Exploring the Microbial Reservoir of Geodia cydonium (Linnaeus, 1767): Insights into Site-Specific Diversity and Biotechnological Potential
Abstract
1. Introduction
2. Results and Discussion
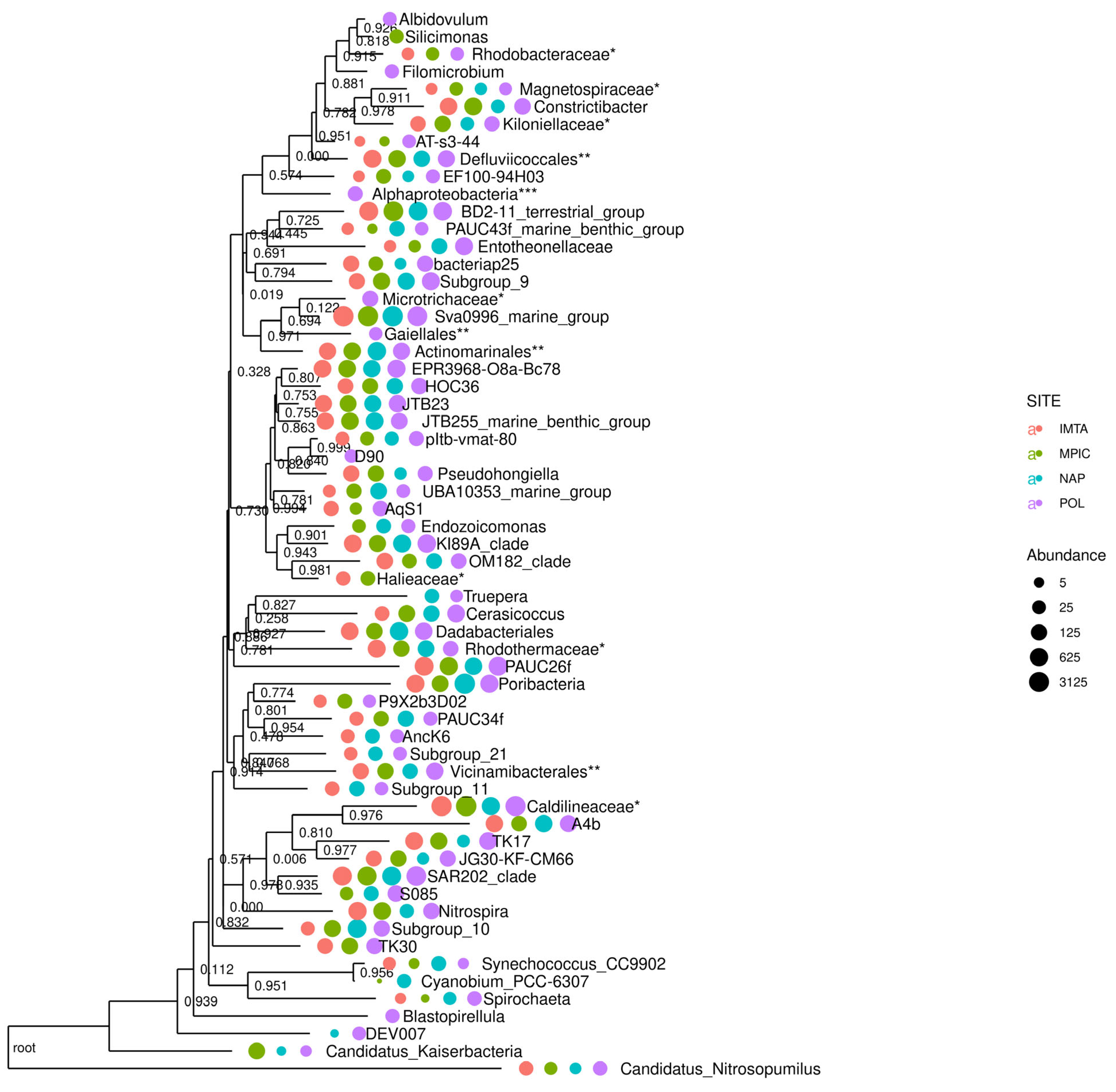
2.1. Taxonomic Identification of G. cydonium Associated Bacterial Community
- i.
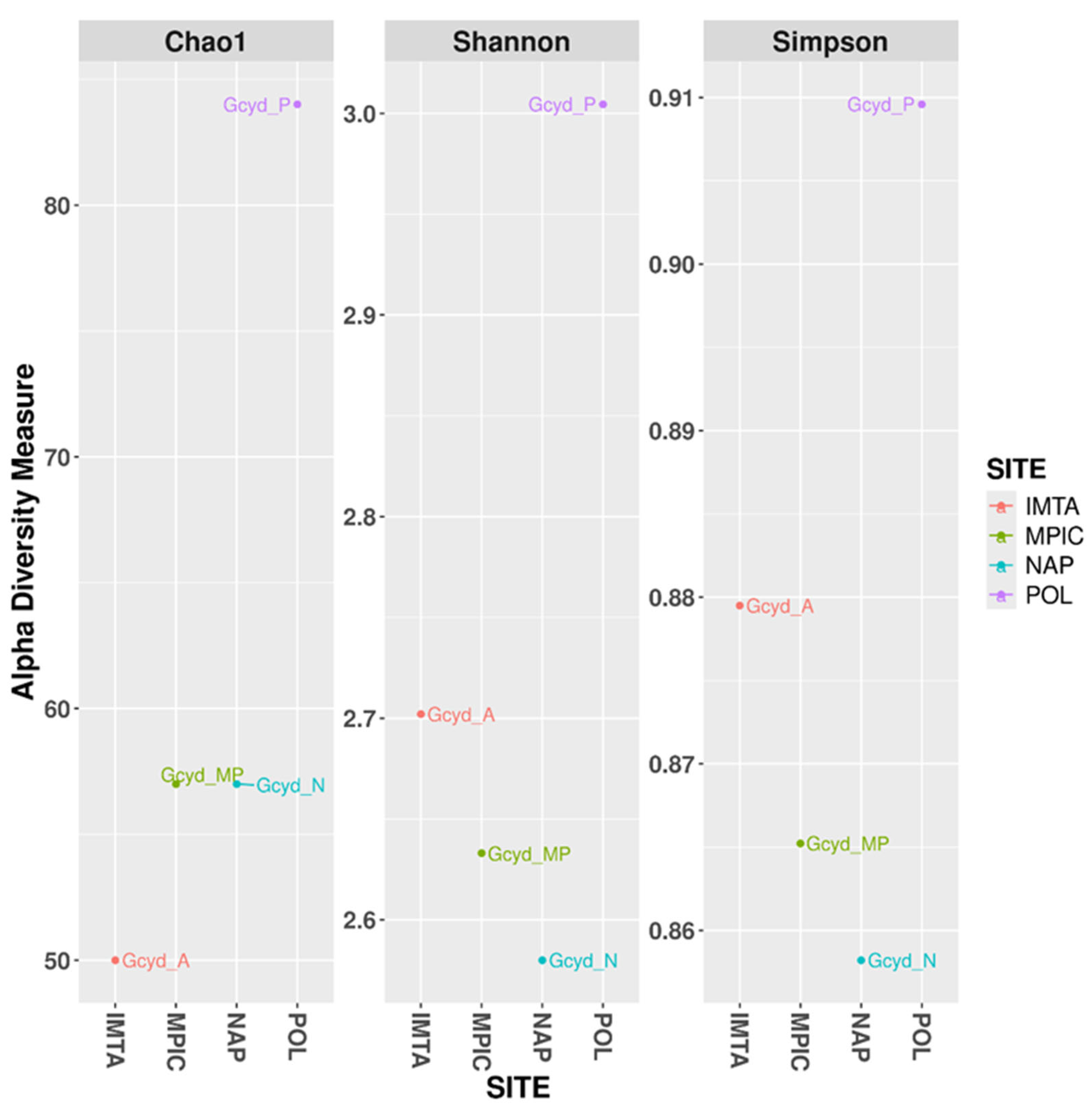
- The largest number of features (300) in G. cydonium collected in Polignano a Mare, especially Anaerolineae (5%), Dehalococcoidia (3%), Acidomicrobiia (3%), Gammaproteobacteria (3%), and Acidobacteriota (1%).
- ii.
- 232 ASVs in G. cydonium sampled in Secca delle Fumose (Naples) with a greater abundance of Acidomicrobiia (6%), Poribacteriota (6%), Gammaproteobacteria (2%), Dehalococcoidia (1%), and Thermoanaerobaculia (1%).
- iii.
- 148 ASVs in the sample of G. cydonium collected in the IMTA system, where Acidomicrobiia (7%), Anaerolineae (7%), Gammaprotebacteria (3%), Dehalococcoidia (2%), Alphaproteobacteria (2%), Acidobacteriota (2%), and BD2-11_terrestrial_group (Gemmatimonadota phylum) were highly represented.
- iv.
- The sample of G. cydonium with the least number of features was sampled in Mar Piccolo and it revealed seven bacterial groups (Anaerolineae (7%), Acidomicrobiia (6%), BD2_11_terrestrial group (3%), Dehalococcoidia, Gammaproteobacteria, Alphaproteobacteria, and Acidobacteriota (2%, each)) (Figures S1 and S2).
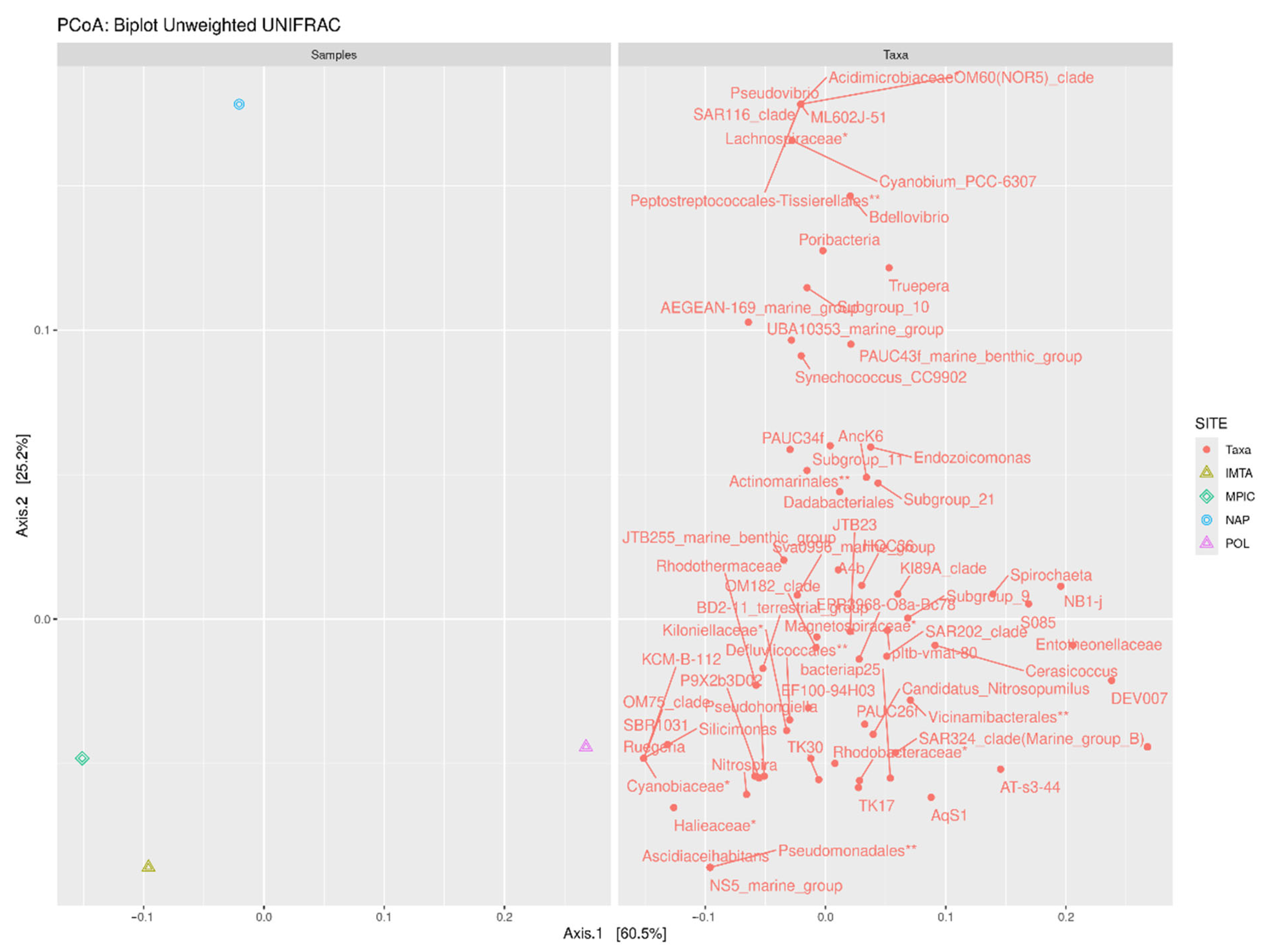
2.2. Relationship Between Abundance and Distribution of the Bacterial Communities Among Sites
3. Materials and Methods
3.1. Studied Species and Collection
3.2. Metataxonomic DNA Extraction and Illumina MiSeq Sequencing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morganti, T.M.; Ribes, M.; Moskovich, R.; Weisz, J.B.; Yahel, G.; Coma, R. In situ Pumping Rate of 20 Marine Demosponges Is a Function of Osculum Area. Front. Mar. Sci. 2021, 8, 583188. [Google Scholar] [PubMed]
- Zhang, X.; Zhang, W.; Xue, L.; Zhang, B.; Jin, M.; Fu, W. Bioremediation of bacteria pollution using the marine sponge Hymeniacidon perlevis in the intensive mariculture water system of turbot Scophthalmus maximus. Biotechnol. Bioeng. 2010, 105, 59–68. [Google Scholar] [PubMed]
- Longo, C.; Cardone, F.; Corriero, G. The co-occurrence of the demosponge Hymeniacidon perlevis and the edible mussel Mytilus galloprovincialis as a new tool for bacterial load mitigation in aquaculture. Environ. Sci. Pollut. Res. 2010, 23, 3736–3746. [Google Scholar]
- Hadas, E.; Marie, D.; Shpigel, M.; Ilan, M. Virus predation by sponges is a new nutrient-flow pathway in coral reef food webs. Limnol. Oceanogr. 2006, 51, 1548–1550. [Google Scholar] [CrossRef]
- Welsh, J.E.; Steenhuis, P.; Ribeiro de Moraes, K.; van der Meer, J.; Thieltges, D.W.; Brussaard, C.P.D. Marine virus predation by non-host organisms. Sci. Rep. 2020, 10, 5221. [Google Scholar] [CrossRef]
- Ribes, M.; Coma, R.; Gili, J.M. Natural diet and grazing rate of the temperate marine sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 1999, 176, 179–190. [Google Scholar]
- Ribes, M.; Coma, R.; Gili, J.M.; Svoboda, A.; Julia, A.; Parera, J. A ‘semi-closed’ recirculating system for the in situ study of feeding and respiration of benthic suspension feeders. Sci. Mar. 2000, 64, 265–275. [Google Scholar]
- Pérez, T.; Sarrazin, L.; Rebouillon, P.; Vacelet, J. First evidence of surfactant biodegradation by marine sponges (Porifera): An experimental study with a linear alkylbenzenesulfonate. Hydrobiologia 2002, 289, 225–233. [Google Scholar] [CrossRef]
- Müller, W.E.G.; Wimmer, W.; Schatton, W.; Böhm, M.; Batel, R.; Filic, Z. Initiation of an aquaculture of sponges for the sustainable production of bioactive metabolites in open systems: Example, Geodia cydonium. Mar. Biotechnol. 1999, 1, 569–579. [Google Scholar] [CrossRef]
