astaxanthin, canthaxanthin and β-carotene differently affect UVA-induced oxidative damage and expression of oxidative stress-responsive enzymes. Exp Dermatol 2009, 18, 222–231. [Google Scholar]
- Jackson, H; Braun, CL; Ernst, H. The chemistry of novel xanthophyll carotenoids. Am J Cardiol 2008, 101, 50D–57D. [Google Scholar]
- Misawa, N. Pathway engineering of plants toward astaxanthin production. Plant Biotechnol 2009, 26, 93–99. [Google Scholar]
- Izumida, H; Adachi, K; Nishizima, M; Endo, M; Miki, W. Akalone: A novel xanthine oxidase inhibitor produced by the marine bacterium, Agrobacterium aurantiacum sp. nov. J Mar Biotechnol 1995, 2, 115–118. [Google Scholar]
- Yokoyama, A; Adachi, K; Shizuri, Y. New carotenoid glycosides, astaxanthin glucoside and adonixanthin glucoside, isolated from the astaxanthin-producing marine bacterium, Agrobacterium aurantiacum. J Nat Prod 1995, 58, 1929–1933. [Google Scholar]
- Lee, JH; Kim, YS; Choi, TJ; Lee, WJ; Kim, YT. Paracoccus haeundaensis sp. nov., a Gram-negative, halophilic, astaxanthin-producing bacterium. Int J Syst Evol Microbiol 2004, 54, 1699–1672. [Google Scholar]
- Takaichi, S; Maoka, T; Akimoto, N; Khan, ST; Harayama, S. Major carotenoid isolated from Paracoccus schoinia NBRC 100637T is adonixanthin diglucoside. J Nat Prod 2006, 69, 1823–1825. [Google Scholar]
- Khan, ST; Takaichi, S; Harayama, S. Paracoccus marinus sp. nov., an adonixanthin diglucoside-producing bacterium isolated from coastal seawater in Tokyo Bay. Int J Syst Evol Microbiol 2008, 58, 383–386. [Google Scholar]
- Harker, M; Hirschberg, J; Oren, A. Paracoccus marcusii sp. nov., an orange gram-negative coccus. Int J Syst Bacteriol 1998, 48, 543–548. [Google Scholar]
- Misawa, N; Nakagawa, M; Kobayashi, K; Yamano, S; Izawa, Y; Nakamura, K; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 1990, 172, 6704–6712. [Google Scholar]
- Armstrong, GA; Alberti, M; Hearst, JE. Conserved enzymes mediate the early reactions of carotenoid biosynthesis in nonphotosynthetic and photosynthetic prokaryotes. Proc Natl Acad Sci USA 1990, 87, 9975–9979. [Google Scholar]
- Hundle, BS; Beyer, P; Kleinig, H; Englert, G; Hearst, JE. Carotenoids of Erwinia herbicola and an Escherichia coli HB101 strain carrying the Erwinia herbicola carotenoid gene cluster. Photochem Photobiol 1991, 54, 89–93. [Google Scholar]
- Nakagawa, M; Misawa, N. Analysis of carotenoid glycosides produced in gram-negative bacteria by introduction of the Erwinia uredovora carotenoid biosynthesis genes. Agric Biol Chem 1991, 55, 2147–2148. [Google Scholar]
- Sandmann, G; Misawa, N. New functional assignment of the carotenogenic genes crtB and crtE with constructs of these genes from Erwinia species. FEMS Microbiol Lett 1992, 90, 253–258. [Google Scholar]
- Math, SK; Hearst, JE; Poulter, CD. The crtE gene in Erwinia herbicola encodes geranylgeranyl diphosphate synthase. Proc Natl Acad Sci USA 1992, 89, 6761–6764. [Google Scholar]
- Neudert, U; Martines-Ferez, IM; Fraser, PD; Sandmann, G. Expression of an active phytoene synthease from Erwinia uredovora and biochemical properties of the enzyme. Biochim Biophys Acta 1998, 1392, 51–58. [Google Scholar]
- Fraser, PD; Misawa, N; Linden, H; Yamano, S; Kobayashi, K; Sandmann, G. Expression in E. coli, purification and reactivation of the recombinant Erwinia uredovora phytoene desturase. J Biol Chem 1992, 267, 19891–19895. [Google Scholar]