- Philp, R.B. Cadmium content of the marine sponge Microciona prolifera, other sponges, water and sediment from the eastern Florida panhandle: Possible effects on Microciona cell aggregation and potential roles of low pH and low salinity. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1999, 124, 41–49. [Google Scholar]
- Pérez, T.; Longet, D.; Schembri, T.; Rebouillon, P.; Vacelet, J. Effects of 12 years’ operation of a sewage treatment plant on trace metal occurrence within a Mediterranean commercial sponge (Spongia officinalis, Demospongiae). Mar. Pollut. Bull. 2005, 50, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Cebrian, E.; Uriz, M.J.; Turon, X. Sponges as biomonitors of heavy metals in spatial and temporal surveys in northwestern Mediterranean: Multispecies comparison. Environ. Toxicol. Chem. 2007, 26, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Amato, A.; Esposito, R.; Federico, S.; Pozzolini, M.; Giovine, M.; Bertolino, M.; Guida, M.; Manfra, L.; Libralato, G.; Zupo, V.; et al. Marine sponges as promising candidates for integrated aquaculture combining biomass increase and bioremediation: An updated review. Front. Mar. Sci. 2024, 10, 1234225. [Google Scholar] [CrossRef]
- Saliu, F.; Becchi, A.; Montalbetti, E.; Isa, V.; Gatti, T.; Riseri, D.; Lasagni, M.; Galli, P.; Seveso, D. Application of marine sponges for biomonitoring active pharmaceutical ingredients (APIs) in coral reefs. Optimization of an SPME and ESI-LC-MS/MS method. Mar. Pollut. Bull. 2024, 205, 116720. [Google Scholar] [CrossRef]
- Simpson, T.L.; Volcani, E.B. Silicon and Siliceous Structures in Biological Systems; Springer: New York, NY, USA, 1981. [Google Scholar]
- Maldonado, M.; López-Acosta, M.; Beazley, L.; Kenchington, E.; Koutsouveli, V.; Riesgo, A. Cooperation between passive and active silicon transporters clarifies the ecophysiology and evolution of biosilicification in sponges. Sci. Adv. 2020, 6, eaba9322. [Google Scholar] [CrossRef]
- de Kluijver, A.; Bart, M.C.; van Oevelen, D.; de Goeij, J.M.; Leys, S.P.; Maier, S.R.; Maldonado, M.; Soetaert, K.; Verbiest, S.; Middelburg, J.J. An integrative model of carbon and nitrogen metabolism in a common deep-sea sponge (Geodia barretti). Front. Mar. Sci. 2021, 7, 596251. [Google Scholar] [CrossRef]
- Maldonado, M.; Bayer, K.; López-Acosta, M. Nitrogen and phosphorus cycling through marine sponges: Physiology, cytology, genomics, and ecological implications. In Frontiers in Invertebrate Physiology: A Collection of Reviews; Apple Academic Press: Waretown, NJ, USA, 2024. [Google Scholar]
- Paul, V.J.; Freeman, C.J.; Agarwal, V. Chemical ecology of marine sponges: New opportunities through “-omics”. Integr. Comp. Biol. 2019, 59, 765–776. [Google Scholar] [CrossRef]
- Anteneh, Y.S.; Yang, Q.; Brown, M.H.; Franco, C.M. Factors affecting the isolation and diversity of marine sponge-associated bacteria. Appl. Microbiol. Biotechnol. 2022, 106, 1729–1744. [Google Scholar] [CrossRef]
- Voser, T.M.; Campbell, M.D.; Carroll, A.R. How different are marine microbial natural products compared to their terrestrial counterparts? Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge-derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef]
- Flemer, B.; Kennedy, J.; Margassery, L.M.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp. J. Appl. Microbiol. 2012, 112, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Lazoglou, G.; Papadopoulos-Zachos, A.; Georgiades, P.; Zittis, G.; Velikou, K.; Manios, E.M.; Anagnostopoulou, C. Identification of climate change hotspots in the Mediterranean. Sci. Rep. 2024, 14, 29817. [Google Scholar] [CrossRef] [PubMed]
- Diffenbaugh, N.S.; Giorgi, F. Climate change hotspots in the CMIP5 global climate model ensemble. Clim. Change 2012, 114, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Yáñez, M.; García, M.J.; Salat, J.; García-Martínez, M.C.; Pascual, J.; Moya, F. Warming trends and decadal variability in the Western Mediterranean shelf. Glob. Planet. Change 2008, 63, 177–184. [Google Scholar] [CrossRef]
- Marbà, N.; Jordà, G.; Agustí, S.; Girard, C.; Duarte, C.M. Footprints of climate change on Mediterranean Sea biota. Front. Mar. Sci. 2015, 2, 56. [Google Scholar] [CrossRef]
- Costello, M.J.; Tsai, P.; Wong, P.S.; Cheung, A.K.L.; Basher, Z.; Chaudhary, C. Marine biogeographic realms and species endemism. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- European Environment Agency. Horizon 2020 Mediterranean Report; European Environment Agency: Copenhagen, Denmark, 2014. [Google Scholar]
- Carballo, J.L.; Bell, J.J. Climate change and sponges: An introduction. In Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization; Springer: Cham, Switzerland, 2017; pp. 1–11. [Google Scholar]
- Barrington, K.; Chopin, T.; Robinson, S. Integrated multi-trophic aquaculture (IMTA) in marine temperate waters. In Integrated Mariculture: A Global Review; FAO Fisheries and Aquaculture Technical Paper 529; FAO: Rome, Italy, 2009; pp. 7–46. [Google Scholar]
- Longo, C.; Pierri, C.; Trani, R.; Mercurio, M.; Nonnis Marzano, C.; Corriero, G.; Aguilo-Arce, J.; Sini, V.; Massari, F.; Zambonin, C.; et al. Toward a green strategy of sponge mariculture and bioactive compounds recovery. Sci. Rep. 2025, 15, 5999. [Google Scholar] [CrossRef]
- Mercurio, M.; Pierri, C.; Cardone, F.; Corriero, G. Temporal and spatial variations of Geodia cydonium (Jameson) (Porifera, Demospongiae) in the Mediterranean confined environments. Diversity 2021, 13, 615. [Google Scholar] [CrossRef]
- Aguilo-Arce, J.; Trani, R.; Schiavo, A.; García-Ordax, M.; Pierri, C.; Longo, C. Marine sponges prefer integrated multitrophic aquaculture environments for growth: Insights from experimental rearing trials in the Mar Grande of Taranto (Mediterranean Sea). Aquaculture 2025, submitted. [Google Scholar]
- Giangrande, A.; Pierri, C.; Arduini, D.; Borghese, J.; Licciano, M.; Trani, R.; Corriero, G.; Basile, G.; Cecere, E.; Petrocelli, A.; et al. An innovative IMTA system: Polychaetes, sponges and macroalgae co-cultured in a southern Italian in-shore mariculture plant (Ionian Sea). J. Mar. Sci. Eng. 2020, 8, 733. [Google Scholar] [CrossRef]
- Aguilo-Arce, J.; Scrascia, M.; Trani, R.; Pazzani, C.; Ferriol, P.; Longo, C. Two demosponges as promising bioremediators of a potential pathogenic Vibrio. Biology 2025, 14, 140. [Google Scholar] [CrossRef]
- Costantini, S.; Guerriero, E.; Trapanese, G.; Libralato, G.; Morroni, L.; Capone, F.; Ruocco, N.; Tedesco, I.; Costantini, M. Anti-inflammatory effects of a methanol extract from the marine sponge Geodia cydonium on the human breast cancer MCF-7 cell line. Mediators Inflamm. 2015, 2015, 204975. [Google Scholar] [CrossRef]
- Ruocco, N.; Esposito, R.; Zagami, G.; Bertolino, M.; De Matteo, S.; Sonnessa, M.; Andreani, F.; Crispi, S.; Zupo, V.; Costantini, M. Microbial diversity in Mediterranean sponges as revealed by metataxonomic analysis. Sci. Rep. 2021, 11, 21151. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Federico, S.; Sonnessa, M.; Reddel, S.; Bertolino, M.; Ruocco, N.; Zagami, G.; Giovine, M.; Pozzolini, M.; Guida, M.; et al. Characterizing the bacterial communities associated with Mediterranean sponges: A metataxonomic analysis. Front. Microbiol. 2024, 14, 1295459. [Google Scholar] [CrossRef] [PubMed]