- Schnurr, G; Misawa, N; Sandmann, G. Expression, purification and properties of lycopene cyclase from. Erwinia uredovora Biochem J 1996, 315, 869–874. [Google Scholar]
- Hundle, BS; O’Brien, DA; Alberti, M; Beyer, P; Hearst, JE. Functional expression of zeaxanthin glucosyltransferase from Erwinia herbicola and a proposed uridine diphosphate binding site. Proc Natl Acad Sci USA 1992, 89, 9321–9325. [Google Scholar]
- Misawa, N; Kajiwara, S; Kondo, K; Yokoyama, A; Satomi, Y; Saito, T; Miki, W; Ohtani, T. Canthaxanthin biosynthesis by the conversion of methylene to keto groups in a hydrocarbon β-carotene by a single gene. Biochem Biophy Res Commun 1995, 209, 867–876. [Google Scholar]
- Misawa, N; Truesdale, MR; Sandmann, G; Fraser, PD; Bird, C; Schuch, W; Bramley, PM. Expression of a tomato cDNA coding for phytoene synthase in Escherichia coli, phytoene formation in vivo and in vitro, and functional analysis of the various truncated gene products. J Biochem 1994, 116, 980–985. [Google Scholar]
- Misawa, N; Satomi, Y; Kondo, K; Yokoyama, A; Kajiwara, S; Saito, T; Ohtani, T; Miki, W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol 1995, 177, 6575–6584. [Google Scholar]
- Sun, Z; Gantt, E; Cunningham, FX, Jr. Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 1996, 271, 24349–24352. [Google Scholar]
- Hannibal, L; Lorquin, J; D’Ortoli, NA; Garcia, N; Chaintreuil, C; Masson-Boivin, C; Dreyfus, B; Giraud, E. Isolation and characterization of the canthaxanthin biosynthesis genes from the photosynthetic bacterium Bradyrhizobium sp. strain ORS278. J Bacteriol 2000, 182, 3850–3853. [Google Scholar]
- Kaneda, K; Kuzuyama, T; Takagi, M; Hayakawa, Y; Seto, H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc Natl Acad Sci USA 2001, 98, 932–937. [Google Scholar]
- Misawa, N. Carotenoids. In Comprehensive Natural Products II Chemistry and Biology; Mander, L, Lui, HW, Eds.; Elsevier: Oxford, UK, 2010; Volume 1; pp. 733–753. [Google Scholar]
- Ye, RW; Stead, KJ; Yao, H; He, H. Mutational and functional analysis of the β-carotene ketolase involved in the production of canthaxanthin and astaxanthin. Appl Environ Microbiol 2006, 72, 5829–5837. [Google Scholar]
- Sieiro, C; Poza, M; de Miguel, T; Villa, TG. Genetic basis of microbial carotenogenesis. Int Microbiol 2003, 6, 11–16. [Google Scholar]
- Fraser, PD; Miura, Y; Misawa, N. In vitro characterization of astaxanthin biosynthetic enzymes. J Biol Chem 1997, 272, 6128–6135. [Google Scholar]
- Fraser, PD; Shimada, H; Misawa, N. Enzymic confirmation of reactions involved in routes to astaxanthin formation, elucidated using a direct substrate in vitro assay. Eur J Biochem 1998, 252, 229–236. [Google Scholar]
- Choi, SK; Nishida, Y; Matsuda, S; Adachi, K; Kasai, H; Peng, X; Komemushi, S; Miki, W; Misawa, N. Characterization of β-carotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Mar Biotechnol 2005, 7, 515–522. [Google Scholar]
- Choi, SK; Matsuda, S; Hoshino, T; Peng, X; Misawa, N. Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl Microbiol Biotechnol 2006, 72, 1238–1246. [Google Scholar]
- Makino, T; Harada, H; Ikenaga, H; Matsuda, S; Takaichi, S; Shindo, K; Sandmann, G; Ogata, T; Misawa, N. Characterization of cyanobacterial carotenoid ketolase CrtW and hydroxylase CrtR by complementation analysis in Escherichia coli. Plant Cell Physiol 2008, 49, 1867–1878. [Google Scholar]