- Radax, R.; Rattei, T.; Lanzen, A.; Bayer, C.; Rapp, H.T.; Urich, T.; Schleper, C. Metatranscriptomics of the marine sponge Geodia barretti: Tackling phylogeny and function of its microbial community. Environ. Microbiol. 2012, 14, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef]
- Gloeckner, V.; Wehrl, M.; Moitinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlik, J.R.; Lindquist, N.L.; Erpenbeck, D.; Wörheide, G.; Hentschel, U. The HMA–LMA dichotomy revisited: An electron microscopical survey of 56 sponge species. Biol. Bull. 2014, 227, 78–88. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- Moitinho-Silva, L.; Seridi, L.; Ryu, T.; Voolstra, C.R.; Ravasi, T.; Hentschel, U. Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ. Microbiol. 2014, 16, 3512–3526. [Google Scholar] [CrossRef]
- Kralj, M.; De Vittor, C.; Comici, C.; Relitti, F.; Auriemma, R.; Alabiso, G.; Del Negro, P. Recent evolution of the physical–chemical characteristics of a Site of National Interest—the Mar Piccolo of Taranto (Ionian Sea)—and changes over the last 20 years. Environ. Sci. Pollut. Res. 2016, 23, 12675–12690. [Google Scholar] [CrossRef]
- Stabili, L.; Giangrande, A.; Arduini, D.; Borghese, J.; Petrocelli, A.; Alabiso, G.; Ricci, P.; Cavallo, R.A.; Acquaviva, M.I.; Narracci, M.; et al. Environmental quality improvement of a mariculture plant after its conversion into a multi-trophic system. Sci. Total Environ. 2023, 904, 163846. [Google Scholar] [CrossRef]
- Matek, A.; Mucko, M.; Casotti, R.; Trano, A.C.; Achterberg, E.P.; Mihanović, H.; Cižmek, H.; Coli, B.C.; Cuculić, V.; Ljubešić, Z. Phytoplankton diversity and co-dependency in a stratified oligotrophic ecosystem in the South Adriatic Sea. Water 2023, 15, 2299. [Google Scholar] [CrossRef]
- Happel, L.; Rondon, R.; Font, A.; González-Aravena, M.; Cárdenas, C.A. Stability of the microbiome of the sponge Mycale (Oxymycale) acerata in the Western Antarctic Peninsula. Front. Microbiol. 2022, 13, 827863. [Google Scholar] [CrossRef] [PubMed]
- Leal, C.V.; Avelino-Alves, D.; Salazar, V.; Omachi, C.; Thompson, C.; Berlinck, R.G.S.; Hajdu, E.; Thompson, F. Sponges present a core prokaryotic community stable across the tropical Western Atlantic. Sci. Total Environ. 2022, 835, 155145. [Google Scholar] [CrossRef] [PubMed]
- Luter, H.M.; Bannister, R.J.; Whalan, S.; Kutti, T.; Pineda, M.C.; Webster, N.S. Microbiome analysis of a disease affecting the deep-sea sponge Geodia barretti. FEMS Microbiol. Ecol. 2017, 93, fix074. [Google Scholar] [CrossRef] [PubMed]
- Couto, R.P.; Rodrigues, A.S.; Neto, A.I. Shallow-water hydrothermal vents in the Azores (Portugal). J. Integr. Coast. Zone Manag. 2015, 15, 495–505. [Google Scholar] [CrossRef]
- Nuianzina-Boldareva, E.N.; Kalashnikov, A.M.; Gaĭsin, V.A.; Sukhacheva, M.V.; Kuznetsov, B.B.; Gorlenko, V.M. Characterization of a new strain of a purple nonsulfur bacterium from a thermal spring. Mikrobiologiia 2014, 83, 170–179. [Google Scholar] [CrossRef]
- Toshchakov, S.V.; Izotova, A.O.; Vinogradova, E.N.; Kachmazov, G.S.; Tuaeva, A.Y.; Abaev, V.T.; Kublanov, I.V. Culture-independent survey of thermophilic microbial communities of the North Caucasus. Biology 2021, 10, 1352. [Google Scholar] [CrossRef]
- Indraningrat, A.A.G.; Steinert, G.; Becking, L.E.; Mueller, B.; de Goeij, J.M.; Smidt, H.; Sipkema, D. Sponge holobionts shift their prokaryotic communities and antimicrobial activity from shallow to lower mesophotic depths. Antonie Van Leeuwenhoek 2022, 115, 1265–1283. [Google Scholar] [CrossRef]
- Márquez, S.L.; Blamey, J.M. Isolation and partial characterization of a new moderate thermophilic Albidovulum sp. SLM16 with transaminase activity from Deception Island, Antarctica. Biol. Res. 2019, 52, 1. [Google Scholar] [CrossRef]
- Zure, M.; Munn, C.B.; Harder, J. Diversity of Rhodopirellula and related planctomycetes in a North Sea coastal sediment employing carB as a molecular marker. FEMS Microbiol. Lett. 2015, 362, 1–9. [Google Scholar] [CrossRef]
- Dewar, M.L.; Arnould, J.P.Y.; Dann, P.; Trathan, P. Interspecific variations in the gastrointestinal microbiota in penguins. Microbiol. Open 2013, 2, 195–204. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef]
- Oliveira, F.A.S.; Colares, G.B.; Hissa, D.C.; Angelim, A.L.; Melo, V.M.M.; Lotufo, T.M.C. Microbial epibionts of the colonial ascidians Didemnum galacteum and Cystodytes sp. Symbiosis 2013, 59, 57–63. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Al Kharusi, S.; Schramm, A.; Robinson, M.D. Bacterial diversity, pigments and nitrogen fixation of biological desert crusts from the Sultanate of Oman. FEMS Microbiol. Ecol. 2010, 72, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Andrew, D.R.; Fitak, R.R.; Munguia-Vega, A.; Racolta, A.; Martinson, V.G.; Dontsova, K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl. Environ. Microbiol. 2012, 78, 7527–7537. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, L.K.; Dupraz, C.; Buckley, D.H.; Spear, J.R.; Pace, N.R.; Visscher, P.T. Microbial species richness and metabolic activities in hypersaline microbial mats: Insight into biosignature formation through lithification. Astrobiology 2009, 9, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Arp, G.; Reimer, A.; Reitner, J.; Daniel, R. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, Central Pacific. PLoS ONE 2013, 8, e66662. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Musat, N.; Musat, F.; Mußmann, M. Structure of microbial communities and hydrocarbon-dependent sulfate reduction in the anoxic layer of a polluted microbial mat. Mar. Pollut. Bull. 2011, 62, 539–546. [Google Scholar] [CrossRef]
- Jensen, S.I.; Kühl, M.; Priemé, A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 2007, 62, 108–117. [Google Scholar] [CrossRef]
- Lage, O.M.; Godinho, O.; García-Domínguez, R.; Øvreås, L.; Devos, D.P. A century of research on the Planctomycetota bacterial phylum, previously known as Planctomycetes. FEMS Microbiol. Rev. 2025, 49, fuaf056. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, X.; Liu, G.; Chen, T.; Zhang, G.; Dong, Z.; Yang, X.; Hu, P. Pyrosequencing reveals bacterial diversity in the rhizosphere of three Phragmites australis ecotypes. Geomicrobiol. J. 2013, 30, 593–599. [Google Scholar] [CrossRef]
- Kaboré, O.D.; Godreuil, S.; Drancourt, M. Planctomycetes as host-associated bacteria: A perspective that holds promise for their future isolations, by mimicking their native environmental niches in clinical microbiology laboratories. Front. Cell. Infect. Microbiol. 2020, 10, 519301. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Wiegand, S.; Heuer, A.; Rensink, S.; Boersma, A.S.; Jogler, M.; Boedeker, C.; Peeters, S.H.; Rast, P.; Jetten, M.S.M.; et al. Blastopirellula retiformator sp. nov., isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie Van Leeuwenhoek 2020, 113, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Panter, F.; Garcia, R.; Thewes, A.; Zaburannyi, N.; Bunk, B.; Overmann, J.; Gutierrez, M.V.; Krug, D.; Müller, R. Production of a dibrominated aromatic secondary metabolite by a planctomycete implies complex interaction with a macroalgal host. ACS Chem. Biol. 2019, 14, 2713–2719. [Google Scholar] [CrossRef]
- Calisto, R.; Søbø, E.F.; Storesund, J.E.; Øvreås, L.; Herfindal, L.; Lage, O.M. Anticancer activity in planctomycetes. Front. Mar. Sci. 2019, 5, 499. [Google Scholar] [CrossRef]