- Steiger, S; Sandmann, G. Cloning of two carotenid ketolase genes from Nostoc punctiforme for the heterologous production of canthaxanthin and astaxanthin. Biotechnol Lett 2004, 26, 813–817. [Google Scholar]
- Mochimaru, M; Masukawa, H; Takaichi, S. The cyanobacterium Anabaena sp. PCC 7120 has two distinct β-carotene ketolases: CrtO for echinenone and CrtW for ketomyxol synthesis. FEBS Lett 2005, 579, 6111–6114. [Google Scholar]
- Takaichi, S; Mochimaru, M; Maoka, T; Katoh, H. Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120, and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol 2005, 46, 497–504. [Google Scholar]
- Kajiwara, S; Kakizono, T; Saito, T; Kondo, K; Ohtani, T; Nishio, N; Nagai, S; Misawa, N. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol Biol 1995, 29, 343–352. [Google Scholar]
- Lotan, T; Hirschberg, J. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett 1995, 364, 125–128. [Google Scholar]
- Huang, JC; Chen, F; Sandmann, G. Stress-related differential expression of multiple β-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J Biotechnol 2006, 122, 176–185. [Google Scholar]
- Giuliano, G; Tavazza, R; Diretto, G; Beyer, P; Taylor, MA. Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 2008, 26, 139–145. [Google Scholar]
- Fernandez-Gonzalez, B; Sandmann, G; Vioque, A. A new type of asymmetrically acting β-carotene ketolase is required for the synthesis of echinenone in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 1997, 272, 9728–9733. [Google Scholar]
- Takaichi, S; Maoka, T; Masamoto, K. Myxoxanthophyll in Synechocyctis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-α-L-fucoside), not rhamnoside. Plant Cell Physiol 2001, 42, 756–762. [Google Scholar]
- Tao, L; Cheng, Q. Novel β-carotene ketolases from non-photosynthetic bacteria for canthaxanthin synthesis. Mol Genet Genomics 2004, 272, 530–537. [Google Scholar]
- Choi, SK; Harada, H; Matsuda, S; Misawa, N. Characterization of two β-carotene ketolases, CrtO and CrtW, by complementation analysis in Escherichia coli. Appl Microbiol Biotechnol 2007, 75, 1335–1341. [Google Scholar]
- Masamoto, K; Misawa, N; Kaneko, T; Kikuno, R; Toh, H. β-carotene hydroxylase gene from the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol 1998, 39, 560–564. [Google Scholar]
- Takaichi, S; Mochimaru, M. Carotenoids and carotenogenesis in cyanobacteria: Unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci 2007, 64, 2607–2619. [Google Scholar]
- Mochimaru, M; Masukawa, H; Maoka, T; Mohamed, HE; Vermaas, WL; Takaichi, S. Substrate specificities and availability of fucosyltransferase and β-carotene hydroxylase for myxol 2′-fucoside synthesis in Anabaena sp. strain PCC 7120 compared with Synechocystis sp. strain PCC 6803. J. Bacteriol 2008, 190, 6726–6733. [Google Scholar]
- Blasco, F; Kauffmann, I; Schmid, RD. CYP175A1 from Thermus thermophilus HB27, the first β-carotene hydroxylase of the P450 superfamily. Appl Microbiol Biotechnol 2004, 64, 671–674. [Google Scholar]
- Tao, L; Rouvière, PE; Cheng, Q. A carotenoid synthesis gene cluster from a non-marine Brevundimonas that synthesizes hydroxylated astaxanthin. Gene 2006, 379, 101–408. [Google Scholar]
- Iwai, M; Maoka, T; Ikeuchi, M; Takaichi, S. 2,2′-β-Hydroxylase (CrtG) is involved in carotenogenesis of both nostoxanthin and 2-hydroxymyxol 2′-fucoside in Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol 2008, 49, 1678–1687. [Google Scholar]