- Sandargo, B.; Jeske, O.; Boedeker, C.; Wiegand, S.; Wennrich, J.P.; Kallscheuer, N.; Jogler, M.; Rohde, M.; Jogler, C.; Surup, F. Stieleriacines, N-acyl dehydrotyrosines from the marine planctomycete Stieleria neptunia sp. nov. Front. Microbiol. 2020, 11, 1408. [Google Scholar] [CrossRef]
- Zhang, F.; Blasiak, L.C.; Karolin, J.O.; Powell, R.J.; Geddes, C.D.; Hill, R.T. Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc. Natl. Acad. Sci. USA 2015, 112, 4381–4386. [Google Scholar] [CrossRef]
- Ou, H.; Li, M.; Wu, S.; Jia, L.; Hill, R.T.; Zhao, J. Characteristic microbiomes correlate with polyphosphate accumulation of marine sponges in South China Sea areas. Microorganisms 2019, 8, 63. [Google Scholar] [CrossRef]
- Schlesner, H. Filomicrobium fusiforme gen. nov., sp. nov., a slender budding, hyphal bacterium from brackish water. Syst. Appl. Microbiol. 1987, 10, 63–67. [Google Scholar] [CrossRef]
- Henriques, A.C.; De Marco, P. Complete genome sequences of two strains of “Candidatus Filomicrobium marinum,” a methanesulfonate-degrading species. Genome Announc. 2015, 3, e00573-15. [Google Scholar] [CrossRef]
- Wu, X.L.; Yu, S.L.; Gu, J.; Zhao, G.F.; Chi, C.Q. Filomicrobium insigne sp. nov., isolated from an oil-polluted saline soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, T.; Al Khaburi, M.; Abed, R.M.M. Fouling microbial communities on plastics compared with wood and steel: Are they substrate- or location-specific? Microb. Ecol. 2019, 78, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Jacquin, J.; Callac, N.; Cheng, J.; Giraud, C.; Gorand, Y.; Denoual, C.; Pujo-Pay, M.; Conan, P.; Meistertzheim, A.-L.; Barbe, V.; et al. Microbial diversity and activity during the biodegradation in seawater of various substitutes to conventional plastic cotton swab sticks. Front. Microbiol. 2021, 12, 604395. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Imhoff, J.F. Phylogenetic identification of bacteria with antimicrobial activities isolated from Mediterranean sponges. Biomol. Eng. 2003, 20, 421–423. [Google Scholar] [CrossRef]
- Bibi, F.; Yasir, M.; Al-Sofyani, A.; Naseer, M.I.; Azhar, E.I. Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EA348. Saudi J. Biol. Sci. 2020, 27, 1139–1147. [Google Scholar] [CrossRef]
- D’Souza, S.R.; Singh, S.; Ravi, L. Secondary metabolites produced from symbiotic microbes. In Microbial Symbionts: Functions and Molecular Interactions on Host; Academic Press: Cambridge, MA, USA, 2023; pp. 803–830. [Google Scholar]
- Elbon, C.E.; Stewart, F.J.; Glass, J.B. Novel Alphaproteobacteria transcribe genes for nitric oxide transformation at high levels in a marine oxygen-deficient zone. Appl. Environ. Microbiol. 2024, 90, e02099-23. [Google Scholar] [CrossRef]
- Albuquerque, L.; França, L.; Rainey, F.A.; Schumann, P.; Nobre, M.F.; Da Costa, M.S. Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov. Syst. Appl. Microbiol. 2011, 34, 595–599. [Google Scholar] [CrossRef]
- Khilyas, I.V.; Sorokina, A.V.; Elistratova, A.A.; Markelova, M.I.; Siniagina, M.N.; Sharipova, M.R.; Shcherbakova, T.A.; D’Errico, M.E.; Cohen, M.F. Microbial diversity and mineral composition of weathered serpentine rock of the Khalilovsky massif. PLoS ONE 2019, 14, e0225929. [Google Scholar] [CrossRef]
- Liu, M.; Huang, H.; Bao, S.; Tong, Y. Microbial community structure of soils in Bamenwan mangrove wetland. Sci. Rep. 2019, 9, 8406. [Google Scholar] [CrossRef]
- Peng, M.; Jia, H.; Wang, Q. The effect of land use on bacterial communities in saline–alkali soil. Curr. Microbiol. 2017, 74, 325–333. [Google Scholar] [CrossRef]
- Steinert, G.; Taylor, M.W.; Schupp, P.J. Diversity of actinobacteria associated with the marine ascidian Eudistoma toealensis. Mar. Biotechnol. 2015, 17, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; He, Y.; Yue, H.; Wang, Q. Microbial structures and community functions of anaerobic sludge in six full-scale wastewater treatment plants as revealed by 454 high-throughput pyrosequencing. Bioresour. Technol. 2015, 186, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Palma Esposito, F.; López-Mobilia, A.; Tangherlini, M.; Casella, V.; Coppola, A.; Varola, G.; Vitale, L.; Della Sala, G.; Tedesco, P.; Montano, S.; et al. Novel insights and genomic characterization of coral-associated microorganisms from Maldives displaying antimicrobial, antioxidant, and UV-protectant activities. Biology 2025, 14, 401. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.A.; Andreakis, N.; Tan, S.; Miller, D.J.; Webster, N.S.; Zhang, G.; Bourne, D.G. Testing cophylogeny between coral reef invertebrates and their bacterial and archaeal symbionts. Mol. Ecol. 2021, 30, 3768–3782. [Google Scholar] [CrossRef]
- O’Brien, P.A.; Robbins, S.J.; Tan, S.; Rix, L.; Miller, D.J.; Webster, N.S.; Bourne, D.G. Comparative genomics identifies key adaptive traits of sponge-associated microbial symbionts. Environ. Microbiol. 2024, 26, e16690. [Google Scholar]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Currie, C.R. Evolution and ecology of actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar]
- Crenn, K.; Serpin, D.; Lepleux, C.; Overmann, J.; Jeanthon, C. Silicimonas algicola gen. nov., sp. nov., a member of the Roseobacter clade isolated from the cell surface of the marine diatom Thalassiosira delicatula. Int. J. Syst. Evol. Microbiol. 2016, 66, 4580–4588. [Google Scholar] [CrossRef]
- Crenn, K.; Bunk, B.; Spröer, C.; Overmann, J.; Jeanthon, C. Complete genome sequence of the Silicimonas algicola type strain, a representative of the marine Roseobacter group isolated from the cell surface of the marine diatom Thalassiosira delicatula. Microbiol. Resour. Announc. 2019, 8, e00232-19. [Google Scholar]
- Xu, M.; Cheng, K.; Xiao, B.; Tong, M.; Cai, Z.; Jong, M.C.; Zhou, J. Bacterial communities vary from different scleractinian coral species and between bleached and non-bleached corals. Microbiol. Spectr. 2023, 11, e04910-22. [Google Scholar] [CrossRef]
- Santucci, R. La Geodia cydonium come centro di associazione biologica. Mem. CIII Com. Talas. It. 1922, 103, 5–19. [Google Scholar]
- Longo, C.; Cardone, F.; Mercurio, M.; Marzano, C.N.; Pierri, C.; Corriero, G. Spatial and temporal distributions of the sponge fauna in southern Italian lagoon systems. Mediterr. Mar. Sci. 2015, 17, 174. [Google Scholar] [CrossRef]
- Forti, P. Processi carsici e speleogenesi. Prima parte. Speleologia 1991, 24, 42–46. [Google Scholar]
- Forti, P. Processi carsici e speleogenesi. Seconda parte. Speleologia 1992, 26, 47–50. [Google Scholar]
- Gimenez, G.; Oddenino, M.; Schiavo, A.; Pierri, C.; Trani, R.; Longo, C. Survey and monitoring of benthic biocenosis of submerged and semi-submerged caves of the Apulian coast in the province of Bari. In Proceedings of the Book of Abstracts of the 8th European Conference on Scientific Diving (ECSD8), Crete, Greece, 22–26 April 2024; p. 65. [Google Scholar]
- Donnarumma, L.; Appolloni, L.; Chianese, E.; Bruno, R.; Baldrighi, E.; Guglielmo, R.; Russo, G.F.; Zeppilli, D.; Sandulli, R. Environmental and benthic community patterns of the shallow hydrothermal area of Secca delle Fumose (Baia, Naples, Italy). Front. Mar. Sci. 2019, 6, 685. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Urbanek, S.; Horner, J. Cairo: R Graphics Device Using Cairo Graphics Library for Creating High-Quality Bitmap (PNG, JPEG, TIFF), Vector (PDF, SVG, PostScript) and Display (X11 and Win32) Output R Package, Version 1.5–12.2; R Project for Statistical Computing: Vienna, Austria, 2020. Available online: https://CRAN.R-project.org/package=Cairo (accessed on 10 December 2025).