- Osawa, A; Harada, H; Choi, SK; Misawa, N; Shindo, K. Production of caloxanthin 3′-β-d-glucoside, zeaxanthin 3,3′-β-d-diglucoside, and nostoxanthin in a recombinant Escherichia coli expressing system harboring seven carotenoid biosynthesis genes, including crtX andcrtG. Phytochemistry 2011, 72, 711–716. [Google Scholar]
- Yokoyama, A; Shizuri, Y; Misawa, N. Production of new carotenoids, astaxanthin glucosides, by Escherichia coli transformants carrying carotenoid biosynthetic genes. Tetrahedron Lett 1998, 39, 3709–3712. [Google Scholar]
- Harada, H; Misawa, N. Novel approaches and achievements in biosynthesis of functional isoprenoids in Escherichia coli. Appl Microbiol Biotechnol 2009, 84, 1021–1031. [Google Scholar]
- Misawa, N. Pathway engineering for functional isoprenoids. Curr Opin Biotechnol 2011. [Google Scholar] [CrossRef]
- Kajiwara, S; Fraser, PD; Kondo, K; Misawa, N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J 1997, 324, 421–426. [Google Scholar]
- Harada, H; Yu, F; Okamoto, S; Kuzuyama, T; Utsumi, R; Misawa, N. Efficient synthesis of functional isoprenoids from acetoacetate through metabolic pathway-engineered. Escherichia coli Appl Microbiol Biotechnol 2009, 81, 915–925. [Google Scholar]
- Kakinuma, K; Dekishima, Y; Matsushima, Y; Eguchi, T; Misawa, N; Takagi, M; Kuzuyama, T; Seto, H. New approach to multiply deuterated isoprenoids using triply engineered Escherichia coli and its potential as a tool for mechanistic enzymology. J Am Chem Soc 2001, 123, 1238–1239. [Google Scholar]
- Newman, JD; Marshall, J; Chang, M; Nowroozi, F; Paradise, E; Pitera, D; Newman, KL; Keasling, JD. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered. Escherichia coli Biotechnol Bioeng 2006, 95, 684–691. [Google Scholar]
- Vadali, RV; Fu, Y; Bennett, GN; San, KY. Enhanced lycopene productivity by manipulation of carbon flow to isopentenyl diphosphate in Escherichia coli. Biotechnol Prog 2005, 21, 1558–1561. [Google Scholar]
- Yoon, SH; Lee, YM; Kim, JE; Lee, SH; Lee, JH; Kim, JY; Jung, KH; Shin, YC; Keasling, JD; Kim, SW. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng 2006, 94, 1025–1032. [Google Scholar]
- Alper, H; Stephanopoulos, G. Uncovering the gene knockout landscape for improved lycopene production in E coli. Appl Microbiol Biotechnol 2008, 78, 801–810. [Google Scholar]
- Hasunuma, T; Miyazawa, S; Yoshimura, S; Shinzaki, Y; Tomizawa, K; Shindo, K; Choi, SK; Misawa, N; Miyake, C. Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 2008, 55, 857–868. [Google Scholar]

© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Misawa, N. Carotenoid β-Ring Hydroxylase and Ketolase from Marine Bacteria—Promiscuous Enzymes for Synthesizing Functional Xanthophylls. Mar. Drugs 2011, 9, 757-771. https://doi.org/10.3390/md9050757
Misawa N. Carotenoid β-Ring Hydroxylase and Ketolase from Marine Bacteria—Promiscuous Enzymes for Synthesizing Functional Xanthophylls. Marine Drugs. 2011; 9(5):757-771. https://doi.org/10.3390/md9050757
Chicago/Turabian StyleMisawa, Norihiko. 2011. "Carotenoid β-Ring Hydroxylase and Ketolase from Marine Bacteria—Promiscuous Enzymes for Synthesizing Functional Xanthophylls" Marine Drugs 9, no. 5: 757-771. https://doi.org/10.3390/md9050757
APA StyleMisawa, N. (2011). Carotenoid β-Ring Hydroxylase and Ketolase from Marine Bacteria—Promiscuous Enzymes for Synthesizing Functional Xanthophylls. Marine Drugs, 9(5), 757-771. https://doi.org/10.3390/md9050757