- Chao, B.F. Interannual length-of-the-day variation with relation to the southern oscillation/El Niño. Geophys. Res. Lett. 1984, 11, 541–544. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423, 623–656. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, W.M. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef]
- Costantini, S.; Guerriero, E.; Teta, R.; Capone, F.; Caso, A.; Sorice, A.; Romano, G.; Ianora, A.; Ruocco, N.; Budillon, A.; et al. Evaluating the effects of an organic extract from the Mediterranean sponge Geodia cydonium on human breast cancer cell lines. Int. J. Mol. Sci. 2017, 18, 2112. [Google Scholar] [CrossRef]
- Di Meo, F.; Esposito, R.; Cuciniello, R.; Favale, G.; Arenga, M.; Ruocco, N.; Costantini, M. Organic extract of Geodia cydonium induces cell cycle block in human mesothelioma cells. Oncol. Lett. 2022, 24, 286. [Google Scholar] [CrossRef]
- Noyer, C.; Casamayor, E.O.; Becerro, M.A. Environmental heterogeneity and microbial inheritance influence sponge-associated bacterial composition of Spongia lamella. Microb. Ecol. 2014, 68, 611–620. [Google Scholar] [CrossRef]
- Federico, S.; Esposito, R.; De Rosa, M.; Sonnessa, M.; Reddel, S.; Laurenzi, G.; Bertolino, M.; Giovine, M.; Pozzolini, M.; Zupo, V.; et al. Comparative metagenomic analyses of the microbiome from three Mediterranean sponges to identify genes involved in biosynthesis of bioactive compounds. Mar. Genom. 2025, 82, 101202. [Google Scholar] [CrossRef]


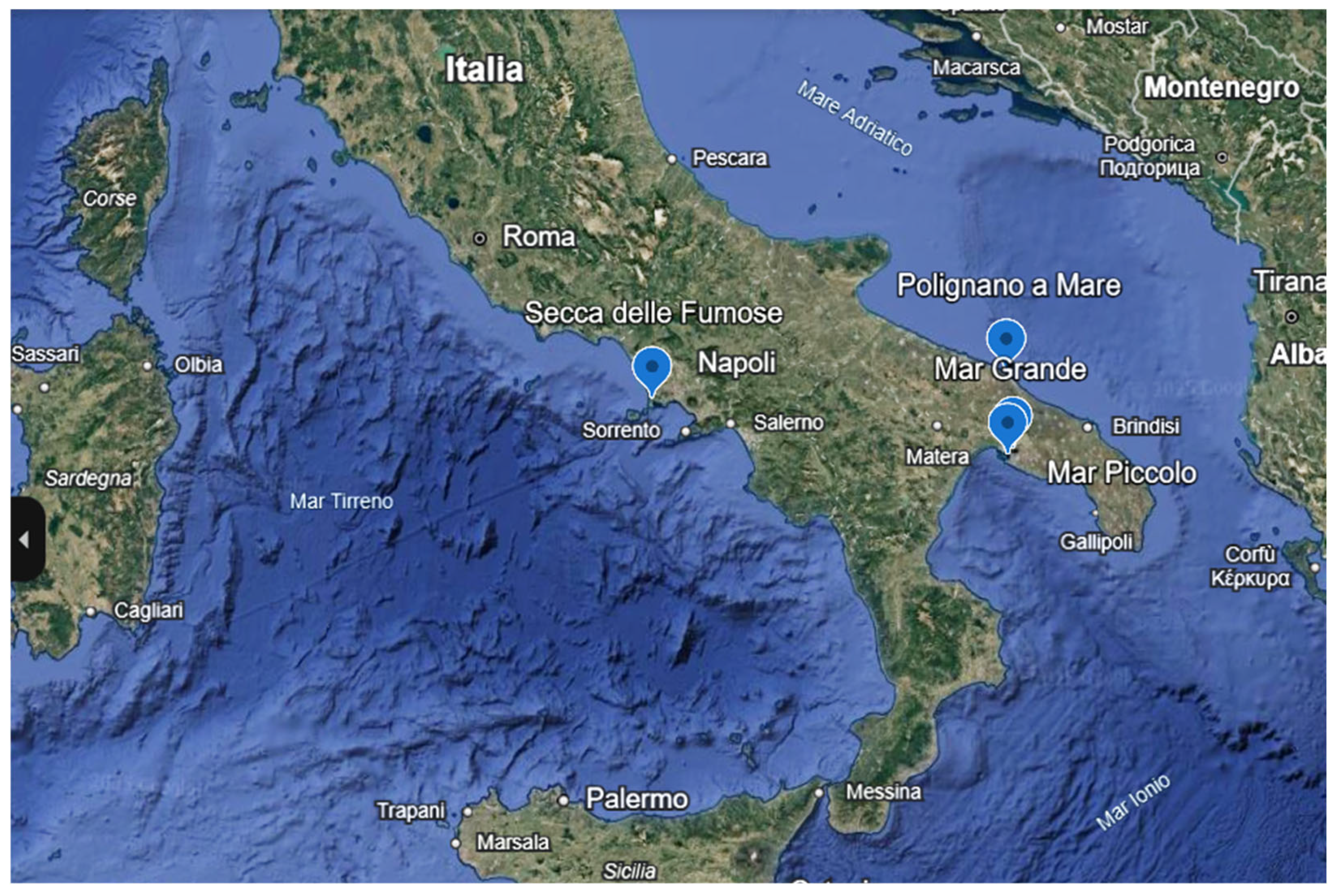
| Sites | Site ID | Coordinates | Depth (m) | Temperature (°C) | pH | Salinity (PSU) |
|---|---|---|---|---|---|---|
| Mar Piccolo | MP | 40°28′ N 17°16′ E | 2.5 | 25 | 8.3 | 39 |
| Mar Grande | IMTA | 40°26′ N 17°14′ E | 3 | 23 | 8.1 | 38.5 |
| Polignano a Mare | POL | 40°59′ N 17°14′ E | 2.5 | 24 | 8.2 | 38 |
| Secca delle Fumose | NAP | 40°49′ N 14°5′ E | 20 | 23.9 | 8.3 | 38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Esposito, R.; Trani, R.; Bertolino, M.; Sonnessa, M.; Laurenzi, G.; Zupo, V.; Longo, C.; Costantini, M. Exploring the Microbial Reservoir of Geodia cydonium (Linnaeus, 1767): Insights into Site-Specific Diversity and Biotechnological Potential. Mar. Drugs 2026, 24, 2. https://doi.org/10.3390/md24010002
Esposito R, Trani R, Bertolino M, Sonnessa M, Laurenzi G, Zupo V, Longo C, Costantini M. Exploring the Microbial Reservoir of Geodia cydonium (Linnaeus, 1767): Insights into Site-Specific Diversity and Biotechnological Potential. Marine Drugs. 2026; 24(1):2. https://doi.org/10.3390/md24010002
Chicago/Turabian StyleEsposito, Roberta, Roberta Trani, Marco Bertolino, Michele Sonnessa, Gaia Laurenzi, Valerio Zupo, Caterina Longo, and Maria Costantini. 2026. "Exploring the Microbial Reservoir of Geodia cydonium (Linnaeus, 1767): Insights into Site-Specific Diversity and Biotechnological Potential" Marine Drugs 24, no. 1: 2. https://doi.org/10.3390/md24010002
APA StyleEsposito, R., Trani, R., Bertolino, M., Sonnessa, M., Laurenzi, G., Zupo, V., Longo, C., & Costantini, M. (2026). Exploring the Microbial Reservoir of Geodia cydonium (Linnaeus, 1767): Insights into Site-Specific Diversity and Biotechnological Potential. Marine Drugs, 24(1), 2. https://doi.org/10.3390/md24010002











